Abstract
Adhesion to the respiratory epithelium plays an important role in Haemophilus influenzae infection. The distribution of H. influenzae adhesins in type b and nontypeable strains has been characterized, but little is known about the prevalence of these factors in non-type b encapsulated strains. We analyzed 53 invasive type a, type e, and type f strains for the presence of hap, hia, hmw, and hif genes; Hap, Hia, and HMW1/2 adhesins; and hemagglutinating pili. The hap gene was ubiquitous, and homologs of hmw and hia were present in 7 of 53 (13.2%) and 45 of 53 (84.9%) strains, respectively. Hap was detected in 28 of 45 (62.2%) hap+ strains, HMW1/2 was detected in 5 of 7 (71.4%) hmw+ strains, and Hia was detected in 31 of 45 (68.8%) hia+ strains. The hif gene cluster was present in 26 of 53 strains (49.1%), and 21 of 26 hif+ strains (80.8%) agglutinated (HA) red blood cells. Nine isolates exhibited HA but lacked the hif gene cluster. The distribution of adhesin genes correlated with the genetic relatedness of the strains. Strains belonging to one type a clonotype and the major type e clonotype possessed hia but lacked the hif cluster. Strains belonging to the second type a clonotype possessed both hia and hif genes. All type f strains belonging to the major type f clonotype possessed hia and lacked hifB. Although the specific complement of adhesin genes in non-type b encapsulated H. influenzae varies, most invasive strains express Hap and Hia, suggesting these adhesins may be especially important to the virulence of these organisms.
In 1931, Pittman classified encapsulated Haemophilus influenzae into six different serotypes (a to f) based on the immunologic reactivity of the capsular polysaccharide (27). H. influenzae type b (Hib) is a major pathogen in children, causing bacteremia, meningitis, epiglottitis, and pneumonia (5). In the past, non-type b encapsulated H. influenzae was regarded as an uncommon cause of serious disease. However, an outbreak of invasive type a infections in immunologically healthy young children was recently described (1). Active bacterial surveillance studies have also documented an increased incidence of non-type b encapsulated H. influenzae infections in Alaska and England in the past decade (25, 29), and other outbreaks of non-type b invasive disease have occurred in The Gambia (15) and other parts of the United States (22, 38, 42). Collectively, these reports suggest that further understanding of the epidemiology and biology of non-type b encapsulated H. influenzae is warranted.
Studies in the 1980s correlating biochemical typing, outer membrane protein (OMP) expression, and multilocus enzyme electrophoresis demonstrated nonrandom associations between these characteristics, suggesting that the population structure of encapsulated H. influenzae is clonal (20, 21). Most invasive Hib disease worldwide is caused by strains of only 3 major OMP or multilocus enzyme electrophoresis types. The analysis of 21 invasive and 213 noninvasive non-type b isolates revealed that strains belonging to serotypes c, e, and f segregate to single divisions and have no close genetic relationships to strains of other serotypes. In contrast, serotypes a and b occur in both primary phylogenetic divisions, suggesting that recombination between these clonal lineages is possible (21). A collection of contemporary invasive isolates of type a, e, and f H. influenzae from the United States was recently analyzed by restriction digest typing (24). Non-type b invasive disease was caused by only 1 or 2 genetically related groups per serotype, suggesting that these invasive strains may be better able to colonize and/or invade the host or that the overall genetic diversity of non-type b encapsulated H. influenzae is limited (24).
In contrast to Hib and nontypeable H. influenzae (NTHi), we know little about virulence factors in invasive non-type b encapsulated strains. H. influenzae initiates infection by colonizing the upper respiratory tract. The pathogenesis of invasive disease involves bacterial translocation across epithelial and endothelial cells, followed by dissemination via the blood to the central nervous system and other tissues (18). Based on studies of Hib and NTHi, bacterial adhesins, lipooligosaccharide, and, in encapsulated strains, the polysaccharide capsule, play a major role in disease (18). It seems likely that these determinants also contribute to the virulence of non-type b encapsulated strains.
At least 5 major adhesins have been identified in Hib and/or NTHi, including Hap, HMW1, HMW2, Hia/Hsf, and hemagglutinating (HA) pili. Hap (Haemophilus adherence and penetration protein) is an autotransporter protein with homology to immunoglobulin A1 (IgA1) protease and is synthesized as a 155-kDa preprotein with three domains. The N-terminal domain is postulated to direct export across the cytoplasmic membrane, the C-terminal domain (Hapβ) translocates the mature protein across the outer membrane, and the serine protease domain (Haps) mediates autoproteolytic cleavage and release from the bacterial surface (12). Hap was originally identified in NTHi and is present in Hib as well, playing an important role in adhesion to epithelial cells (12, 34). In addition, Hap promotes bacterial aggregation and microcolony formation, potentially conferring resistance to bacteriostatic compounds present in human respiratory secretions (12).
Three other nonpilus adhesins, HMW1, HMW2, and Hia, have been described in NTHi (2, 4). Approximately 75% of NTHi strains express HMW1-like and HMW2-like proteins, and most of the remaining NTHi strains express Hia (3, 31, 36). Roughly 5% of NTHi lack both HMW and Hia and are minimally adherent to Chang epithelial cells (31, 36). HMW1 and HMW2 are encoded by two separate loci, designated hmw1 and hmw2, respectively. Each locus consists of three genes encoding the adhesin (hmwA), an integral OMP required for the translocation of HMW1 and HMW2 across the outer membrane (hmwB) (35), and a cytoplasmic protein that may stabilize the adhesin prior to export from the cytoplasm (hmwC) (35). Overall, the predicted amino acid sequences of HMW1 and HMW2 share 71% identity and 80% similarity (2). HMW1 binds glycoprotein receptors containing N-linked oligosaccharide chains with sialic acid in an α2-3 configuration on cultured human epithelial cells (30, 31) while the receptor structure recognized by HMW2 remains unknown. Hia is a high-molecular-weight autotransporter protein that shares 72% identity and 81% similarity with Hsf (Haemophilus surface fibril), an adhesin expressed by Hib that is associated with the expression of short, thin, surface fibrils (4, 32-33). These proteins display the same binding patterns to different epithelial cell lines, suggesting that hia and hsf are allelic (33).
HA pili are expressed by nearly all Hib and a subset of NTHi strains and are encoded by the hif gene cluster, which contains 5 genes (designated hifA to hifE). The pilus structure consists of a major structural subunit (HifA) and two minor subunits (HifD and HifE). Pilus assembly requires the HifB periplasmic chaperone and the HifC outer membrane usher (17, 40). The organization of the hif gene cluster varies significantly, with 9 different arrangements in 20 H. influenzae strains analyzed to date (6, 16-17). Mhlanga-Mutangadura and coworkers have proposed a model for the evolution of these genes, postulating that a progenitor strain acquired the entire gene cluster by horizontal transfer and that other variants arose by deletion (17). The presence of pili can be detected by the ability of bacteria to agglutinate erythrocytes expressing the blood group AnWj antigen (7). Pili promote bacterial adherence to oropharyngeal cells through an interaction involving sialic-containing lactosylceramide structures on the eukaryotic cells (39).
Adhesins are critical to the ability of H. influenzae to colonize and invade the human respiratory tract and produce disease. The distribution of these virulence factors is highly variable in Hib and NTHi. To begin to understand what role these proteins may play in the pathogenesis of infections caused by non-type b encapsulated H. influenzae, we examined the prevalence, distribution, and expression of adhesins in a collection of recent non-type b invasive isolates.
(This work was presented in part at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Canada, 17 to 20 September 2000 [C. A. Rodriguez, J. W. St. Geme III, A. L. Smith, and E. E. Addersen, 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 751908, 2000] and at the Pediatric Academic Societies Annual Meeting, Baltimore, Md., April 2001 [C. A. Rodriguez, J. W. St. Geme III, and E. E. Adderson, abstr. 243A, Pediatr. Res., 2001]).
MATERIALS AND METHODS
Bacterial strains.
Non-type b encapsulated H. influenzae isolates were obtained from state public health departments and other investigators as previously described (23-24). All invasive strains were isolated from blood or cerebrospinal fluid, and most were recovered between 1998 and 2000. Serotypes were confirmed by capsular genotyping with the pU038 probe (provided by J. S. Kroll, Imperial College of Medicine, London, United Kingdom) (23). Only type a (n = 19), e (n = 12), and f (n = 22) strains were considered for final analysis, as only 3 type c and 1 type d isolate were present in the collection. Four control Hib, 15 control NTHi isolates and 6 noninvasive type a isolates were obtained from state public health laboratories, the American Type Culture Collection (Manassas, Va.), and K. Korgenski (Primary Children's Medical Center, Salt Lake City, Utah).
H. influenzae strains were grown on chocolate II agar (Edge Biological, Memphis, Tenn.) or in brain heart infusion broth (Difco, Sparks, Md.) supplemented with 10 μg of hemin/ml and 2 μg of β-NAD (Sigma, St Louis, Mo.)/ml (BHIs). Frozen stocks were stored at −80°C in 100% skim milk.
Escherichia coli strains were grown on Luria-Bertani (LB) agar or in LB broth (Difco) and were stored at −80°C in LB broth with 20% glycerol. For selection, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml (Sigma).
Genomic DNA preparation.
Chromosomal DNA from each bacterial strain was isolated as previously described (13). Briefly, bacteria were grown overnight in BHIs, washed, and resuspended in a buffer containing 10 mM Tris (pH 7.4), 10 mM EDTA, 150 mM NaCl, 0.4% sodium dodecyl sulfate (SDS), and 1 mg of proteinase K (Roche Molecular Biochemicals, Indianapolis, Ind.)/ml. Cells were digested overnight at 37°C, then genomic DNA was extracted with phenol, chloroform, and isoamyl alcohol (Gibco BRL, Life Technologies Inc., Rockville, Md.), treated with RNase A, precipitated, and resuspended in 10 mM Tris (pH 8.0). The final DNA concentration was determined by spectrophotometry.
Southern and dot blots.
Probes for hmw1/2, hia, hifB, and hifE were generated as previously described (2, 4, 37). A hap probe was derived from pJS104, a plasmid derivative of pT7-7 containing a 6.7-kb PstI insert including the full-length hap gene from H. influenzae N187 (26). A 1.4-kb EcoRI-BglII intragenic fragment was used for Southern hybridization.
Probes for hifA, hifC, and hifD were amplified from genomic DNA by PCR. All amplifications were performed in a reaction buffer containing 1× NH4 buffer (Bioline, London, United Kingdom), 0.2 mM concentrations of deoxynucleoside triphosphates (dNTPs), 3 mM MgCl2, 5 U of DNA polymerase (Bioline), and 500 ng of genomic DNA. Chromosomal DNA from a HA-positive H. influenzae type a (strain UT03) was used as template. The following primer pairs were employed: for hifA, 5′ sense TATTCGTAAGCAATTTGGAAATC and 3′ antisense AACACTTCTTGGTAGCTTAATT; for hifC, 5′ sense ATGCTGGATTTGATGGATGAAG and 3′ antisense GCGGTTTTGCATTGAATATCGTG; for hifD, 5′ sense AAATTAACCGCGCTTTTCCATC and 3′ antisense TCACCTGTGGCGTAATAACG. PCR conditions consisted of denaturation at 94°C for 1 min, annealing at 48°C for 1 min, and extension at 72°C for 2.5 min. Thirty-five cycles of amplification were performed for each reaction. Amplification products were cloned into pBSII KS (Stratagene, La Jolla, Calif.), or pCR2.1 (Invitrogen Corporation, Carlsbad, Calif.), and ligated plasmids were transformed into competent E. coli DH5α or topo10F′ cells (Invitrogen). The nucleotide sequence of each probe was confirmed. Restriction endonuclease digestions and gel electrophoresis were performed according to standard techniques (28).
For Southern blots, 5 μg of genomic DNA was digested overnight with EcoRI or BglII restriction endonuclease (Gibco) in the supplied buffer. Restriction fragments were separated by electrophoresis through a 0.8% Tris-borate-EDTA agarose gel, transferred to a nylon membrane (Hybond N+, Amersham Pharmacia Biotech, Inc., Piscataway, N.J.), and cross-linked by exposure to UV. Probes were isolated from agarose gels and labeled with [32P]dCTP (Random Primed DNA labeling kit; Roche Molecular Biochemicals) or fluorescein-conjugated dNTPs (Gene Images CPD-Star; Amersham Pharmacia Biotech, Inc.), according to the manufacturers' directions. Membranes were prehybridized in 5× SSC (3.0 M NaCl, 0.3 M sodium citrate, 0.1% SDS) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 5% dextran sulfate overnight and hybridized with labeled probes according to the manufacturer's protocol. For dot blots, genomic DNA was denatured in 2 M NaCl, 0.1 M NaOH, and 5% ethidium hydroxide, neutralized in 1 M ammonium acetate, and transferred to a nylon membrane by using a 96-well vacuum manifold. The membrane was cross-linked, prehybridized, and hybridized as described above.
PCR amplification of hia.
Amplification of the hia gene was performed by using the 5′ sense TAAATTGCCGTTCCCTTTTGCCTAAAACCTGCTT and 3′ antisense CCAAACTTTACCACTGGTAACCAACACCAGCTGC primers, extending from 0.4 kb upstream of the hia/hsf start codon to the 3′ terminus of the coding sequence, respectively. Amplifications were performed with 500 ng of chromosomal DNA, 15 pM (each) forward and reverse primers, 350 nM dNTPs, 5 μl of the provided buffer with 17.5 mM MgCl2, and 3.5 U of Expand Long Template enzyme mixture (Roche) in a final volume of 50 μl per reaction mixture. PCR conditions consisted of denaturation at 95°C for 40 s, annealing at 50°C for 40 s, and extension at 68°C for 6 min for 30 cycles of amplification.
Analysis of hmw loci.
In NTHi strain 12, the hmw1 locus is located downstream of open reading frame (ORF) HI1679 and the hmw2 locus is downstream of ORF HI1598 (in relation to the Rd reference sequence) (GenBank NC000917.1). Non-type b encapsulated strains that were positive by Southern analysis for hmw genes were analyzed by PCR for the number and physical location of hmw loci. Reaction mixtures were set up with chromosomal DNA and primers corresponding to the 3′ end of ORF HI1679 (5′-TCTTTTGCTGTGGCTGATGCCCCTA) plus the 5′ end of hmw1A (5′-AGTAACATAGCGGAAAGTGGCTTTA) or the 3′ end of ORF HI1598 (5′-CACTGATAGGTTGCTCATATTCGCC) plus the 5′ end of hmw2A (5′-AGTAACATAGCGGAAAGTGGCTTTA). PCR conditions consisted of denaturation at 92°C for 2 min, annealing at 60°C for 1 min, and extension at 72°C for 1.5 min, for a total of 30 cycles, followed by a final extension at 72°C for 7 min. Amplification products were resolved by electrophoresis on a 1% agarose gel and were visualized by staining with ethidium bromide. NTHi strain 12 was used as a positive control, and type e strain NC04 was used as a negative control.
Whole-cell (dot) immunoblots.
Bacteria were grown to an optical density at 600 nm (OD600) of 0.800, washed with phosphate-buffered saline (PBS), fixed for 30 min at room temperature with 4% paraformaldehyde in PBS, and resuspended in PBS to an OD600 of 0.500. Aliquots (100 μl) were applied to nitrocellulose filters by using a vacuum manifold. Following blocking of nonspecific binding for 2 h with 5% skim milk in PBS-0.1% Tween, surface proteins were detected by using guinea pig polyclonal antiserum GP75 raised against recombinant HMW1 and reactive with HMW1 and HMW2, at a 1:1,500 dilution. Horseradish peroxidase-conjugated anti-guinea pig IgG (Sigma) was used as secondary antibody, and detection was performed according to the manufacturer's directions (ECL Western blotting analysis system; Amersham).
Western blotting.
To detect Hap and HMW1, bacteria were grown to mid-log phase in BHIs and pelleted by centrifugation. Pellets were washed, resuspended in PBS with 0.1% phenylmethylsulfonyl fluoride (PMSF) (Boehringer Mannheim, Indianapolis, Ind.), and sonicated. After centrifugation at 12,000 × g for 20 min at 4°C, the protein concentration of purified cell lysates was measured by a UV spectrophotometer at 280 nm. One hundred micrograms of protein was subjected to SDS-polyacrylamide gel electrophoresis on 4 to 15% Tris-HCl polyacrylamide gels (Bio-Rad), and Western blot analysis was performed as described previously (28). For Hap, polyclonal guinea pig antisera against Haps, GP74, was used at a dilution of 1:2,000. For HMW1, GP75 antiserum was used at a 1:1,500 dilution. For Hia, bacteria were grown to an OD600 of 0.800 and pelleted by centrifugation. The bacterial pellet was resuspended in 0.5 ml of cold 10 mM HEPES (pH 7.4) with 10 μg of PMSF/ml and sonicated to clarity on ice. Sonicates were centrifuged for 10 min at 2,040 × g at 4°C. The resulting pellets were resuspended in 0.5 ml of HEPES-PMSF, and 50 μl was incubated with 200 μl of formic acid (Sigma) for 10 min at 25°C. Samples were quickly frozen in a dry ice-ethanol bath, dried in a lyophilizer, resuspended in 1× loading buffer, and subjected to SDS-polyacrylamide gel electrophoresis on 4 to 15% Tris-HCl polyacrylamide gels. For protein detection, rabbit polyclonal antiserum 40C, raised against recombinant Hia and absorbed with whole-cell extracts of E. coli and a hia-negative strain of H. influenzae, was used at a 1:1,000 dilution. Anti-guinea pig IgG (Sigma) or anti-rabbit IgG (Amersham) horseradish peroxidase conjugates were used as secondary antibodies.
NCHA.
The presence of HA pili was determined by nitrocellulose hemadsorption assay (NCHA) as previously described (7). Briefly, bacterial cultures were serially diluted to obtain approximately 300 colonies per plate. Colonies were transferred to nitrocellulose membranes and blocked by incubation with 3% bovine serum albumin. After washing, filters were incubated in a 5% suspension of O-negative red blood cells (RBCs). HA colonies appear as red dots on the membrane and were quantified as a percentage of total colonies. For isolates possessing the hif gene cluster that did not HA upon initial testing, enrichment of piliation was performed by selection of HA colonies after incubation with O-negative RBCs and passage on a 0.3 M sucrose gradient, as previously described (7).
RESULTS
hap gene is ubiquitous in H. influenzae.
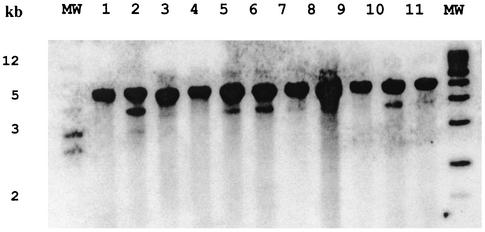
Based on dot blot hybridization, all invasive non-type b encapsulated H. influenzae and NTHi strains analyzed possessed a homolog of hap (Table 1). To confirm these results, we performed Southern blot analysis on a subset of these strains (Fig. 1). In all cases we observed a strongly hybridizing band approximately 5 kb in size. In 10 of the 16 strains there was also a weakly hybridizing smaller-molecular-weight band, suggesting either that two hap alleles may exist or that a second gene with moderate homology to hap may be present in these strains. Overall, Hap protein expression was observed in 28 of 45 (62.2%) strains that possessed the hap gene (data not shown).
TABLE 1.
Presence of H. influenzae adhesins in encapsulated non-type b strainsa
| Strain name | Type | Result for:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hap | Hap (WB) | hmw1A/1B/1C | HMW dot blot | HMW (WB) | hia (kb) | Hia (WB) | hifA, C, D, E | hifB | SA | NCHA | ||
| UT01 | a | + | + | − | − | − | +/+ (7.5) | + | + | + | + | + |
| UT02 | a | + | + | − | − | ND | +/+ (7.5) | + | + | + | + | + |
| UT03 | a | + | + | − | − | ND | +/+ (7.5) | + | + | + | + | + |
| CA03 | a | + | − | − | − | ND | +/+ (7.5) | + | + | + | + | + |
| CO01 | a | + | − | − | − | − | +/+ (7.5) | + | + | + | + | + |
| CA01 | a | + | + | − | − | − | +/+ (8.0) | + | − | − | − | − |
| CA02 | a | + | + | − | − | ND | +/+ (8.0) | + | − | − | − | − |
| UT04 | a | + | − | − | − | ND | +/+ (8.0) | + | − | − | − | + |
| UT05 | a | + | ND | − | − | ND | +/+ (8.0) | + | − | − | − | + |
| CA04 | a | + | − | − | − | ND | +/+ (8.0) | + | − | − | − | − |
| CA05 | a | + | − | − | − | ND | +/+ (8.0) | + | − | − | − | − |
| CA08 | a | + | − | − | − | ND | +/+ (8.0) | + | − | − | − | − |
| R421 | a | + | − | − | − | ND | +/+ (4.0) | + | − | − | − | − |
| MO01 | a | + | − | + | − | − | − | − | + | + | − | − |
| MO02 | a | + | − | + | − | − | − | − | + | + | − | − |
| AT01 | a | + | + | + | + | + | − | − | − | − | − | − |
| NC01 | a | + | + | + | + | + | − | − | − | − | − | − |
| TN01 | a | + | + | + | + | − | − | − | − | − | − | − |
| UT06 | a | + | ND | − | − | − | − | − | − | − | − | − |
| UT07 | e | + | − | − | − | ND | +/+ (5.5) | + | − | − | − | − |
| MN03 | e | + | − | − | − | ND | +/+ (7.5) | − | − | − | − | − |
| AL03 | e | + | + | − | − | ND | +/+ (5.5) | + | − | − | − | − |
| MO03 | e | + | + | − | − | ND | +/+ (7.5) | + | − | − | + | + |
| MO04 | e | + | + | − | − | ND | +/+ (7.5) | + | − | − | + | + |
| MO05 | e | + | + | − | − | ND | +/+ (5.5) | − | − | − | + | + |
| WI01 | e | + | − | − | − | ND | +/+ (5.5) | − | − | − | + | + |
| CO02 | e | + | ND | − | − | ND | +/+ (5.5) | − | + | + | + | + |
| IA02 | e | + | ND | − | − | ND | +/+ (5.5) | + | − | − | − | − |
| IA03 | e | + | + | − | − | ND | +/+ (5.5) | + | − | − | − | − |
| AL02 | e | + | + | + | + | + | − | − | − | − | − | − |
| OK04 | e | + | − | − | − | ND | +/+ (3.5) | + | − | − | − | − |
| MO06 | f | + | + | − | − | ND | +/+ (7) | − | + | − | − | − |
| MO07 | f | + | + | − | − | ND | +/+ (7) | − | + | − | + | + |
| MO08 | f | + | + | − | − | ND | +/+ (7) | − | + | − | + | + |
| MO09 | f | + | + | − | − | ND | +/+ (7) | − | + | − | + | + |
| CO03 | f | + | + | − | − | ND | +/+ (7) | + | + | − | + | + |
| CO04 | f | + | + | − | − | ND | +/+ (7) | + | + | − | + | + |
| AL04 | f | + | ND | − | − | ND | +/+ (7) | + | + | − | + | + |
| AL05 | f | + | ND | ND | ND | ND | +/+ (7) | + | + | − | + | + |
| NC02 | f | + | + | − | − | − | +/+ (7) | + | + | − | + | + |
| IA07 | f | + | ND | − | − | ND | +/+ (7) | + | + | − | + | + |
| IA08 | f | + | + | − | − | ND | +/+ (7) | + | + | − | + | + |
| CT01 | f | + | + | − | − | − | +/+ (7) | − | + | − | + | + |
| WI03 | f | + | + | − | − | ND | +/+ (7) | + | + | − | + | + |
| WI04 | f | + | + | − | − | ND | +/+ (7) | + | + | − | + | + |
| WI05 | f | + | + | − | − | ND | +/+ (7) | + | + | − | + | + |
| OK05 | f | + | − | − | − | − | +/+ (7) | − | + | − | − | − |
| OK06 | f | + | ND | − | − | ND | +/+ (7) | + | + | − | − | − |
| IA11 | f | + | + | − | − | − | +/+ (7) | + | + | − | + | + |
| IA10 | f | + | − | − | − | − | +/+ (7) | + | − | − | + | + |
| IA05 | f | + | − | − | − | ND | +/+ (7) | − | − | − | + | + |
| MN04 | f | + | − | − | − | ND | +/+ (7) | − | − | − | − | − |
| NC03 | f | + | + | + | + | + | − | − | − | − | + | + |
Shown is the strain, serotype, and presence (+) or absence (−) of hap, Hap, hmw1A/B/C, HMW1/2, hia, Hia, hifA/B/C/D/E, and positive (+) or negative (−) slide and nitrocellulose HA assays. The length of the PCR fragment corresponding to hia is shown in parentheses. ND, not done; WB, western blotting; SA, slide agglutination.
FIG. 1.
hap homologs in type a H. influenzae. Chromosomal DNA was digested with EcoRI, separated by agarose electrophoresis, transferred to nylon membranes, and hybridized with a hap probe. Lanes 1, 2, 9, and 10, invasive isolates; lanes 3 to 8, noninvasive type a isolates; lane 11, NTHi. Molecular sizes are noted on the left. A second 4-kb hybridizing band is present in lanes 2, 5, 6, and 10, suggesting the presence of two alleles of hap or a second homologous gene in these strains.
hmw and hia genes are mutually exclusive in encapsulated H. influenzae.
In previous studies, hmw genes have been found exclusively in NTHi (36). Interestingly, Southern hybridization revealed that 7 of 53 (13.2%) non-type b encapsulated strains in this study possessed homologs of the hmw gene cluster, including 5 type a, 1 type e, and 1 type f strain (Table 1). PCR analysis revealed 2 hmw loci in each of these strains, in all cases one downstream of ORF HI1598 and the other downstream of ORF HI1679, analogous to the location in nontypeable strains (A. Buscher and J. St. Geme III, unpublished data). In all 7 of these strains, Southern blot analysis also demonstrated the presence of hmwB and hmwC homologs (Table 1). Western blot analysis demonstrated immunoreactive HMW proteins in 5 of the 7 (71.4%) hmw+ strains (data not shown).
Length of the hia ORF varies among different isolates.
Forty-five of 53 (84.9%) encapsulated non-type b strains possessed a homolog of hia (Table 1). As in NTHi, none of the non-type b encapsulated strains hybridized with probes for both hmw and hia (36). One (1.9%) strain lacked both hmw and hia (36). The hia ORF is 3,296 bp long in NTHi strain 11 (GenBank U38617). In contrast, the hsf ORF is 7,062 bp long in the type b isolate C54 (GenBank U41852). With this information in mind, we performed PCR amplification of the hia gene in all hia+ non-type b encapsulated H. influenzae strains (Table 1). An identical 7.0-kb PCR product was amplified from all but one type f strain. An 8.0-kb product was amplified from all but one type a strain belonging to the major clonotype, and a 7.5-kb fragment was amplified from members of the other major type a clonotype. All but one type e strain belonging to the major type e clonotype yielded fragments of 5.5 (7 of 10) or 7.5 (3 of 10) kb; the remaining type e strain yielded a 3.5-kb PCR product. Amplification of hia/hsf from one noninvasive type a strain and two nontypeable strains that were positive by colony hybridization for hia produced 3.5-, 3.0-, or 2.5-kb fragments, respectively (data not shown). Western blot analysis detected immunoreactive Hia in 31 of 45 (68.8%) hia+ strains (data not shown). The mass of Hia corresponded to the size of the hia ORF, ranging from approximately 120 kDa for OK04 Hia (3.5 kb) to 275 kDa for CA01 Hia (8.0 kb).
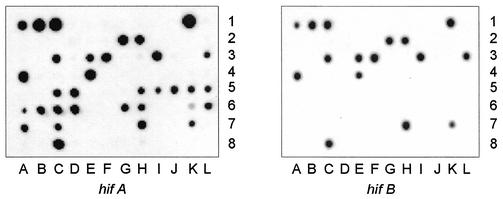
hif gene cluster is present in 49% of non-type b encapsulated strains.
Overall, 26 of 53 (49.0%) strains had genes homologous with some or all members of the hif gene cluster (Fig. 2). hifB was uniformly absent from the 22 type f strains, including 18 isolates that possessed hifA,hifC,hifD, and hifE homologs (Fig. 2) (17).
FIG. 2.
hif homologs in non-type b encapsulated H. influenzae. Colony hybridization was performed with 29 type a (wells A1 to C2 and E2 to F3), 4 type b (wells D2 and G3 to I3), 3 type c (wells J to L3), 1 type d (well A4), 13 type e (wells B4 to B5), 24 type f (wells C5 to C7), and 15 NTHi (wells D7 to F8) strains by using probes for hifA (left) and hifB (right). Type f strains contain a deletion of hifB. The entire hif gene cluster was absent in most type e strains and in one of the two major clonotypes of type a isolates in this collection.
HA assays and presence of hif homologs.
Overall, 30 of the 53 (56.6%) exhibited a variable degree of HA in the NCHA assay, with a range of 0.1 to 80% of HA colonies (Table 1). After performing enrichment to select HA phase variants, we were able to isolate positive HA colonies from 5 of 8 (62.5%) previously hif+/HA− strains. Therefore, 21 of 26 (80.8%) strains that possess some or all members of the hif gene cluster exhibited positive HA. The majority of type f isolates, 18 of 22 (81.8%), were HA, compared to 5 of 12 (41.7%) type e isolates and 7 of 19 (36.8%) type a strains. Interestingly, 9 isolates were able to agglutinate RBCs, despite the complete absence of the hif gene cluster.
In order to determine whether HA strains that appeared to be lacking hif sequences might fail to hybridize with hif probes because of a low degree of homology to the probe sequences, we repeated Southern blots under low stringency conditions. However, this process failed to identify additional hif+ strains.
DISCUSSION
Previous studies have characterized the prevalence of specific adhesins among type b and nontypeable strains of H. influenzae. In contrast, knowledge of the distribution of adhesins and other virulence genes in non-type b encapsulated strains is incomplete. The increasing importance of these bacteria as human pathogens prompted this study.
We found that the hap gene is universally present in H. influenzae, suggesting that Hap plays a major role in the ability of H. influenzae strains to colonize the nasopharynx. As previously described for NTHi strains, hmw and hia are mutually exclusive in non-type b encapsulated strains, with hia being most common. Homologs of Hap and Hia have been recently described in Neisseria meningitidis, underscoring the potential importance of these proteins in colonization and disease (41). A homolog of hmw1/2, previously reported to be present only in NTHi, was found in 13.2% of the invasive non-type b encapsulated H. influenzae strains analyzed in this study. Interestingly, each of these strains is genetically dissimilar from the predominant clonotype of each serotype, based on restriction digest pattern typing. Three of these strains failed to hybridize with cap or IS1016 probes (23). It is possible, therefore, that these isolates are actually true NTHi strains. Four strains, however, could be genotyped by using a cap probe, suggesting that these are previously encapsulated strains that have lost the ability to express capsules. Alternatively, these hmw+ strains may have originated from a nonencapsulated precursor by acquisition of the encapsulation locus or precursor hmw-deficient strains may have lost the hmw locus and diverged from hmw+ encapsulated precursors at a very early stage in clonal evolution. We favor the first hypothesis, as multiple-locus sequencing typing analysis of housekeeping genes (10) suggests that these strains are more closely related to nontypeable strains (unpublished data).
H. influenzae evades host defenses by altering surface antigens. Antigenic diversity has been described for many H. influenzae surface structures, including HMW, P1, P2, P5, Hif, IgA protease, pilus, capsule, and lipooligosaccharide. A variety of molecular mechanisms are responsible for this antigenic variation, including point mutations, gene amplification, phase variation, horizontal gene transfer, homologous recombination, or a combination of these events (8, 11, 43). Variation in pili expression, for example, has been attributed to point mutation, phase variation, and horizontal gene transfer and recombination (11).
In our study only 5 of 7 (71.4%) hmw+ strains expressed HMW1/2 by Western blot analysis. The lack of detectable protein may be a manifestation of phase variation, resulting from slipped-strand mispairing of 7-bp tandem repeats in the hmw promoter region (8). Although HMW adhesins facilitate bacterial colonization, invasive isolates may down-regulate HMW expression in order to avoid recognition by the immune system (8). Alternatively, these strains may contain truncated hmw genes, detectable by Southern analysis but unable to encode stable proteins.
The hif gene cluster consists of the hifA, hifB, hifC, hifD, and hifE genes and is flanked by direct repeats, rendering it susceptible to deletion by homologous recombination. Additional interstrain and intertype differences have been postulated to arise by multiple-sequence rearrangements that occur in intergenic dyad sequences (17). For example, some type f strains have a deletion extending from the region upstream of hifB through the middle part of hifC (17). The absence of hifB in all type f strains in this study suggests that deletion of this gene may have occurred early during evolution of type f H. influenzae. However, the absence of hifB does not impair the ability to HA RBCs in the majority of these strains, suggesting that other structures may be responsible for HA in type f strains. Alternatively, another chaperone may be involved in pilus assembly in this serotype. Interestingly, the entire hif gene cluster was absent from the majority of type e strains and from all members of one of the two major genetically related groups of type a isolates in this study. Thus, absence of the hif locus is common among non-type b encapsulated H. influenzae and appears to have little effect on epithelial colonization and invasion by these strains.
Not all strains possessing the intact hif gene cluster agglutinate RBCs. Bloodstream invasion may favor nonpiliated strains and select for phase variants. Although helpful in promoting epithelial colonization, these surface proteins may be recognized by innate and specific host immune responses and may be down-regulated to avoid immune recognition (17, 44). During natural infection with Hib, nasopharyngeal isolates are often piliated, whereas their isogenic counterparts from systemic sites are usually nonpiliated, consistent with this hypothesis (26). The ability of some hif-negative strains to agglutinate RBCs suggests that alternative nonpilus structures are capable of mediating HA. Other investigators have described three nontypeable strains that had HA activity, despite the absence of the hifA gene (17). Nonpilus bacterial HAs have also been identified in E. coli (9).
Only one isolate in this study lacked both hmw and hia, suggesting that the HMW and Hia adhesins have an important role in colonization. Consistent with this hypothesis, hmw- and hia-negative NTHi are uncommon and adhere poorly to epithelial cells (36). However, the absence of these adhesins, as well as the hif gene cluster, in one of the invasive non-type b encapsulated isolates in this study suggests that Hap and/or other adhesive structures permitted sufficient adherence to promote epithelial colonization and subsequent invasion.
We have used restriction digest patterns to define the population structure of non-type b encapsulated H. influenzae isolates (24) (Fig. 3). The distribution of adhesin genes correlates with this genetic classification. Most type f isolates causing invasive disease belong to a single, genetically related clonotype, and most type e strains also belong to a single genetically related group. The majority of invasive type a strains belong to one of two major clonotypes, which also correspond to cap a(N) and a(T) genotypes. In a(N) and a(T) strains, the cap locus is duplicated, but in a(T) strains the second gene copy is partially deleted (15, 21). Strains belonging to the largest type a clonotype [cap a(T)] and the major type e clonotype possess hia but lack hif. Strains belonging to the second largest type a clonotype [cap a(N)] possess both hia and an intact hif gene cluster. All type f strains from the major clonotype possess hia and hifA, hifC, hifD, and hifE (Fig. 3).
FIG. 3.
Dendrogram based on SmaI restriction digest patterns (right) of encapsulated H. influenzae isolates. Shown on the left are serotypes (in parentheses) and strain designations. To the left are hybridization results with hia, hmw, and hifA to hifE adhesin probes. +, positive; −, negative; †, COO2 was the only serotype e strain that possess the hif gene cluster; ∗, strains not included in this study. A genetic similarity of 1.0 indicates identical restriction digest patterns.
The hsf and hia genes are allelic and are located in the same region of the chromosome. The size of the hsf-hia locus also correlates with the genetic relatedness of invasive strains. Type a strains belonging to the two major clonotypes have either an 8.0- or a 7.5-kb locus, type e strains contain either a 5.5- or a 7.0-kb locus, and type f strains contain a 7.0-kb locus (33). Invasive encapsulated strains, therefore, contain a large gene more similar to Hib hsf than to NTHi hia. It has been postulated that in encapsulated strains, a larger protein is necessary to protrude beyond the polysaccharide capsule (33).
It has been proposed that some invasive non-type b encapsulated H. influenzae strains might have originated from Hib strains that underwent capsular switching, thus acquiring virulence factors normally present in virulent Hib strains. This phenomenon may occur through homologous recombination facilitated by the common organization of the H. influenzae capsulation locus (14-15). Hib and non-type b encapsulated H. influenzae strains share the same ecological niche, favoring this possibility. Although in vitro experiments suggest that exchange of encapsulation loci is possible, the clonal population structure of these bacteria suggests that this is uncommon. Additionally, we found that hmcA (19), the structural gene encoding the type b-specific bacteriocin haemocin was absent in all non-type b encapsulated isolates, suggesting that none of these strains was related to Hib (data not shown).
The correlation between the presence of genes encoding important adhesins and the genetic relatedness of invasive non-type b encapsulated H. influenzae strains by restriction digest pattern typing confirms the validity of this genetic classification system. Our data also suggest that particular combinations of adhesins, perhaps Hap and Hia, and/or as-yet-unidentified virulence factors contribute to the ability of genetically related strains of H. influenzae to colonize or to cause invasive disease. Comparison of particular genotypes and phenotypes of H. influenzae will shed light on the role of these bacterial factors in the pathogenesis of human disease.
Acknowledgments
We are grateful for the excellent technical assistance of Juliana Anyanwu and Yan Wang.
This work was supported by Cancer Center CORE grant P30 CA21765 (to E.E.A.), the American Lebanese Syrian Associated Charities (ALSAC) (E.E.A. and C.A.R), Public Health Service grants AI-44167 (to J.W.S) and DC-02873 (to J.W.S).
Editor: J. T. Barbieri
REFERENCES
- 1.Adderson, E. E., C. L. Byington, L. Spencer, A. Kimball, M. Hindiyeh, K. Carroll, S. Mottice, E. K. Korgenski, J. C. Christenson, and A. T. Pavia. 2001. Invasive serotype a Haemophilus influenzae infections with a virulence genotype resembling Haemophilus influenzae type b: Emerging pathogen in the vaccine era? Pediatrics 108:E18.. [DOI] [PubMed] [Google Scholar]
- 2.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenkamp, S. J., and J. W. St. Geme III. 1994. Genes encoding high molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 62:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 5.Broome, C. V. 1987. Epidemiology of Haemophilus influenzae type b infections in the United States. Pediatr. Infect. Dis. J. 6:779-782. [DOI] [PubMed] [Google Scholar]
- 6.Clemans, D. L., C. Marrs, M. Patel, M. Duncan, and J. R. Gilsdorf. 1998. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect. Immun. 66:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connor, E. M., and M. R. Loeb. 1983. A hemadsorption method for detection of colonies of Haemophilus influenzae type b expressing fimbriae. J. Infect. Dis. 148:855-860. [DOI] [PubMed] [Google Scholar]
- 8.Dawid, S., S. J. Barenkamp, and J. W. St. Geme III. 1999. Variation in expression of Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc. Natl. Acad. Sci. USA 96:1077-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duguid, J. P., S. Clegg, and M. I. Wilson. 1979. The fimbrial and nonfimbrial haemagglutinins of Escherichia coli. J. Med. Microbiol. 12:213-227. [DOI] [PubMed] [Google Scholar]
- 10.Feil, E. J., E. C. Holmes, D. E. Bessen, M.-S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilsdorf, J. R. 1998. Antigenic diversity and gene polymorphisms in Haemophilus influenzae. Infect. Immun. 66:5053-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrixson, D. R., and J. W. St. Geme III. 1998. The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol. Cell 2:841-850. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann, B. G., and A. M. Frischauf. 1987. Isolation of genomic DNA, p. 180-182. In S. L. Berger and A. R. Kimmel (ed.), Methods in enzymology. Guide to molecular cloning techniques. Academic Press, Inc., San Diego, Calif.
- 14.Kroll, J. S., B. Loynds, and E. R. Moxon. 1991. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol. Microbiol. 5:1549-1560. [DOI] [PubMed] [Google Scholar]
- 15.Kroll, J. S., E. R. Moxon, and B. M. Loynds. 1994. Natural genetic transfer of a putative virulence-enhancing mutation to Haemophilus influenzae type a. J. Infect. Dis. 169:676-679. [DOI] [PubMed] [Google Scholar]
- 16.McCrea, K. W., J. L. Sauver, C. F. Marrs, D. Clemans, and J. R. Gilsdorf. 1998. Immunologic and structural relationships of the minor pilus subunits among Haemophilus influenzae isolates. Infect. Immun. 66:4788-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mhlanga-Mutangadura, T., G. Morlin, A. L. Smith, A. Eisenstark, and M. Golomb. 1998. Evolution of the major pilus gene cluster of Haemophilus influenzae. J. Bacteriol. 180:4693-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moxon, E. R. 1992. Molecular basis of invasive Haemophilus influenzae type b disease. J. Infect. Dis. 165:S77-81. [DOI] [PubMed] [Google Scholar]
- 19.Murley, Y. M., T. D. Edlind, P. A. Plett, and J. J. LiPuma. 1998. Cloning of the haemocin locus of Haemophilus influenzae type b and assessment of the role of haemocin in virulence. Microbiology 144:2531-2538. [DOI] [PubMed] [Google Scholar]
- 20.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, G. Hammond, E. A. Hoiby, K. E. Jonsdottir, M. Kabeer, I. Kallings, W. N. Khan, M. Kilian, K. Knowles, H. J. Koornhof, B. Law, K. I. Li, J. Montgomery, P. E. Pattison, J.-C. Piffaretti, A. K. Takala, M. L. Thong, R. A. Wall, J. I. Ward, and R. K. Selander. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 22.Nitta, D. M., M. A. Jackson, V. F. Burry, and L. C. Olson. 1995. Invasive Haemophilus influenzae type f disease. Pediatr. Infect. Dis. J. 14:157-160. [PubMed] [Google Scholar]
- 23.Ogilvie, C., A. Omikunle, Y. Wang, J. W. St. Geme III, C. A. Rodriguez, and E. E. Adderson. 2001. Capsulation loci of non-serotype b encapsulated Haemophilus influenzae. J. Infect. Dis. 184:144-149. [DOI] [PubMed] [Google Scholar]
- 24.Omikunle, A., S. Takahashi, C. L. Ogilvie, Y. Wang, C. A. Rodriguez, J. W. St. Geme III, and E. E. Adderson. 2002. Limited genetic diversity of recent invasive isolates of non-serotype b encapsulated Haemophilus influenzae. J. Clin. Microbiol. 40:1264-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perdue, D. G., L. R. Bulkow, B. G. Gellin, M. Davidson, K. M. Petersen, R. J. Singleton, and A. J. Parkinson. 2000. Invasive Haemophilus influenzae disease in Alaskan residents aged 10 years and older before and after infant vaccination programs. JAMA 283:3089-3094. [DOI] [PubMed] [Google Scholar]
- 26.Pichichero, M. E., M. Loeb, P. Anderson, and D. H. Smith. 1982. Do pili play a role in pathogenicity of Haemophilus influenzae type b? Lancet 2:960-962. [DOI] [PubMed] [Google Scholar]
- 27.Pittman, M. 1931. Variation and type specificity in the bacterial species Haemophilus influenzae. J. Exp. Med. 53:471-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Slack, M. P., H. J. Azzopardi, R. M. Hargreaves, and M. E. Ramsay. 1998. Enhanced surveillance of invasive Haemophilus influenzae disease in England, 1990 to 1996: impact of conjugate vaccines. Pediatr. Infect. Dis. J. 17:S204-S207. [DOI] [PubMed] [Google Scholar]
- 30.St. Geme, J. W., III. 1994. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun 62:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St. Geme, J. W., III. 2000. The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19:S41-S50. [DOI] [PubMed] [Google Scholar]
- 32.St. Geme, J. W., III, and D. Cutter. 2000. The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182:6005-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St. Geme, J. W., III, D. Cutter, and S. J. Barenkamp. 1996. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 178:6281-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.St. Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 35.St. Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 36.St. Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St. Geme, J. W., III, J. S. Pinkner III, G. P. Krasan, J. Heuser, E. Bullitt, A. L. Smith, S. J. Hultgren. 1996. Haemophilus influenzae pili are composite structures assembled via the HifB chaperone. Proc. Natl. Acad. Sci. USA 15:11913-11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urwin, G., J. A. Krohn, K. Deaver-Robinson, J. D. Wender, and M. M. Farley. 1996. Invasive disease due to Haemophilus influenzae serotype f: clinical and epidemiologic characteristics in the Haemophilus influenzae serotype b vaccine era. The Haemophilus influenzae Study Group. Clin. Infect. Dis. 22:1069-1076. [DOI] [PubMed] [Google Scholar]
- 39.van Alphen, L., L. Geelen-van den Broek, L. Blaas, M. van Ham, J. Dankert. 1991. Blocking of fimbria-mediated adherence of Haemophilus influenzae by sialyl gangliosides. Infect. Immun. 59:4473-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Ham, S. M., L. van Alphen, F. R. Mooi, and J. P. van Putten. 1994. The fimbrial gene cluster of Haemophilus influenzae type b. Mol. Microbiol. 13:673-684. [DOI] [PubMed] [Google Scholar]
- 41.van Ulsen, P., L. van Alphen, C. T. Hopman, A. van der Ende, J. Tommassen. 2001. In vivo expression of Neisseria meningitidis proteins homologous to the Haemophilus influenzae Hap and Hia autotransporters. FEMS Immunol. Med. Microbiol. 32:53-64. [DOI] [PubMed] [Google Scholar]
- 42.Waggoner-Fountain, L. A., J. O. Hendley, E. J. Cody, V. A. Perriello, L. G. Donowitz. 1995. The emergence of Haemophilus influenzae types e and f as significant pathogens. Clin. Infect. Dis. 21:1322-1324. [DOI] [PubMed] [Google Scholar]
- 43.Weiser, J. N. 2000. The generation of diversity by Haemophilus influenzae. Trends Microbiol. 8:433-435. [DOI] [PubMed] [Google Scholar]
- 44.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30:767-775. [DOI] [PubMed] [Google Scholar]