Abstract
Prostaglandin (PG) D2 is the most abundant prostanoid produced in the central nervous system of mammals and has been implicated in the modulation of neural functions such as sleep induction, nociception, regulation of body temperature, and odor responses. We generated gene-knockout mice for lipocalin-type PGD2 synthase (L-PGDS) and found that the intrathecal administration of PGE2, an endogenous pain-producing substance, failed to elicit allodynia (touch-evoked pain), which is one typical phenomenon of neuropathic pain, whereas it evoked thermal hyperalgesia, in L-PGDS−/− mice. We also found that the allodynic response induced by the γ-aminobutyric acid (GABA)A receptor antagonist bicuculline was selectively abolished in the L-PGDS−/− mice, among excitatory and inhibitory agents that induced allodynia in wild-type mice. Interestingly, simultaneous injection of a femtogram amount of PGD2 with PGE2 or bicuculline induced allodynia in L-PGDS−/− mice to the same extent as in wild-type mice. The PGE2- or bicuculline-evoked allodynia in wild-type and in PGD2-supplemented L-PGDS−/− mice was blocked by a PGD2 receptor antagonist given in a femtogram amount. These results reveal that endogenous PGD2 is essential for both PGE2- and bicuculline-induced allodynia.
Keywords: gene targeting, hyperalgesia
Prostaglandin (PG) D2 is the major prostanoid produced in the central nervous system (CNS) of mammals (1, 2) and has been shown to be involved in a variety of central actions, e.g., it induces sleep, decreases body temperature, and modulates odor and pain responses (3–5). Among the three enzymes catalyzing the conversion of PGH2 to PGD2 (6, 7), lipocalin-type PGD synthase (L-PGDS) is considered to be responsible for the biosynthesis of PGD2 in the CNS. L-PGDS is present mainly in the leptomeninges, choroid plexus, and oligodendrocytes of the CNS (8, 9), and is secreted into the cerebrospinal fluid to become β-trace (10, 11), a major protein component of the cerebrospinal fluid (12–14). The receptor for PGD2, the D type of PG (DP) receptor (15, 16), also is localized in the leptomeninges of the brain (17, 18). Therefore, PGD2 is assumed to be produced in the membranous tissues and oligodendrocytes of the CNS; to circulate through the cerebrospinal fluid in the ventricular system, subarachnoidal space, and extracellular space in the CNS; to interact with DP receptors in the meninges; and to act as a neurohormone or an informational substance for global regulation of various CNS functions (5).
Prolonged tissue damage or injury often leads to chronic pain states such that noxious stimuli evoke hyperalgesia and innocuous tactile stimuli evoke pain (allodynia). In relation to clinically relevant hyperalgesic states, such as inflammation and neuropathic pain, there is considerable interest in the neurochemical mechanisms of hyperalgesia and allodynia (19–21). Since Vane (22) reported that aspirin-like drugs prevented the development of inflammation by blocking the synthesis of PGs, it has been widely accepted that PGs are involved in inflammation and pain.
PGs are crucial for the processing of pain not only by sensitizing peripheral terminals of primary afferent nociceptors but also by augmenting processing of pain information at the spinal level (23–25). We and others (26) previously reported that intrathecal (i.t.) administration of PGD2 and PGE2 into mice induced hyperalgesia to noxious stimuli. In addition, we found that i.t. administration of PGE2 and PGF2α induced allodynia; the mice showed squeaking, biting, and scratching movements in response to low-threshold stimuli (27–29). Whereas PGD2 itself did not induce allodynia, it blocked the PGE2-evoked allodynia but not the PGF2α-evoked one (25). Although antinociceptive actions of nonsteroidal anti-inflammatory drugs administered i.t. have suggested spinal roles of PGs in nociception, the respective role(s) of a given PG in pain transmission and mutual interactions among PGs produced in situ remain to be clarified. Therefore, to investigate the physiological and pathophysiological roles of PGD2 and L-PGDS in vivo, we generated mice homozygous for a null mutation of L-PGDS by using the gene targeting technique. We then examined the responses for thermal hyperalgesia and allodynia at the spinal level in L-PGDS-deficient mice as compared with those in wild-type mice and found that endogenous PGD2 is essential for the PGE2-induced allodynia but not for the hyperalgesia. Futhermore, we also found that endogenous PGD2 is essential for the induction of allodynia by bicuculline, the γ-aminobutyric acid (GABA)A receptor antagonist, among the excitatory and inhibitory agents in pain transmission.
MATERIALS AND METHODS
Gene Targeting.
The genomic clone (15.3 kb) containing the entire gene for mouse L-PGDS was isolated from a 129/Sv genomic phage library (Stratagene) by use of the cDNA and the gene for rat L-PGDS as probes (30, 31). The 15.3-kb genomic fragment was ligated into a Bluescript KS(+) vector (Stratagene) without subcloning and sequenced by a Li-Cor 4000L automated DNA sequencer (Li-Cor, Lincoln, NE). The genomic fragment contained the entire gene for L-PGDS (3,037 bp), a 5′ upstream region of 9,587 bp, and a 3′ downstream region of 2,640 bp. The coding region existed in exons I–VI: exon I encoded the signal peptide of L-PGDS; and exon II, the active site (Cys65) of the enzyme (32) and two N-glycosylation sites (Asn51 and Asn78) (30).
The targeting vector was constructed by replacing a 1.84-kb fragment containing parts of exons II–V with the neomycin-resistance gene (Neor). The herpes simplex virus thymidine kinase gene (HSV-tk) was inserted upstream. The targeting vector was linearized with NotI and introduced into E14–1 embryonic stem (ES) cells from 129/Ola mice by electroporation. G418- and gancyclovir-resistant clones were isolated and screened by PCR amplification using two primer pairs: P1 (5′-AGGCACCTGCTCTGCTCTGAGCAAAT-3′) and P2 (5′-CTGATGCTCTTCGTCCAGATCATCCTGA-3′); and P3 (5′-ATCGCCTTCTATCGCCTTCTTGACGAGT-3′) and P4 (5′-TCTTGAGAGTGCACAGAGCAAAGGAGTC-3′) for homologous recombination. The former PCR product was 400 bp, and the latter 640 bp. The mutated ES cells then were confirmed by Southern blot hybridization with the external probe.
Enzyme Assay.
Under pentobarbital anesthesia, animals were perfused through the left ventricle of the heart with Krebs–Ringer solution at 4°C. The brain was quickly removed, weighed, and homogenized in 3 vol of PBS (10 mM sodium phosphate, pH 7.4, and 154 mM NaCl) with a Polytron homogenizer. The homogenate was centrifuged at 4°C for 15 min at 400,000 g. The supernatant was used for detection of L-PGDS activity as described (33).
Pain Responses.
Mice weighing 20 ± 2 g were used. Drugs (in vehicle; 5 μl) were slowly injected into the subarachnoid space between the L5 and L6 vertebrae (i.t.). Subsequent behaviors were analyzed as described (25). For hyperalgesia, mice were placed on a hot plate (55.0 ± 0.1°C), and the time elapsed until the mice showed the first avoidance response was measured starting 10 and 30 min after i.t. injection of PGD2 and PGE2, respectively. Allodynia was assessed every 5 min over a 50-min period after i.t. injection, by light stroking of the flanks of the mice with a paintbrush, and the allodynic response was ranked as 0–2. The maximum possible cumulative score for allodynia of six mice was 12 in any 5-min period and 120 over the 50-min experimental period.
All animal experiments were performed according to the Japanese Law for the Protection of Experimental Animals and also conformed with the regulations issued by the National Institutes of Health and the Society for Neuroscience.
Immunohistochemistry.
Under pentobarbital anesthesia, animals were perfused at 4°C with 4% paraformaldehyde and 4% sucrose in sodium phosphate, pH 7.4, followed by the same fixative adjusted with acetic acid to pH 3.5–4.0. The isolated spinal cord was postfixed with the acidic fixative containing 20% sucrose at 4°C overnight. Cryosections of this tissue at a 15-μm thickness were mounted on 3-aminopropyltriethoxysilane-coated slides and incubated with rat monoclonal anti-mouse L-PGDS antibody (1 μg/ml) in PBS containing 0.1% normal goat serum and 0.1% Triton X-100 overnight at room temperature. As a control, preimmune rat IgG was used as the primary antibody. The immunoreactivity was visualized with the avidin-biotinylated enzyme complex (VECTASTAIN Elite ABC kit) and hydrogen peroxide-supplemented 3,3′-diaminobenzidine tetrahydrochloride. The sections then were counterstained with cresil violet.
Statistical Analysis.
Data for hyperalgesia were analyzed by parametric ANOVA, and statistical significance (P < 0.05) was further examined by Duncan’s test. Data for allodynia were analyzed by nonparametric ANOVA, and statistical significance (P < 0.05) was further examined by Steel’s or Williams’ test for multiple comparison.
RESULTS
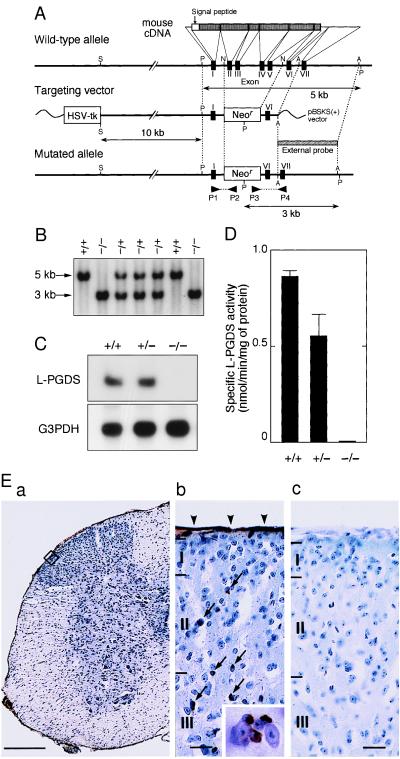
We isolated the mouse gene encoding L-PGDS and constructed a targeting vector used for homologous recombination to produce the null mutation of the gene (Fig. 1A). Three independent clones (nos. 23, 36, and 169) of mutated ES cells were obtained, and each clone was used to produce chimeric males that were bred to C57BL/6 female animals to generate heterozygous mutant F1 mice on a mixed background. Interbreeding of the heterozygotes produced homozygous L-PGDS−/− mice at a ratio expected from the mendelian frequency, i.e., 19 (+/+), 47 (+/−), and 15 (−/−) males and 22 (+/+), 44 (+/−), and 18 (−/−) females, indicating that the deficiency of the L-PGDS gene does not lead to fetal death. The deficiency of this gene was confirmed by PCR and Southern (Fig. 1B) and Northern (Fig. 1C) blot analyses. No L-PGDS activity was detected in the brain of L-PGDS−/− mice, and the activity of L-PGDS+/− mice was about half of that of the wild-type mice (0.860 ± 0.03 nmol/min per mg of protein; Fig. 1D). The L-PGDS-immunoreactivity was detected in the leptomeninges and oligodendrocytes of the spinal cord (Fig. 1E-a and b) and the brain (data not shown) of wild-type mice, but not in those of L-PGDS−/− mice (Fig. 1E-c). There were no differences between the wild-type and L-PGDS−/− mice as judged by the visual observation of the behavior and the growth curves of body weight up to 40 weeks of age. L-PGDS−/− mice had no apparent morphological or histological abnormalities.
Figure 1.
Generation of the L-PGDS mutation. (A) Restriction maps for the wild-type L-PGDS allele (Top), the targeting vector (Middle), and the mutated allele (Bottom). Restriction sites: S, SalI; P, PvuII; N, NcoI; A, AcyI. HSV-tk, herpes simplex virus thymidine kinase gene; Neor, neomycin resistance gene. (B) Southern blot analysis of PvuII-digested genomic DNA (20 μg/lane) from tails of heterozygous offsprings (F2 generation), using the 32P-labeled external probe shown in A. (C) Northern blot analysis of total RNA (20 μg/lane) in the brain using 32P-labeled cDNAs for mouse L-PGDS and glyceraldehyde-3-phosphate dehydrogenase (G3PDH). (D) L-PGDS activity in the brain. +/+, wild-type; +/−, heterozygote; −/−, homozygote of F2 mice. Each bar represents the mean ± SEM (n = 5). (E) Immunohistochemical staining of L-PGDS in the lumbar spinal cord of wild-type (a and b) and L-PGDS−/− mice (c). b depicts a high-magnification view of the superficial laminae (I, II, III) of the spinal cord (squared in a). Arrowheads and arrows indicate the immunoreactivity in the leptomeninges and oligodendrocytes, respectively. The inset in b shows a 4-fold higher-magnification view of the immunoreactive oligodendrocytes. [Scale bars: (a), 500 μm; (b) and (c), 50 μm.]
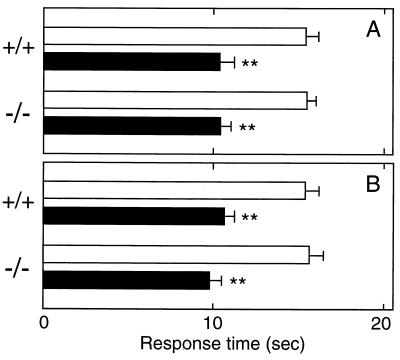
To clarify the physiological role of PGD2 and L-PGDS in pain transmission at the spinal level, we compared pain responses between wild-type and L-PGDS−/− mice. We first examined thermal hyperalgesia evoked by PGD2 and PGE2 by the hot plate test. The response time of mice to the hot plate (55°C) was measured at 10 and 30 min after i.t. injection of PGD2 and PGE2, respectively, the points of the maximum effects achieved. Exogenous PGD2 (5 ng/mouse) administered i.t. to wild-type and L-PGDS−/− mice reduced the latency period from 15.2 ± 1.0 s and 15.2 ± 0.6 s (mean ± SEM, n = 10) to 10.1 ± 0.7 s and 10.2 ± 0.6 s, respectively (Fig. 2A). Therefore, the hyperalgesic reaction of the same extent between wild-type and L-PGDS−/− mice indicated that the expression of DP receptor may not be significantly altered in the spinal cord of the L-PGDS−/− mice. Similar results were obtained with PGE2 (10 ng)-evoked hyperalgesia (Fig. 2B).
Figure 2.
Hyperalgesia evoked by PGD2 and PGE2 in wild-type and L-PGDS−/− mice. PGD2 (5 ng, filled bars in A), PGE2 (10 ng, filled bars in B), or saline (empty bars in A and B) was injected into the subarachnoid space of wild-type and L-PGDS−/− mice. Thermal hyperalgesia was assessed 10 and 30 min after i.t. injection of PGD2 and PGE2, respectively. Each bar represents the mean ± SEM (n = 10). ∗∗, P < 0.01 compared with the saline-injected control group.
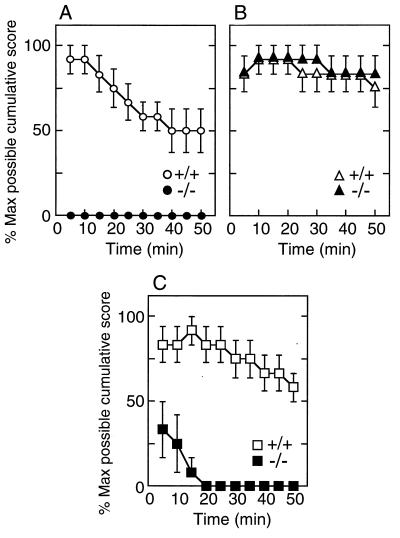
When PGE2 (10 ng/mouse) was administered i.t. to wild-type mice, innocuous tactile stimuli applied to the flank of the mice evoked allodynic responses 5 min after i.t. injection, and the mechanical allodynia gradually decreased over the 50-min experimental period (Fig. 3A). Unexpectedly, i.t. injection of PGE2 (10 ng) could not induce allodynia at all in L-PGDS−/− mice (Fig. 3A). In contrast, i.t. injection of PGF2α (1 μg) induced allodynia for 50 min to a similar extent in wild-type and L-PGDS−/− mice (Fig. 3B). These results suggest that PGD2 is essential for the induction of allodynia by PGE2, but not by PGF2α.
Figure 3.
Time courses of allodynia induced by PGE2, PGF2α, and bicuculline in wild-type and L-PGDS−/− mice. PGE2 (10 ng, A), PGF2α (1 μg, B), or bicuculline (25 ng, C) was injected into the subarachnoid space of wild-type (○, ▵, □) and L-PGDS−/− mice (•, ▴, ■) mice. Assessment of allodynia was made as described in Materials and Methods. The value (mean ± SEM) represents the percent of the maximum possible cumulative score of 5–6 mice evaluated every 5 min.
Both excitatory and inhibitory neurotransmitters are important in sensory processing in the spinal dorsal horn. Either the blockade of inhibitory neurotransmission by GABA and glycine (34, 35) or augmentation of pain transmission by the excitatory transmission by the glutamate-NO system (36, 37) is known to induce allodynia. As shown in Fig. 3C, i.t. administration of bicuculline [GABA type A (GABAA)] receptor antagonist, 25 ng/mouse) induced allodynia over a 50-min experiment in wild-type mice. Intrathecal administration of strychnine (a strychnine-sensitive glycine receptor antagonist, 250 ng), glutamate receptor agonists α-amino-3-hydroxy-5-methyl-4-isoxalole-4-propionic acid (10 ng) and N-methyl-d-aspartate (10 ng), the substrate of NO synthase arginine (5 μg), or the NO donor sodium nitroprusside (10 ng) also induced allodynia in wild-type mice (Table 1). When the scores of allodynia obtained for the overall 50 min were cumulated and expressed as a percent of the maximum possible score, the allodynic score of any one agent was beyond 70%. In L-PGDS−/− mice, however, the allodynic response induced by bicuculline was markedly reduced after the first 15 min and disappeared thereafter (Fig. 3C). When the 50-min cumulative scores were compared (Table 1), it became evident that the reduction of allodynia in L-PGDS−/− mice was specific to bicuculline (6.7 ± 4.0%), besides PGE2 (0.0%). The i.t. injection of PGD2 alone did not induce allodynia in either wild-type or L-PGDS−/− mice at doses up to 100 ng. These results demonstrate that PGD2 may be involved in the GABAA receptor pathway in the spinal cord.
Table 1.
Allodynia induced by various agents in wild-type and L-PGDS−/−mice
| Agent | Allodynia, %
|
|
|---|---|---|
| +/+ | −/− | |
| PGD2, 100 ng | 0.0 | 0.0 |
| PGE2, 10 ng | 67.5 ± 7.7 | 0.0* |
| PGF2α, 1 μg | 85.0 ± 8.9 | 87.5 ± 8.1 |
| Bicuculline, 25 ng | 76.7 ± 8.4 | 6.7 ± 4.0* |
| Strychnine, 250 ng | 79.0 ± 9.8 | 75.0 ± 8.9 |
| AMPA, 10 ng | 81.0 ± 10.1 | 78.3 ± 8.7 |
| NMDA, 10 ng | 74.0 ± 8.7 | 65.8 ± 8.2 |
| Arginine, 5 μg | 80.0 ± 9.9 | 74.2 ± 9.1 |
| SNP, 10 ng | 73.8 ± 11.4 | 75.8 ± 9.4 |
Data (mean ± SEM, n = 5–6) are expressed as a percent of the maximum possible cumulative score over the 50-min experiment. ∗, P < 0.01 compared to wild-type (+/+) mice. AMPA, α-amino-3-hydroxy-5-methyl-4-isoxalole-4-propionic acid; NMDA, N-methyl-d-aspartate; SNP, sodium nitroprusside.
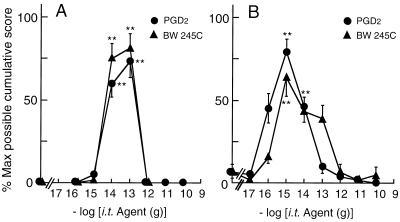
To test whether endogenous PGD2 affected the PGE2- and bicuculline-evoked allodynia, we examined the effect of exogenous PGD2 and its analogue on allodynia in wild-type and L-PGDS−/− mice. Simultaneous i.t. injection of PGD2 with 10 ng of PGE2 evoked allodynia in L-PGDS−/− mice at 10 and 100 fg (Fig. 4A). Similarly, PGD2 reproduced bicuculline (25 ng)-evoked allodynia in L-PGDS−/− mice over a range of dosage from 0.1 to 10 fg (Fig. 4B). The extent (75.0 ± 9.8%) of allodynic response induced by PGE2 with 100 fg of PGD2 and that (79.2 ± 8.0%) by bicuculline with 1 fg of PGD2 in L-PGDS−/− mice were almost the same as those (67.5 ± 7.7% and 76.7 ± 8.4%) by PGE2 and bicuculline alone in wild-type mice. The PGE2- and bicuculline-induced allodynia also were reproduced by the simultaneous injection of the DP agonist BW 245C (3) in a similar dose-dependent manner (Fig. 4 A and B). Although EP2 subtype of PGE2 receptor and PGI2 receptor as well as DP receptor are coupled to stimulation of adenylate cyclase (38), neither the EP2 agonist butaprost nor the PGI2 receptor agonist cicaprost evoked allodynia by bicuculline in L-PGDS−/− mice over a wide range of dosage from 0.01 fg to 10 ng (data not shown), demonstrating that the synergistic effect of PGD2 with bicuculline and PGE2 is selective among PGs examined.
Figure 4.
Effects of PGD2 and the DP receptor agonist BW 245C on allodynia evoked by PGE2 or bicuculline in L-PGDS−/− mice. PGE2 (10 ng, A) or bicuculline (25 ng, B) was injected simultaneously with various doses of PGD2 (•) or BW 245C (▴) into the subarachnoid space of L-PGDS−/− mice. The value (mean ± SEM) represents the percent of the maximum possible cumulative score of 5–6 mice evaluated for 50 min. ∗∗, P < 0.01 compared with the PGE2- or bicuculline-injected group.
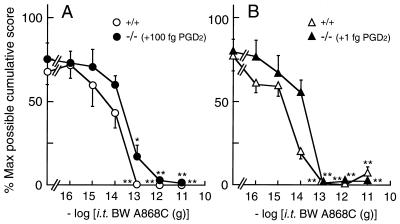
To further confirm the involvement of endogenous PGD2 in the PGE2- and bicuculline-evoked allodynia, we examined the effect of the DP antagonist BW A868C (3) on them (Fig. 5). As expected, allodynia evoked by the simultaneous injection of PGE2 (10 ng) with 100 fg of PGD2 in L-PGDS−/− mice was dose-dependently reversed by BW A868C with an ID50 (95% confidence limits) of 33.2 fg (7.20–141 fg) (Fig. 5A). In the case of bicuculline (25 ng) with 1 fg of PGD2 in L-PGDS−/− mice, allodynia was also dose-dependently reversed by BW A868C, with an ID50 of 11.9 fg (2.43–55.7 fg) (Fig. 5B). Furthermore, allodynia evoked by PGE2 or bicuculline in wild-type mice also was blocked by BW A868C with an ID50 of 13.1 fg (4.90–36.7 fg) and 2.17 fg (0.11–14.4 fg), respectively. These observations on wild-type and L-PGDS−/− mice strongly suggest that endogenous PGD2 at femtogram levels is involved in the actions of PGE2 and bicuculline, possibly via the GABAA receptor, at the spinal level.
Figure 5.
Effect of the DP receptor antagonist BW A868C on allodynia evoked by PGE2 (A) or bicuculline (B) in wild-type and L-PGDS−/− mice. Various doses of BW A868C were injected simultaneously with 10 ng PGE2 or 25 ng bicuculline in wild-type mice (○, ▵) or 10 ng PGE2 + 100 fg PGD2 or 25 ng bicuculline + 1 fg PGD2 into the subarachnoid space of L-PGDS−/− mice (•, ▴). The value (mean ± SEM) represents the percent of the maximum possible cumulative score of 5–6 mice evaluated for 50 min. ∗, P < 0.05; ∗∗, P < 0.01 compared with the PGE2- or bicuculline-injected group.
DISCUSSION
The present study with L-PGDS−/− mice first of all revealed that endogenous PGD2 is essential for the PGE2-evoked allodynia and that exogenous PGD2 injected in a femtogram amount is able to compensate for the loss of function in L-PGDS−/− mice (Figs. 3A and 4A). We also showed that exogenous PGD2 in the amount of higher than picogram suppressed the PGE2-evoked allodynia in both wild-type and L-PGDS−/− mice (Fig. 4A). This finding is in agreement with our previous results on wild-type mice showing that PGD2 blocked the PGE2-induced allodynia in a picogram to nanogram amount and that the DP receptor antagonist BW A868C alone induced allodynia, which was blocked by the further addition of PGD2 (24). PGD2 and PGE2 are positional isomers and have been shown to exhibit occasionally opposite biological activities in the CNS (3–5). For example, PGD2 and PGE2 are two of the major endogenous sleep-regulating substances, the former promoting sleep and the latter wakefulness (3, 4). PGD2 lowers body temperature, whereas PGE2 elevates it (3, 4). However, in the case of allodynia, the relationship between PGD2 and PGE2 is considered to be more complex than the previous prediction.
PGs are ubiquitously distributed in virtually all mammalian tissues and organs and produce a broad range of biological actions through their binding to specific receptors on plasma membranes. The diversity of PGE2 actions is the result of the coupling of EP receptor subtypes EP1, EP2, EP3, and EP4 to different signal transduction pathways (38). The PGE2-induced allodynia is mediated by the EP1 subtype, and the PGE2-induced thermal hyperalgesia, by the EP2 and EP3 subtypes (39, 40). This finding that L-PGDS−/− mice did not show the PGE2-induced allodynia (Fig. 3A), yet displayed the PGE2-induced hyperalgesia to the same extent as wild-type mice (Fig. 2B), indicates that PGD2 is essential for the EP1-mediated allodynia. Both PGD2- and PGE2-binding proteins were shown to be present at high density in the superficial dorsal horn in autoradiographic studies (41, 42). Because PGs act in the vicinity of their sites of formation via an autocrine or a paracrine process, the balance of PGD2 and PGE2 might be a pivotal factor for induction and suppression of allodynia in the spinal cord.
Among excitatory and inhibitory agents that induced allodynia in wild-type mice, only the bicuculline-evoked allodynia was abolished in L-PGDS−/− mice (Table 1, Fig. 3C). GABAergic neurons are present in laminae I-III of the spinal cord, and many of the neurons with somata in laminae I-III are inhibitory interneurons containing GABA (43). We previously suggested that both PGE2- and bicuculline-induced allodynia were mediated through pathways that include the glutamate receptor-NO system (35–37). The induction of allodynia was considered to result from removal of tonic or evoked inhibition from pathways relaying information about innocuous stimuli and augmentation of pain transmission by the excitatory transmission by the glutamate-NO system. However, all activators of the glutamate receptor-NO system induced allodynia in wild-type and L-PGDS−/− mice in a similar manner (Table 1), suggesting that the site(s) of PGD2 involvement in the induction of allodynia by PGE2 or bicuculline lies upstream of the glutamate-NO system. Although the precise site(s) of action of PGD2 and the relationship between PGE2 and bicuculline in the induction of allodynia are not known, the present study clearly demonstrates that PGD2 affects the GABAA receptive system. GABA is also a major inhibitory neurotransmitter of higher centers of the mammalian brain. The control of the excitability of GABAergic and GABAA receptive neurons is proposed to play a critical role in sleep-wakefulness (44), body temperature (45), and odor responses (46). The elucidation of the involvement of PGD2 in the GABAA receptor system in the spinal cord may clarify the action mechanism for other neural functions of PGD2 in the brain.
Acknowledgments
We are grateful to Dr. R. Kühn for E14-1 ES cells; Dr. P. M. Raper for BW 245C and BW A868C; Prof. T. Kaneko (Kyoto University) for morphological analysis; and Dr. K. Toyoshima (Osaka Medical Center for Cancer and Cardiovascular Diseases), Prof. H. Okayama (University of Tokyo), and Prof. T. Kishimoto (Osaka University) for their comments. We also thank Y. Nouka, J. Wiley, Y. Sakaguchi, and S. Takei for DNA sequence; and Y. Ishikawa, S. Matsumoto, Y. Kuwahata, K. Shinoda, D. Irikura, A. Maekawa, and Y. Ono for technical assistance. This work was supported by the Japan Science and Technology Corporation (N.E. and Y.U.); the Ministry of Education, Science, Sports, and Culture of Japan (06508003 to O.H., 07457033 and 07558108 to Y.U., 08770111 to Y.K., 09480168 to S.I. and 09771194 to T.M.); Suntory Institute for Bioorganic Research (N.E.); Takeda Science Foundation (N.E.); Yamanouchi Foundation for Research on Metabolic Disorders (N.E.); the Science Research Promotion Fund of the Japan Private School Promotion Foundation (S.I.); Jinsenkai Foundation of Osaka Medical College (T.M.); and Naito Foundation (T.M.).
ABBREVIATIONS
- PG
prostaglandin
- L-PGDS
lipocalin-type PGD synthase
- GABA
γ-aminobutyric acid
- CNS
central nervous system
- i.t.
intrathecal(ly)
- ES
embryonic stem
- DP
D type of PG
- EP
E type of PG
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. D83329).
References
- 1.Narumiya S, Ogorochi T, Nakao K, Hayaishi O. Life Sci. 1982;31:2093–2103. doi: 10.1016/0024-3205(82)90101-1. [DOI] [PubMed] [Google Scholar]
- 2.Ogorochi T, Narumiya S, Mizuno N, Yamashita K, Miyazaki H, Hayaishi O. J Neurochem. 1984;43:71–82. doi: 10.1111/j.1471-4159.1984.tb06680.x. [DOI] [PubMed] [Google Scholar]
- 3.Ito S, Narumiya S, Hayaishi O. Prostaglandins Leukotrienes Essent Fatty Acids. 1989;37:219–234. doi: 10.1016/0952-3278(89)90033-1. [DOI] [PubMed] [Google Scholar]
- 4.Hayaishi O. FASEB J. 1991;5:2575–2581. [PubMed] [Google Scholar]
- 5.Urade Y, Hayaishi O, Matsumura H, Watanabe K. J Lipid Mediators Cell Signaling. 1996;14:71–82. doi: 10.1016/0929-7855(96)01511-8. [DOI] [PubMed] [Google Scholar]
- 6.Urade Y, Watanabe K, Hayaishi O. J Lipid Mediators Cell Signaling. 1995;12:257–273. doi: 10.1016/0929-7855(95)00032-l. [DOI] [PubMed] [Google Scholar]
- 7.Ujihara M, Urade Y, Eguchi N, Hayashi H, Ikai K, Hayaishi O. Arch Biochem Biophys. 1988;260:521–531. doi: 10.1016/0003-9861(88)90477-8. [DOI] [PubMed] [Google Scholar]
- 8.Urade Y, Kitahama K, Ohishi H, Kaneko T, Mizuno N, Hayaishi O. Proc Natl Acad Sci USA. 1993;90:9070–9074. doi: 10.1073/pnas.90.19.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashima T, Sakuda K, Tohma Y, Yamashita J, Oda H, Irikura D, Eguchi N, Beuckmann C T, Kanaoka Y, Urade Y, Hayaishi O. J Neurosci. 1997;17:2376–2382. doi: 10.1523/JNEUROSCI.17-07-02376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann A, Conradt H S, Gross G, Nimtz M, Lottspeich F, Wurster U. J Neurochem. 1993;61:451–456. doi: 10.1111/j.1471-4159.1993.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe K, Urade Y, Mäder M, Murphy C, Hayaishi O. Biochem Biophys Res Commun. 1994;203:1110–1116. doi: 10.1006/bbrc.1994.2297. [DOI] [PubMed] [Google Scholar]
- 12.Clausen J. Proc Soc Exp Biol Med. 1961;107:170–172. doi: 10.3181/00379727-107-26569. [DOI] [PubMed] [Google Scholar]
- 13.Kuruvilla A P, Hockwald G M, Ghiso J, Castano E M, Pizzolato M, Frangione B. Brain Res. 1991;565:337–340. doi: 10.1016/0006-8993(91)91666-o. [DOI] [PubMed] [Google Scholar]
- 14.Zahn M, Mäder M, Schmidt B, Bollensen E, Felgenhauer K. Neurosci Lett. 1993;154:93–95. doi: 10.1016/0304-3940(93)90179-o. [DOI] [PubMed] [Google Scholar]
- 15.Hirata M, Kakizuka A, Aizawa M, Ushikubi F, Narumiya S. Proc Natl Acad Sci USA. 1994;91:11192–11196. doi: 10.1073/pnas.91.23.11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boie Y, Sawyer N, Slipetz D M, Metters K M, Abramovitz M. J Biol Chem. 1995;270:18910–18916. doi: 10.1074/jbc.270.32.18910. [DOI] [PubMed] [Google Scholar]
- 17.Oida H, Hirata M, Sugimoto Y, Ushikubi F, Ohishi H, Mizuno N, Ichikawa A, Narumiya S. FEBS Lett. 1997;417:53–56. doi: 10.1016/s0014-5793(97)01253-2. [DOI] [PubMed] [Google Scholar]
- 18.Gerashchenko D, Beuckmann C T, Kanaoka Y, Eguchi N, Gordon W C, Urade Y, Bazan N G, Hayaishi O. J Neurochem. 1998;71:937–945. doi: 10.1046/j.1471-4159.1998.71030937.x. [DOI] [PubMed] [Google Scholar]
- 19.Yaksh T L, Aimone L. In: Textbook of Pain. 2nd Ed. Wall P D, Melzack R, editors. Edinburgh: Churchill Livingstone; 1989. pp. 283–320. [Google Scholar]
- 20.Woolf C J. In: Textbook of Pain. 3rd Ed. Wall P D, Melzack R, editors. Edinburgh: Churchill Livingstone; 1994. pp. 101–112. [Google Scholar]
- 21.Dray A, Urban L, Dickenson A. Trends Pharmacol Sci. 1994;15:190–197. doi: 10.1016/0165-6147(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 22.Vane J R. Nat New Biol. 1971;231:232–235. doi: 10.1038/newbio231232a0. [DOI] [PubMed] [Google Scholar]
- 23.Yaksh T L, Malmberg A B. In: Textbook of Pain. 3rd Ed. Wall P D, Melzack R, editors. Edinburgh: Churchill Livingstone; 1994. pp. 165–200. [Google Scholar]
- 24.Cervero F, Laird J M A. Pain. 1996;68:13–23. doi: 10.1016/S0304-3959(96)03165-X. [DOI] [PubMed] [Google Scholar]
- 25.Minami T, Okuda-Ashitaka E, Mori H, Ito S, Hayaishi O. J Pharmacol Exp Ther. 1996;278:1146–1152. [PubMed] [Google Scholar]
- 26.Uda R, Horiguchi S, Ito S, Hyodo M, Hayaishi O. Brain Res. 1990;510:26–32. doi: 10.1016/0006-8993(90)90723-o. [DOI] [PubMed] [Google Scholar]
- 27.Minami T, Uda R, Horiguchi S, Ito S, Hyodo M, Hayaishi O. Pain. 1992;50:223–229. doi: 10.1016/0304-3959(92)90166-9. [DOI] [PubMed] [Google Scholar]
- 28.Minami T, Uda R, Horiguchi S, Ito S, Hyodo M, Hayaishi O. Pain Res. 1992;7:129–134. doi: 10.1016/0304-3959(92)90166-9. [DOI] [PubMed] [Google Scholar]
- 29.Minami T, Uda R, Horiguchi S, Ito S, Hyodo M, Hayaishi O. Pain. 1994;57:217–223. doi: 10.1016/0304-3959(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 30.Urade Y, Nagata A, Suzuki Y, Fujii Y, Hayaishi O. J Biol Chem. 1989;264:1041–1045. [PubMed] [Google Scholar]
- 31.Igarashi M, Nagata A, Toh H, Urade Y, Hayaishi O. Proc Natl Acad Sci USA. 1992;89:5376–5380. doi: 10.1073/pnas.89.12.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urade Y, Tanaka T, Eguchi N, Kikuchi M, Kimura H, Toh H, Hayaishi O. J Biol Chem. 1995;270:1422–1428. doi: 10.1074/jbc.270.3.1422. [DOI] [PubMed] [Google Scholar]
- 33.Urade Y, Fujimoto N, Hayaishi O. J Biol Chem. 1985;260:12410–12415. [PubMed] [Google Scholar]
- 34.Yaksh T L. Pain. 1989;37:111–123. doi: 10.1016/0304-3959(89)90160-7. [DOI] [PubMed] [Google Scholar]
- 35.Onaka M, Minami T, Nishihara I, Ito S. Anesthesiology. 1996;84:1215–1222. doi: 10.1097/00000542-199605000-00024. [DOI] [PubMed] [Google Scholar]
- 36.Minami T, Nishihara I, Uda R, Ito S, Hyodo M, Hayaishi O. Pain. 1994;57:225–231. doi: 10.1016/0304-3959(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 37.Minami T, Nishihara I, Ito S, Sakamoto K, Hyodo M, Hayaishi O. Pain. 1995;61:285–290. doi: 10.1016/0304-3959(94)00183-F. [DOI] [PubMed] [Google Scholar]
- 38.Coleman R A, Smith W L, Narumiya S. Pharmacol Rev. 1994;46:205–229. [PubMed] [Google Scholar]
- 39.Minami T, Nishihara I, Uda R, Ito S, Hyodo M, Hayaishi O. Br J Pharmacol. 1994;112:735–740. doi: 10.1111/j.1476-5381.1994.tb13139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minami T, Nishihara I, Sakamoto K, Ito S, Hyodo M, Hayaishi O. Br J Pharmacol. 1995;115:73–76. doi: 10.1111/j.1476-5381.1995.tb16321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe Yu, Watanabe Y, Hayaishi O. In: Biomedical Imaging. Hayaishi O, Torizuka K, editors. Tokyo: Academic; 1986. pp. 227–238. [Google Scholar]
- 42.Matsumura K, Watanabe Yu, Imai-Matsumura K, Connolly M, Koyama Y, Onoe H, Watanabe Y. Brain Res. 1992;581:292–298. doi: 10.1016/0006-8993(92)90720-t. [DOI] [PubMed] [Google Scholar]
- 43.Malcangio M, Bowery N G. Trends Pharmacol Sci. 1994;15:190–197. [Google Scholar]
- 44.Kim U, Sanchez-Vives M V, McCormick D A. Science. 1997;278:130–134. doi: 10.1126/science.278.5335.130. [DOI] [PubMed] [Google Scholar]
- 45.Sancibrian M, Serrano J S, Minano F J. Gen Pharmacol. 1991;22:259–262. doi: 10.1016/0306-3623(91)90443-a. [DOI] [PubMed] [Google Scholar]
- 46.Yokoi M, Mori K, Nakanishi S. Proc Natl Acad Sci USA. 1995;92:3371–3375. doi: 10.1073/pnas.92.8.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]