Abstract
Yersinia enterocolitica strains comprise an important group of bacterial enteropathogens that cause a broad range of gastrointestinal syndromes. Three groups are distinguishable within this bacterial species, namely, the nonpathogenic group (biotype 1A strains), the low-pathogenicity, non-mouse-lethal group (biotypes 2 to 5), and the high-pathogenicity, mouse-lethal group (biotype 1B). To date, the presence of the high-pathogenicity island (HPI), a chromosomal locus that encodes the yersiniabactin system (involved in iron uptake), defines essentially the difference between low-pathogenicity and high-pathogenicity Y. enterocolitica strains, with the low-pathogenicity strains lacking the HPI. Using the powerful tool of representational difference analysis between the nonpathogenic 1A strain, NF-O, and its high-pathogenicity 1B counterpart, WA-314, we have identified a novel type II secretion gene cluster (yts1C-S) occurring exclusively in the high-pathogenicity group. The encoded secreton, designated Yts1 (for Yersinia type II secretion 1) was shown to be important for virulence in mice. A close examination of the almost completed genome sequence of another high-pathogenicity representative, Y. enterocolitica 8081, revealed a second putative type II secretion cluster uniformly distributed among all Y. enterocolitica isolates. This putative species-specific cluster (designated yts2) differed significantly from yts1, while resembling more closely the putative type II cluster present on the genome of Y. pestis. The Yts1 secreton thus appears to have been additionally acquired by the high-pathogenicity assemblage for a virulence-associated function.
The genus Yersinia comprises an important group of bacterial pathogens, with Yersinia enterocolitica, Y. pseudotuberculosis, and Y. pestis representing the species of interest. Y. pestis is the etiologic agent of plague, whereas Y. pseudotuberculosis and Y. enterocolitica are enteropathogens that cause a broad range of gastrointestinal syndromes ranging from acute gastroenteritis to mesenteric lymphadenitis. Central to the pathogenicity of the Yersinia is the presence of a 70-kb pYV virulence plasmid whose products mediate, among other things, the resistance of Yersinia to phagocytosis by polymorphonuclear leukocytes and macrophages (41). In addition, the chromosomally encoded Ail (for attachment invasion loci), Myf (for mucoid Yersinia factor), and Inv (invasin) proteins have been implicated in the virulence of the species to various degrees (3, 38).
Two groups have been identified among the Y. enterocolitica strains, which is a focus of the present study: nonpathogenic strains comprising mainly biotype 1A organisms and pathogenic strains carrying the virulence plasmid pYV. The latter group is further subdivided into a low-pathogenicity (LP), non-mouse-lethal group represented by biotypes 2 to 5 and a high-pathogenicity (HP), mouse-lethal group exemplified by biotype 1B strains (8). As a mouse virulence determinant, an HP island (HPI) has been identified, which encodes for synthesis of the siderophore yersiniabactin, an iron-sequestering low-molecular-weight compound invaluable in the iron-limiting environment of the host (8, 25, 36). The presence of the HPI has also been demonstrated in Y. pseudotuberculosis and Y. pestis (4, 5, 14, 18). The members of the LP group typically lack the HPI but possess all other known virulence factors. In terms of geographical distribution, the LP and HP Y. enterocolitica species exhibit some preferences, with the HP organisms being more frequently isolated in the United States (so-called New World strains), whereas the LP isolates (so-called Old World strains) are predominantly isolated in Europe and Japan (9, 10).
We report here the application of representational difference analysis to map out novel chromosomal loci unique to HP Y. enterocolitica of serotype O:8 biotype 1B.
This powerful method of suppressive subtractive hybridization has been successfully applied in mapping out crucial genomic differences between closely related bacterial genomes and even isolates of the same species (7, 11, 17, 34). We describe the discovery of a novel type II secretion apparatus with impact on the virulence of the HP Y. enterocolitica species. The distribution of this novel secreton is highly similar to that of the recently described chromosomally encoded type III secretion system (TTSS) that was found to be present only in the HP group of Y. enterocolitica species (19, 22). This novel transport cluster designated Yts1 (for Yersinia type II secretion 1) shares the highest homology with the Eps protein transport cluster of Vibrio cholerae (44), the latter mediating the extracellular transport of protease, the cholera toxin, and chitinase.
MATERIALS AND METHODS
The bacterial strains and plasmids used in the present study are listed in Table 1. Typically, bacterial strains were cultivated in Luria-Bertani (LB) medium or on LB agar plates (Difco Laboratories, Detroit, Mich.) at 27°C for Yersinia or 37°C for E. coli. The following antibiotic concentrations were used for plasmid maintenance and selection: ampicillin at 100 μg ml−1 (300 μg ml−1 for Yersinia strains), tetracyline at 40 μg ml−1, kanamycin at 100 μg ml−1, streptomycin at 100 μg ml−1, and chloramphenicol at 20 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Y. enterocolitica | ||
| WA-314 | Y. enterocolitica O:8 biotype 1B with pYV virulence plasmid | 25 |
| WA-C | Plasmidless derivative of strain WA-314; spontaneously resistant to nalidixic acid | 25 |
| WA-CS | Derivative of WA-C; spontaneous Smr mutant | This study |
| 8081 | Clinical isolate, O:8 biotype 1B | 40 |
| WA-CS yts1E::Kanr | Derivative of WA-CS; Nalr, Kanr Smr (insertional inactivation of yts1E by a kanamycin cassette) | This study |
| Ye1209-79 | Clinical isolate, O:13 biotype 1B | 25 |
| Ye1223-75-1 | Clinical isolate, O:20 biotype 1B | 25 |
| Ye737 | Clinical isolate, O:21 biotype 1B | 25 |
| Y-108 | Clinical isolate, O:3 biotype 4 | 25 |
| Y-96 | Clinical isolate, O:9 biotype 2 | 25 |
| NF-O | Clinical isolate, O:5 biotype 1A | 42 |
| MRS-40 | Clinical isolate, O:9 biotype 2 | 25 |
| Y. pseudotuberculosis | ||
| H141/84 | Serotype O:1a | S. Aleksic |
| H457/86 | Serotype O:2a | S. Aleksic |
| IP3295 | Serotype O:1 | E. Carniel |
| 346 | Serotype O:3 | S. Aleksic |
| Y. pestis | ||
| KIM | Biotype Mediaevalis | R. R. Brubaker |
| KUMA | Biotype Antiqua | R. R. Brubaker |
| TS | Biotype Orientalis | R. R. Brubaker |
| E. coli | ||
| C-4441 | Enteroaggregative, O:128 | 47 |
| 12860 | Enteroinvasive, O:124 | 47 |
| DH5α | endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 (φ80lacZΔM15) | 23 |
| S17-1λpir | pir+tra+ | 48 |
| XL1-Blue MR | Δ(mcrA) 183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lacc | Stratagene |
| MOSBlue | endA1 hsdR17 (rK12− mK12+) supE44 thi-1recA1 gyrA96 relA1 lac [F′ proA+B+ lacIqZΔM15:Tn10(Tcr)] | Amersham |
| Plasmids | ||
| Supercos1 | Cloning vector; Apr Neor | Stratagene |
| ScF yts1E::Kanr | BglII/BamHI self-religated Supercos1 fragment carrying the mutated yts1E gene | This study |
| pKAS 32 | Cloning vector with dominant rpsL gene | 49 |
| pKAS 32 yts1E::Kanr | pKAS32 carrying a fragment from the putative type II yts1E gene inactivated by a kanamycin cassette | This study |
| pACYC184 | Cloning vector; Cmr Tcr | New England Biolabs |
| pUC4K | pUC vector carrying the kanamycin cassette from Tn903; Kanr Apr | Amersham |
| pMOSBlue | Cloning vector; Apr | Amersham |
Nalr, nalidixic acid resistant; Tcr, tetracycline resistant; Cmr, chloramphenicol resistant.
Molecular genetic techniques.
Plasmid DNA was prepared by the modified nucleic acid extraction kits from Nucleobond. Chromosomal DNA was prepared as previously described (1). Construction of the cosmid gene library from Y. enterocolitica WA-314 serotype O:8 biotype 1B (mouse lethal) was performed with chromosomal DNA Sau3AI digested to yield DNA fragments ranging in size from 35 to 40 kb, which were subsequently ligated into the BamHI site of the cosmid vector Supercos1. Ligated DNAs were packaged in vitro into viable phage heads according to manufacturer's instructions (Stratagene) and transduced into Escherichia coli XL1-Blue MR. High-stringency Southern blot hybridizations were performed with digoxigenin -labeled probes at 68°C according to standard procedures. To determine the presence of the yts1D and yts2D genes among different Yersinia isolates, the following oligonucleotide primers were employed: Yts1D.for (5′-GCAGTAAAAG-GCAACATCAGCG-3′) and Yts1D.rev (5′-AAACAACGCGCATGACGACTTC-3′) for amplification of the yts1D gene and 2D.for (5′-GTAGGATATGCGGAGAACTTC-3′) and 2D.rev (5′-CGTTTTGCGGGAATAACTTTT-3′) for amplification of yts2D.
Construction and analysis of subtractive hybridization library.
The procedure of subtractive hybridization was performed as described in the PCR select genomic subtraction kit from Clontech. In the present study, Y. enterocolitica WA-314 genomic DNA was used as the tester, and genomic DNA of Y. enterocolitica serotype O:5, biotype 1A (NF-O, nonpathogen), was used as the driver. Briefly, chromosomal DNA was isolated (1) from both tester and driver strains and was RsaI digested. The tester DNA was then ligated with two different oligonucleotide adapters in two separate reactions, and then each adapter-ligation reaction was mixed with an excess of the driver DNA population and subjected to successive rounds of hybridization at 63°C. The subtracted fragments were then enriched through two rounds of PCR-amplification with adapter-specific primers. The secondary PCR products generated, enriched for tester specific sequences, were mobilized into the highly efficient pMOSBlue cloning vector (Amersham) and transformed into E. coli MOSBlue-competent cells (Amersham) for sequence analysis. To analyze the efficiency of the subtraction, the entire subtracted fragments were concentrated, and the pool was labeled as a probe to screen the NF-O DNA and DNA representative of other yersiniae in a high-stringency Southern blot hybridization reaction. In a second analysis, NF-O driver DNA was labeled as a probe and used to screen more than 50 subtracted clones (data not shown). Selected subtracted fragments of interest were digoxigenin-labeled and used to probe the WA-314 cosmid library in high-stringency colony blot reactions. Hybridizing cosmids were isolated and subsequently sequenced (1).
DNA sequencing and analysis.
DNA Sequencing was performed by the dideoxy-chain terminating method with an automated ABI Prism DNA sequencer. The T7 forward primer was used to initiate sequence reactions with the subtracted clones. Analysis of subtracted fragments was carried out with the BLASTN and BLASTX programs provided by the National Center for Biotechnology Information, The Institute for Genomic Research BLAST Center, and the Y. pestis genome sequence from the Sanger Center.
Reverse transcription analysis.
RNA isolation was carried out with Trizol reagent (Gibco) according to the manufacturer's instructions. RNA was isolated from bacterial cells grown in LB medium at various temperatures (27 and 37°C). As positive controls, the inv and ail genes were probed in parallel for transcription.
Construction of a Y. enterocolitica yts1E mutant.
To inactivate the yts1 genes, a site-directed insertional mutation in yts1E was generated in Y. enterocolitica WA-314. A 1.2-kb EcoRI-NotI PCR product was amplified from the original subtracted fragment (n28) that showed homology to the epsE gene in V. cholerae and cloned into the EcoRI-NotI site of a BglII/BamHI self-ligated Supercos1 vector fragment (designated ScF). A kanamycin cassette amplified from the pUC 4K plasmid and containing flanking BamHI restriction sites was introduced into the BamHI site of the amplified n28 fragment. The resulting n28 insert containing the kanamycin cassette was subsequently ligated into the suicide vector, pKAS 32. pKAS 32 harbors the dominant rpsL gene which encodes the S12 protein of the ribosomes (37). Thus, insertion of this suicide vector into the chromosome confers a streptomycin-sensitive (Sms) phenotype to a previously streptomycin-resistant (Smr) strain. The construct was transformed into E. coli S17-1λpir and subsequently mobilized into Y. enterocolitica WA-CS through conjugation. Selection for Smr strains indicated the presence of transconjugants that have excised the plasmid sequences through double allelic exchange. Transconjugants (mutants) were selected on LB agar plates containing kanamycin and streptomycin. The presence of the kanamycin cassette was confirmed through PCR, sequencing, and Southern blot hybridization.
In another experiment, the original n28 fragment was cloned into pKAS32, and the construct was introduced into Y. enterocolitica WA-314 yts1E::Kanr (kanamycin-resistant) strain using the procedure described above. Selection of Strr, Kans, and ampicillin-sensitive (Aps) clones indicated clones where double allelic exchange had excised the kanamycin cassette and restored the parental wild-type allele. This strain was subsequently designated reconstituted wild type. For transcomplementation, cosmid n28 or cosmid 1D (both harboring complete copies of the Yts1 secreton) were introduced into electrocompetent Y. enterocolitica WA-314 yts1E mutant by using standard procedures (1).
Animal experiments.
Female BALB/c mice (6 weeks old) were infected (groups of five) with yersiniae orogastrically at a dosage of 5 × 109 CFU. Mice were monitored twice daily; 48 h after infection the animals were sacrificed, and the average bacterial loads from infected organs (i.e., the liver, spleen, and Peyer's patches) were extrapolated. For the intestinal lavage, 5 ml of ice-cold sterile phosphate-buffered saline was flushed through the small intestine and then recovered, and bacterial loads were determined. For intravenous mouse inoculation, 104 CFU were inoculated into the mice, which were subsequently monitored for 2 to 3 days before sacrifice.
Nucleotide sequence accession number.
The sequence of the Y. enterocolitica WA-314 yts1C-S genes has been submitted to the EMBL database under the accession number AJ344214.
RESULTS
Construction of a library of subtracted fragments unique to Y. enterocolitica WA-314 and absent from the genome of Y. enterocolitica NF-O.
Subtraction of total genomic DNA of Y. enterocolitica NF-O (biogroup 1A, nonpathogen) from the genome of its highly pathogenic counterpart, WA-314, was performed by hybridization by using the PCR select genomic subtraction kit from Clontech. The secondary PCR products generated were cloned into pMOSBlue vector (Amersham), and a total of 200 subtracted clones were analyzed. These subtracted fragments, ranging in size from 200 to 1,500 bp, were sequenced and compared at both the nucleotide and the amino acid levels for homologies with sequences in databanks (provided by the National Center for Biotechnology Information, the Sanger Center, and the Institute for Genomic Research). Generally, the fragments fell into three categories: (i) fragments with sequence similarities to known genes from the Yersinia species and other organisms (67%), (ii) fragments with similarities to phages and mobile genetic elements (transposases, etc.) (13%), and (iii) fragments with unknown sequences (20%).
To verify the efficiency of the subtraction procedure, the generated library of subtracted fragments was probed with primers specific for known virulence markers among pathogenic Yersinia species. Genomic probes specific for the HPI and the chromosomally encoded ail and inv genes were utilized in high-fidelity PCRs. The PCRs were positive for the fyuA, irp2, ybtA, and irp1 gene probes (located on the HPI) and also for the ail and inv genes, indicating that the subtraction library was sufficiently enriched for tester-specific virulence markers.
Identification of a novel putative type II secretion pathway in the HP Y. enterocolitica WA-314 strain.
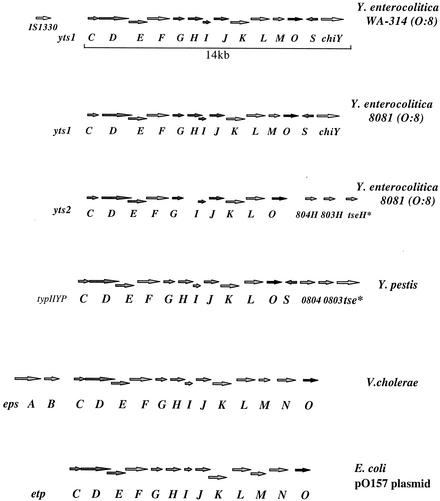
The subtracted fragment n28 was highly homologous to the epsE gene of V. cholerae (44), part of the type II secretion apparatus in this organism. To determine whether this high homology was continuous, the n28 fragment was used to isolate hybridizing cosmids from the cosmid library of Y. enterocolitica WA-314 described previously. Two hybridizing cosmids were identified. Cosmid n28 was fully sequenced and its analysis revealed 13 open reading frames (ORFs) with homology to proteins identified in type II secretion clusters. Also, a gene, designated chiY, which encodes a putative chitin-binding protein, was identified downstream of this novel type II cluster. The type II secretion gene cluster was designated yts1C-S; these letters correspond to the generally accepted nomenclature for the type II secretion pathway identified in different members of the Enterobacteriaceae (see first cluster in Fig. 1). Upstream of the yts1 cluster are sequences highly homologous to the chromosomally encoded Ysa TTSS.
FIG. 1.
The yts1 gene cluster of the HP Y. enterocolitica biotype 1B strains and its comparison to related secretons: the eps cluster in V. cholerae, the etp cluster in E. coli, and the secreton of Y. pestis (designated here typIIYP). 0804 and 0803 correspond to ORFs predicted from the genome sequence of Y. pestis. 804H and 803H are homologues of 0804 and 0803 on the Y. enterocolitica genome. tseH* is a homologue of the Y. pestis tse gene on the Y. enterocolitica genome. tse* is the serine sensor receptor.
The Yts1 secretion apparatus demonstrates significantly high similarity (for example, 59% homology of Yts1E to the EpsE protein of V. cholerae) to the secreton of V. cholerae (EpsC-N) required for outer membrane biogenesis and secretion of cholera toxin, along with a protease and a chitinase. The similarity to other homologous secretons was highest for Yts1D-G, an observation that confirms previous findings that the yts1D, yts1E, yts1F, and yts1G genes typically share the highest identity among the species (60 to 80%) with comparatively less homology (25 to 40%) among the other genes (45, 46). The putative yts1 cluster described here lacks a yts1N gene, in contrast to its counterpart in V. cholerae. This is, however, not surprising since many functional type II clusters described to date do not possess a yts1N gene, which several studies have shown not to be essential for protein secretion (45).
To determine whether the genes on the yts1 cluster are transcribed, reverse transcription analysis was carried out for the yts1D gene for bacteria grown in LB medium at 27 and at 37°C. As positive controls, the ail and inv genes (optimally transcribed at 37 and 23°C, respectively) were analyzed in parallel. The yts1D gene yielded a positive transcript for culture grown at 37°C, whereas very basal transcription occurred at 27°C (see Fig. 2).
FIG. 2.
Reverse transcription analysis of the yts1D and yts2D genes at 27 and at 37°C. Lanes: 1, marker; 2, ail at 37°C; 4, inv at 27°C; 6, yts1D at 27°C; 8, yts1D at 37°C; 10, yts2D at 27°C; 12, yts2D at 37°C. Lanes 3, 5, 7, 9, 11, and 13, the corresponding negative controls (without reverse transcription).
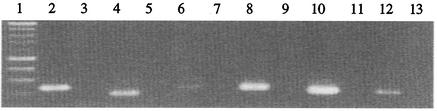
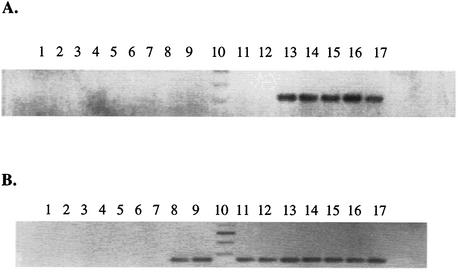
Next, we determined the presence of the yts1 genes in diverse Yersinia strains. As seen in Fig. 3, it is evident that the Yts1 secreton is exclusive to the HP Y. enterocolitica strains (Y. enterocolitica serotypes O:8, O:13, O:20, and O:21). The tested LP isolates (Y. enterocolitica serotypes O:3 and O:9) and several Y. pestis and Y. pseudotuberculosis strains did not hybridize to the yts1E probe. This observation was consistent with results from high-stringency PCRs employing primers for several genes on the cluster: among the different yersiniae, only the HP Y. enterocolitica strains tested positive (Fig. 4A).
FIG. 3.
Distribution of yts1 secretion cluster among different Yersinia species determined by Southern blot hybridization employing yts1E as probe. Lanes: 1, Y. pestis TS; 2, Y. pestis KIM; 3, Y. pestis KUMA; 4, Y. pseudotuberculosis O:1a; 5, Y. pseudotuberculosis O:2a; 6, Y. pseudotuberculosis O:3; 7, Y. enterocolitica O:8 (WA-314); 8, Y. enterocolitica WA-314 yts1E::Kanr; 9, Y. enterocolitica O:5 (NF-O); 10, Y. enterocolitica O:9; 11, Y. enterocolitica O:8 8081; 12, Y. enterocolitica O:21; 13, Y. enterocolitica O:20; 14, Y. enterocolitica O:13; 15, Y. enterocolitica O:9; 16, Y. enterocolitica O:3; 17, E. coli C-4441; 18, E. coli 12860.
FIG. 4.
Presence of the yts1D (A) and yts2D (B) genes among different Yersinia representatives. PCR was performed on purified chromosomal DNA with oligonucleotide primers specific for the yts1D and yts2D genes, respectively. Lanes: 1, Y. pseudotuberculosis H141/84; 2, Y. pseudotuberculosis H457/86; 3, Y. pseudotuberculosis IP3295; 4, Y. pseudotuberculosis 346; 5, Y. pestis KIM; 6, Y. pestis KUMA; 7, Y. pestis TS; 8, Y. enterocolitica O:5 (NF-O); 9, Y. enterocolitica O:9; 10, 100-bp ladder; 11, Y. enterocolitica O:3; 12, Y. enterocolitica O:9; 13, Y. enterocolitica O:8 (WA-314); 14, Y. enterocolitica O:8 8081; 15, Y. enterocolitica O:13; 16, Y. enterocolitica O:20; 17, Y. enterocolitica O:21
Interestingly, the closely related Y. enterocolitica serotype O:8 strains, WA-314 and 8081, showed differences in gene arrangement bordering the yts1 cluster. In WA-314, a novel IS element, IS1330 (29), is upstream of the yts1 cluster, whereas in strain 8081, it is lacking in the proximity of the yts1 cluster (see Fig. 1).
A species-specific type II secretion cluster distributed among all Y. enterocolitica serotypes.
Interest in identifying a second secretion cluster that would uniformly cut across the Y. enterocolitica strains without preference to the pathogenic species led us to look closely at the almost-completed sequence genome of Y. enterocolitica 8081. A search in the Sanger Center Y. enterocolitica database GenBank revealed a putative second secretion cluster designated yts2 here (Fig. 1). Genomic probing of representative Yersinia strains through PCRs revealed the presence of this putative type II secretion cluster among all tested Y. enterocolitica strains (nonpathogenic, LP and HP strains alike), whereas Y. pseudotuberculosis and Y. pestis tested negative for the presence of the genes of this cluster (Fig. 4B). Reverse transcription analysis of the yts2D gene (using RNA isolated from cells grown in LB medium at 27 and 37°C) revealed a positive transcript at 27°C, whereas transcription was basal at 37°C (Fig. 2). The ORFs encoded by this cluster bear homology to those previously described except for the absence of some proteins, notably homologues of the H, M, and S proteins. Figure 1 presents an overview of the two type II secretion clusters identified here and a comparison with other closely related systems in V. cholerae, E. coli, and Y. pestis.
The G+C content of the genes on this species-specific yts2 cluster differs from that of the yts1C-S cluster (restricted to HP strains), having a 33% versus a 48% G+C content, respectively. Strikingly, the yts2 cluster shared the greatest homology (50 to 65%) with the type II secretion system (designated in Fig. 1 as typIIYP) identified on the Y. pestis CO-92 genome (35). For example, the yts2D gene was 63% homologous to the Y. pestis typIIYPD gene (in contrast, the yts1D gene was only 48% homologous to its counterpart on the Y. pestis genome). The G+C content of the typIIYP cluster was also unusually low (34%); however, only the yts1M gene homologue was absent from this cluster, whereas the yts1H and yts1S gene homologues were present.
Role of the secreton in virulence.
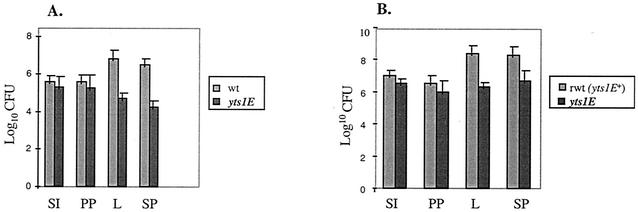
In order to determine the impact of the Yts1 protein secretion apparatus on the virulence of strain WA-314, the average numbers of bacterial CFU recovered from infected organs were compared for the wild type and the WA-314 yts1E mutant in perorally inoculated mice. The results obtained are summarized in Fig. 5A. As depicted in the figure, the bacterial CFU recovered from the small intestine and Peyer's patches were only marginally different for wild-type WA-314 and yts1E. In contrast, the bacterial CFU recovered from liver and spleen were 100-fold higher in WA-314 compared to the yts1E mutant, representing a highly significant attenuation in the yts1E mutant. As an example, the bacterial CFU recovered from infected liver was 6.3 × 106 CFU for WA-314 compared to 5.2 × 104 CFU for yts1E.
FIG. 5.
Pathogenicity of the Yts1 secreton for mice (oral inoculation). BALB/c mice (five per group) were orally challenged with 5 × 109 Y. enterocolitica. At 48 h after infection, mice were sacrificed, and the bacterial load was determined by plating the intestinal lavage (SI) and tissue homogenates from Peyer's patches (PP), liver (L), and spleen (SP). (A) Comparison of mice infected with strain WA-314 (wt) and yts1E mutant; (B) comparison of mice infected with reconstituted wild-type strain (rwt) harboring an intact yts1E gene (after gene replacement of the kanamycin-inactivated yts1E gene with the wild-type parental allele) and with the yts1E mutant. Values are means for five animals, with the standard error of the means indicated by an error bar.
In a parallel experiment, survival of yts1E mutant bacteria were compared with the reconstituted wild-type strain (in which the mutated yts1E gene was replaced with the wild-type allele), the results of which are depicted in Fig. 5B. Typically, the mutant strains presented with significant attenuation in spleen and liver, a finding comparable to the results described above. For example, 2.3 × 108 bacterial CFU were recovered from infected liver for the reconstituted wild type compared to the 1.9 × 106 CFU for the yts1E mutant, the latter representing a 120-fold attenuation in virulence by the mutant bacteria. In contrast, no significant differences in bacterial CFU were observed when the intravenous route of inoculation was used. In a mouse virulence assay, BALB/c mice (five animals per group) were inoculated intravenously with 104 bacteria. At 48 h after challenge, the mice were sacrificed and the bacterial loads were determined by plating tissue homogenates from the liver and spleen. The results (means ± the standard deviation) were as follows: for strain WA-314, (1.0 ± 0.14) × 107 CFU (liver) and (7.5 ± 0.7) × 107 CFU (spleen); and for the yts1E mutant strain, (7.2 ± 1.0) × 106 CFU (liver) and (4.5 ± 0.2) × 107 CFU (spleen).
DISCUSSION
The potential to secrete proteins into the extracellular medium is generally regarded as an important attribute of pathogenic bacteria. Typically, six bacterial pathways for protein secretion to the extracellular milieu have been documented (2, 6, 12, 15, 24, 26, 30, 31, 45). These include the signal sequence independent pathway (type I), the main terminal branch of the general secretion pathway (type II), the contact-dependent pathway (type III), the type IV pathway related to bacterial conjugation systems, the two-partner secretion system exemplified by Bordetella pertussis, and the autotransporter pathway. Among pathogenic Yersinia species, four TTSSs have been discovered, namely, the Ysc TTSS responsible for Yops secretion, the Ysa TTSS of Y. enterocolitica species required for the extracellular secretion of a set of proteins designated Ysps, the chromosomally encoded TTSS of Y. pestis, and lastly the flagellar biosynthetic system (22, 32, 35, 41, 52, 53). Although the type II secretion pathway has been well described among many representatives of the Enterobacteriaceae (45), it has not been previously demonstrated among Yersinia species. In the present study, we describe the presence of a secreton, the Yts1 secretion apparatus unique to the HP Y. enterocolitica species. The yts1 transport cluster shares significant homology with the eps cluster of V. cholerae, with declining identities to the putative type II secretion cluster in Y. pestis and the secretons identified in Erwinia carotovora, Erwinia chrysanthemi, Klebsiella pneumoniae, and Pseudomonas aeruginosa.
Originally discovered in Klebsiella oxytoca (16), in which it was found to be essential for extracellular secretion of the lipoprotein pullulanase, the type II transport cluster has been identified to date in a broad variety of gram-negative bacteria (31, 43, 44). In the classical studies by Francetic et al. (20, 21), an intact secreton was even identified in the widely used laboratory strain E. coli K-12 (nonpathogen). Typically, the interaction of between 12 to 15 gene products have been shown to be essential for protein secretion through this pathway. The encoded proteins form a complex network of interactions spanning the cytoplasmic, inner, and outer membranes. In the Yts1 secreton described here (Fig. 1), all of the interacting partners for a functional secretion are represented, namely, the cytoplasmic, energy-transducing Yts1E protein (which interacts with protein L); the integral inner membrane proteins comprising Yts1C, Yts1M, Yts1K, and Yts1F; the four type 4 pilin-like proteins (pseudopilins), Yts1G, Yts1H, Yts1I, and Yts1J; the two outer member proteins or secretins, Yts1D and Yts1S; and finally, the Yts1O protein, a putative prepilin-like peptidase (51). This is in contrast to the system in Y. pestis, in which a Yts1M homologue does not exist (Fig. 1).
The reverse transcription data presented here suggest that the yts1 genes are preferentially transcribed at 37°C. This is quite interesting since most of the virulence genes unique to Yersinia species come under thermoregulation. Transcription of the chromosomally encoded ail and many virulence pYV-encoded genes, for example, are temperature induced (3, 13, 39, 50). In contrast, the chromosomally encoded invasin is maximally produced at reduced temperatures (23°C), which makes sense since invasin, which functions as an adhesin, is important in the early stages of infection, and it would therefore be advantageous for it to be already present on ingested Yersinia (3, 28, 37). That the yts1 genes are preferentially transcribed at 37°C speaks for a possible role of the secreton during the later stages of infection after invasion.
In order to determine the impact of this novel Yts1 secreton on the virulence of the species, mouse virulence tests were conducted that compared the wild-type WA-314 species with a yts1E secretion-defective mutant. The results confirm that the chromosomally encoded Yts1 secreton does enhance the virulence of the Y. enterocolitica species as evidenced by a 100-fold attenuation in virulence observed in the yts1E mutant when mice were perorally inoculated. In contrast, the numbers of bacterial CFU after intravenous inoculation of mice were not significantly different for the wild type and the secretion mutant. These results strikingly resemble the observations made by Haller et al. (22) with respect to the Ysa TTSS. These authors reported that the Ysa secretion system appears to play a role in the virulence of Y. enterocolitica strains when the bacteria are inoculated into mice perorally, whereas bacteria inoculated intraperitoneally appeared not to benefit from this transport system. Haller et al. concluded that the Ysa TTSS possibly played a role in virulence after the initial invasion of the Peyer's patches but before spread to deeper tissues. In contrast, the Yts1 secreton may be important in the further dissemination of bacteria into deeper tissues (liver and spleen) after initial invasion and colonization of Peyer's patches. The mouse virulence data presented here confirm this hypothesis since the attenuation in virulence for the yts1E mutant was observed only with respect to colonization of liver and spleen, whereas the bacterial counts extrapolated from Peyer's patches were not significantly different between the wild type and the secretion-deficient mutant. Interestingly, as discussed above, preliminary reverse transcription analysis indicating preferential transcriptability of the yts1 genes at 37°C would support the hypothesis for a virulence-related role of the Yts1 secreton occurring later in the course of infection. More-detailed studies, however, should shed light on the nature of the molecular interaction between the Yts1 secreton and the cognate host tissue it recognizes during the course of bacterial infection.
Sequence analysis of the genome of Y. enterocolitica 8081 revealed a second chromosomal locus with great similarity to a type II protein secretion locus (designated Yts2) that was confirmed to be uniformly distributed among all tested Y. enterocolitica strains, including both nonpathogenic LP and HP strains (Fig. 4B). Future studies should reveal whether this common, generic Yts2 secreton is functional and will define the possible substrates that could rely on it for secretion.
Comparison of the yts1 type II cluster with its counterpart on the Y. pestis genome reveals a divergence between these two systems with respect to both gene arrangement and sizes of the putative encoded ORFs. The Y. pestis secreton, however, shared greater homology with the Yts2 species-specific type II gene cluster common to all Y. enterocolitica strains in terms of both G+C content (a low 33 to 34%) and the arrangement of genes bordering the two clusters (see Fig. 1). This finding could argue for a common origin for these two secretion clusters, whereas the Yts1 transport system was independently acquired by the HP Y. enterocolitica 1B strains.
The genes encoding certain secretion systems have been assumed to be products of horizontal gene transfer, as evidenced by their different G+C content compared to the average G+C content of the organism (27, 33). The genes of the Yts1 protein secretion apparatus, however, have an average G+C content of 48%, a finding characteristic of the Yersinia species and a codon usage in line with the species. This could argue against horizontal acquisition of the transport system or might point to transfer from an organism with similar G+C content. Interestingly, in WA-314, a copy of an IS10-like element, IS1330, flanks upstream the yts1 cluster, which might be indicative of an insertional event, and analysis of the sequences downstream of the flanking chiY gene will yield additional information in this regard. Future studies will therefore shed light on what mechanisms played a role in the acquisition of this additional type II secretion system by the HP Y. enterocolitica species.
The Yts1 protein secretion system shares the highest homology with the Eps transport system in V. cholerae responsible for the secretion of the cholera toxin, a protease, and a chitinase (44). It is therefore tempting to propose the existence of an additional virulence factor such as a toxin that could rely on the transport cluster for export. Future studies will focus on defining the substrates that are recognized by the Yts1 secretion cluster, a potential candidate being the product encoded by the chiY gene that flanks the cluster, and the heat-stable enterotoxin.
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinscahft (He 1297/8-4). Work carried out at the Lawrence Livermore Laboratory was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory under contract W-7405-Eng-48.
We thank G. Wilharm for critical reading of the manuscript.
Editor: B. B. Finlay
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2000. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Binet, R., S. Letoffe, J. M. Ghigo, P. Delepelaire, and C. Wandersman. 1997. Protein secretion by gram-negative bacterial ABC exporters: a review. Gene 192:7-11. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, A. P., and G. Y. Cornelis. 2001. Yersinia, p. 227-253. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 4.Buchrieser, C., M. Prentice, and E. Carniel. 1998. The 102-kb unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which unergoes internal rearrangement. J. Bacteriol. 180:2321-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchrieser, C., R. Brosch, S. Bach., A. Guiyoule, and E. Carniel. 1998. The high-pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol. Microbiol. 30:965-978. [DOI] [PubMed] [Google Scholar]
- 6.Burns, D. L. 1999. Biochemistry of type IV secretion. Curr. Opin. Microbiol. 2:25-29. [DOI] [PubMed] [Google Scholar]
- 7.Calia, K. E., M. K. Waldor, and S. B. Calderwood. 1998. Use of representational difference analysis to identify genomic differences between pathogenic strains of Vibrio cholerae. Infect. Immun. 66:849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carniel, E. 2002. Plasmids and pathogenicity islands of Yersinia. Curr. Top. Microbiol. Immunol. 264:89-108. [PubMed] [Google Scholar]
- 9.Carniel, E., A. Guiyoule, I. Guilvout, and O. Mercereau-Puijalon. 1992. Molecular cloning, iron-regulation and mutagenesis of the irp2 gene encoding HMWP2, a protein specific for the highly pathogenic Yersinia. Mol. Microbiol. 6:379-388. [DOI] [PubMed] [Google Scholar]
- 10.Carter, P. B. 1975. Pathogenicity of Yersinia enterocolitica for mice. Infect. Immun. 11:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, J. Y., C. D. Sifri, B. C. Goumnerov, L. G. Rahme, F. M. Ausubel, and S. B. Calderwood. 2002. Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis. J. Bacteriol. 184:952-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type-III protein secretion system in Salmonella: a review. Gene 192:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis, G. R., C. Sluiters, C. L. Lambert de Rouvroit, and T. Michels. 1989. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Almeida, A. M. P., A. Guiyoule, I. Guilvout, I. Iteman, G. Baranton, and E. Carniel. 1993. Chromosomal irp2 gene in Yersinia: distribution, expression, deletion, and impact on virulence. Microb. Pathog. 14:9-21. [DOI] [PubMed] [Google Scholar]
- 15.de Groot, A., G. Gerritse, J. Tommassen, A. Lazdunski, and A. Filloux. 1999. Molecular organization of the xcp gene cluster in Pseudomonas putida: absence of an xcpX (gspK) homologue. Gene 226:35-40. [DOI] [PubMed] [Google Scholar]
- 16.d'Enfert, C., A. Ryter, and A. P. Pugsley. 1987. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization, and secretion of the lipoprotein pullulanase. EMBO J. 6:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diatchenko, L., Y.-F. C. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fetherston, J. D., V. J. Bertolino, and R. D., Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 19.Foultier, B., P. Troisfontaines, S. Müller, F. R. Opperdoes, and G. R. Cornelis. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 55:37-51. [DOI] [PubMed] [Google Scholar]
- 20.Francetic, O., D. Belin, C. Badaut, and A. P. Pugsley. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francetic, O., C. Badaut, S. Rimsky, and A. P. Pugsley. 2000. The ChiA (YheB) protein of Escherichia coli K-12 is an endochitinase whose gene is negatively controlled by the nucleoid-structuring protein H-NS. Mol. Microbiol. 35:1506-1517. [DOI] [PubMed] [Google Scholar]
- 22.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1983. Studies of transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:577-580. [DOI] [PubMed] [Google Scholar]
- 24.He, S. Y., M. Lindeberg, A. K., Chatterjee, and A. Collmer. 1991. Cloned Erwinia chysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc. Natl. Acad. Sci. USA 88:1079-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heesemann, J. 1987. Chromosomal-encoded siderophores are required for mouse virulence of enteropathogenic Yersinia species. FEMS Microbiol. Lett. 48:229-233. [Google Scholar]
- 26.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 27.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isberg, R. R., A. Swain, and S. Falkow. 1988. Analysis of expression and thermoregulation of the Yersinia pseudotuberculosis inv gene with hybrid proteins. Infect. Immun. 56:2133-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwobi, A., A. Rakin, E. Garcia, and J. Heesemann. 2002. Representational difference analysis uncovers a novel IS10-like insertion element unique to pathogenic strains of Yersinia enterocolitica. FEMS Microbiol. Lett. 210:251-255. [DOI] [PubMed] [Google Scholar]
- 30.Jacob-Dubuisson, F., C. El Hamel, N. Saint., S. Guedin., E. Willery, G. Molle, and C. Locht. 1999. Channel formation by FhaC, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 274:37731-37735. [DOI] [PubMed] [Google Scholar]
- 31.Jahagirdar, R., and S. P. Howard. 1994. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J. Bacteriol. 176:6819-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mecsas, J., and E. J. Strauss. 1996. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg. Infect. Dis. 2:271-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrow, B. J., J. E. Graham, and R. Curtiss III. 1999. Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect. Immun. 67:5106-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 36.Pelludat, C., A. Rakin, C. A., Jacobi, S. Schubert, and J. Heesemann. 1998. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 38.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis: etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierson, D. E., and S. Falkow. 1993. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect. Immun. 61:1846-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Portnoy, D. A., and S. Falkow. 1981. Virulence-associated plasmids from Yersinia enterocolitica and Y. pestis. J. Bacteriol. 148:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Portnoy, D. A., S. L. Moseley, and S. Falkow. 1981. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect. Immun. 31:775-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratnam, S., E. Merser, B. Picco, S. Parsons, and R. Butler. 1982. A nosocomial outbreak of diarrheal disease due to Yersinia enterocolitica serotype O:5, biotype 1. J. Infect. Dis. 145:242. [DOI] [PubMed]
- 43.Russell, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 44.Sandkvist, M., L. O. Michel, L. P. Hough, V. M. Morales, M. Bagdasarian, M. Koomey, V. J. DiRita, and M. Bagdasarian. 1997. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 179:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 47.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, R., U. Preifer, and A. Pühler. 1988. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-785. [Google Scholar]
- 49.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 50.Skurnik, M., and P. Toivanen. 1992. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 174:2047-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stathopoulos, C., D. R. Hendrixson, D. G. Thanassi, S. J. Hultgren, J. W. St. Geme III, and R. Curtiss III. 2000. Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 2:1061-1072. [DOI] [PubMed] [Google Scholar]
- 52.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]