Abstract
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) possess a filamentous type III secretion system (TTSS) employed to deliver effector proteins into host cells. EspA is a type III secreted protein which forms the filamentous extension to the TTSS and which interacts with host cells during early stages of attaching and effacing (A/E) lesion formation. By immunofluorescence, a polyclonal antibody previously raised to EspA from EPEC strain E2348/69 (O127:H6) stained ∼12-nm-diameter EspA filaments produced by this strain but did not stain similar filaments produced by EHEC serotype O157:H7. Similarly, an antibody that we subsequently raised to EHEC strain 85-170 (O157:H7) EspA stained ∼12-nm-diameter EspA filaments produced by strain 85-170 but did not stain E2348/69 EspA filaments. Given such heterogeneity between EPEC and EHEC EspA filaments, we examined polymorphisms of functional EspA filaments among different EPEC and EHEC serotypes. With use of the EPEC EspA antiserum, EspA filaments were observed only with EPEC serotypes O127:H6 and O55:H6, serotypes which encode an identical EspA protein. When stained with the EHEC EspA antiserum, EspA filaments were detected only on EHEC strains belonging to serotype O157:H7; the EHEC antiserum did, however, stain EspA filaments produced by the closely related EPEC serotype O55:H7 but not filaments of any other EPEC serotype tested. Such polymorphisms among functional EspA filaments of EPEC and EHEC would be expected to have important implications for the development of broad-range EspA-based vaccines.
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) are two classes of extracellular diarrheagenic E. coli that communicate with the eukaryotic cytoskeleton while adhering intimately to the outer surface of the intestinal epithelial cell plasma membrane (7, 22). Intimate bacterial attachment is mediated through tight interaction between the outer bacterial membrane adhesion molecule, intimin (8, 10), and Tir (12), a receptor for intimin that is delivered to the host cell membrane via a type III secretion system (TTSS) (12). The TTSS, Tir, and intimin are all expressed from a pathogenicity island known as the locus of enterocyte effacement (LEE) (6, 19).
Following infection and assembly of the TTSS, EPEC and EHEC translocate a range of effector proteins to the eukaryotic cell (7). The injected proteins target different cellular compartments; EspF (20) and EspG (5) are believed to remain cytosolic, and Map targets the mitochondria (13), while Tir is delivered and inserted into the host cell plasma membrane such that its amino and carboxy termini are intracellular and its central domain is exposed on the outer cell surface and serves as a receptor for intimin (9, 11). Binding of intimin to Tir triggers dramatic intracellular changes including reorganization of cytoskeletal proteins, actin polymerization at the site of intimate bacterial contact (15), and formation of a characteristic attaching and effacing (A/E) lesion (16, 21). Recently, based on sequence variation at the receptor binding carboxy-terminal 280 amino acids of intimin, it was reported that intimin could be classified into several distinct intimin types (1).
EspA, a protein also expressed from the LEE pathogenicity island and essential for A/E lesion formation (14), is one of the TTSS translocator proteins and a major if not the only component of a filamentous structure which extends from the basic needle complex of the secretion apparatus and connects the pathogen to the plasma membrane of the host cell (4, 17). EspB and EspD are additional translocator proteins believed to form a translocation pore in the host cell membrane at the distal end of the EspA filament (26, 29). Under growth conditions that induce LEE gene expression, antibodies made against a recombinant EspA from EPEC strain E2348/69 stained the ∼12-nm-diameter EspA filaments (17) with, on average, each bacterium producing 12 EspA filaments (3). Although by scanning electron microscopy EHEC O157 was reported elsewhere to produce filaments resembling EspA filaments of EPEC (24), no positive identification of EHEC EspA filaments has yet been made. The aim of this study was to confirm and positively identify expression of EspA filaments by EHEC and to examine polymorphisms of functional EspA filaments between different EPEC and EHEC strains.
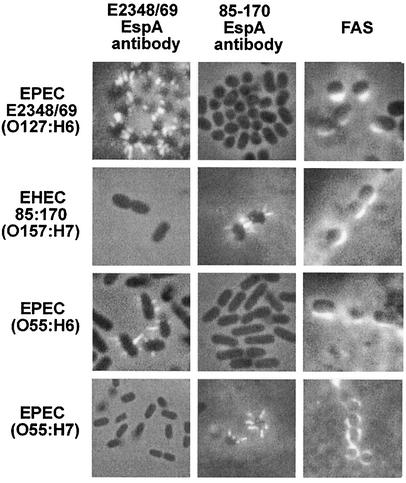
EspA filaments can readily be detected by immunofluorescence during early stages of A/E lesion formation (17). Accordingly, we infected HEp-2 cells with EHEC strain 85-170 (a Shiga toxin-negative O157:H7 strain) (25) and with EPEC strain E2348/69 (O127:H6) (18) as a control for 3 h, fixed the cells in formalin, and performed indirect immunofluorescence with a rabbit EspAE2348/69 polyclonal antiserum (17) and a goat anti-rabbit Alexa 488 fluorescence conjugate (Molecular Probes). This revealed positive staining of the EPEC E2348/69 EspA filaments but no staining of corresponding structures produced by EHEC 85-170 (Fig. 1). At later stages of infection bacteria induced actin accretion that could readily be detected by fluorescent actin staining (FAS) with fluorescein-conjugated phalloidin (15). Indeed, a positive FAS reaction was observed following infection with both EPEC E2348/69 and EHEC 85-170 (Fig. 1). It is well documented that a positive FAS reaction, a correlate of A/E lesion formation, is dependent on formation of EspA filaments (14, 17). Accordingly, we hypothesized that our inability to detect the EHEC 85-170 EspA filaments using the EPEC antisera was due to EspA polymorphisms between the two strains.
FIG. 1.
Immunofluorescence staining of EspA filaments with EPEC EspAE2348/69 (column 1) and EHEC EspA85-170 antisera (column 2); HEp-2 cells were infected with EPEC and EHEC strains for 3 h, and bacteria were visualized by phase contrast. The EPEC antibody stained EspA filaments produced by EPEC E2348/69 (O127:H6) (row 1) and EPEC O55:H6 (row 3) while the EHEC antibody stained EspA filaments produced by EHEC 85-170 (O157:H7) (row 2) and EPEC O55:H7 (row 4). A positive FAS assay (column 3) indicated that all four strains produced A/E lesions on HEp-2 cells.
In order to investigate possible polymorphisms, EHECEspA was cloned, following PCR amplification with Deep Vent DNA polymerase (New England Biolabs), the primer pair espA-F (5′ TATCATATGGATACATCAAATGCAACATCCGTT 3′) and espA-R (5′ TATGGATCCTTATTTACCAAGGGATATTGCTGAAATAG 3′), and EHEC genomic DNA from the prototype strain 85-170 (O157:H7) as DNA template, into NdeI/BamHI-digested pET28-a, generating plasmid pICC207 for expression as a His-tagged protein.
His6-EspA fusions were expressed in BL21(DE3)pLysS(pICC207). An overnight culture was diluted 1:100 in 100 ml of LB (30 μg of kanamycin/ml, 30 μg of chloramphenicol/ml, and 0.2% glucose), grown to an optical density at 600 nm of 0.4 to 0.8 at 37°C with shaking, and induced with the addition of 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Following a further 4-h (30°C) incubation, bacterial cells were pelleted, resuspended in cold binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.9), and sonicated. Following removal of cell debris by centrifugation (45,000 × g for 30 min), the cell extracts were filtered through a 0.45-μm-pore-size filter device and His6-EspA was purified on a 2.5-ml nickel-charged column as recommended by the manufacturer (Novagen) and described before (17). An 80- to 100-μg quantity of the recombinant His6-EspA was used to immunize, subcutaneously in complete Freund's adjuvant, female Sandy half-lop rabbits. The animals were boosted twice with the same antigen in incomplete Freund's adjuvant at 3-week intervals before exsanguinations.
Employing this EHEC 85-170 EspA antiserum on infected epithelial cells revealed staining of EspA filaments specifically in association with EHEC 85-170 bacteria, whereas no staining of the EspA filaments was seen in association with EPEC E2348/69 (Fig. 1). This observation not only provided the first positive identification of EspA filaments on EHEC O157:H7 but supported the possible existence of polymorphisms between different EspA filaments.
The fact that no cross-reactivity was seen between the EPEC E2348/69 and EHEC 85-170 EspA antisera prompted us to determine their reactivities with different EPEC and EHEC clinical isolates from our strain collection (Table 1). Significantly, the EspA85-170 antiserum stained EspA filaments expressed by all three EHEC O157:H7 strains tested but did not stain EspA filaments produced by other EHEC strains, e.g., EHEC O26:H11 (Table 1). In addition, when tested against different EPEC serotypes, the EspA85-170 antiserum stained only EspA filaments of the atypical EPEC serotype O55:H7 (Table 1; Fig. 1). Of note is the fact that O55:H7 EPEC is believed to be the ancestral serotype from which EHEC O157:H7 emerged (27, 28). This observation is intriguing as the level of identity between the amino acid sequences of O55:H7 and O157:H7 EspA polypeptides (80%) is not significantly greater than the identity between E2348/69 and EHEC EspA proteins (79%). The ∼20% variant amino acids are dispersed throughout the protein (23), and the structure of the EspA protein has not been determined. Hence, it is unclear if a particular surface-exposed epitope is responsible for these observed differences in cross-reactivity.
TABLE 1.
EPEC and EHEC strains examined by FAS and for EspA filaments by using the EPEC EspAE2348/69 and EHEC EspA85-170 antisera following a 3-h infection of HEp-2 cells
| Strain | Serotype | EspA filaments
|
FAS | |
|---|---|---|---|---|
| EPECE2348/69 antibody | EHEC85-170 antibody | |||
| E2348/69 | O127:H6 | + | − | + |
| CH 26/1 | O127:H6 | + | − | + |
| AJ 7/6 | O127:H6 | + | − | + |
| 85-170 | O157:H7 | − | + | + |
| 813 (Sakai) | O157:H7 | − | + | + |
| 87-23 | O157:H7 | − | + | + |
| TT2B | O157:H7 | − | + | + |
| CH7/6 | O26:H− | − | − | + |
| YI29/3 | O26:H− | − | − | + |
| BK32 | O26:H11 | − | − | + |
| H19 | O26:H11 | − | − | + |
| 39 | O55:H− | − | − | + |
| 35 | O55:H− | − | − | + |
| 17 | O55:H6 | + | − | + |
| 21 | O55:H6 | + | − | + |
| 30 | O55:H6 | + | − | + |
| WC155 | O55:H7 | − | + | + |
| WC211 | O55:H7 | − | + | + |
| WC416 | O55:H7 | − | + | + |
| 261-88 | O86:H34 | − | − | + |
| 236-80 | O86:H34 | − | − | + |
| G10 | O111:H2 | − | − | + |
| G19 | O111:H2 | − | − | + |
| 112 | O119:H2 | − | − | + |
| 38 | O119:H6 | − | − | + |
| 19 | O119:H6 | − | − | + |
| 280/84 | O127:H40 | − | − | + |
| 53-85 | O142:H34 | − | − | + |
| 226-83 | O142:H34 | − | − | + |
In contrast to the EspA85-170 antiserum, the EspAE2348/69 antiserum specifically stained EspA filaments of EPEC serotypes O127:H6 and O55:H6. The antiserum did not react with EspA filaments expressed by any of the other EPEC and EHEC strains tested (Table 1; Fig. 1). The cross-reactivity of the E2348/69 antiserum with O55:H6 strains was not unexpected, however, as the amino acid sequence of the EspA protein is identical between the two serotypes (23).
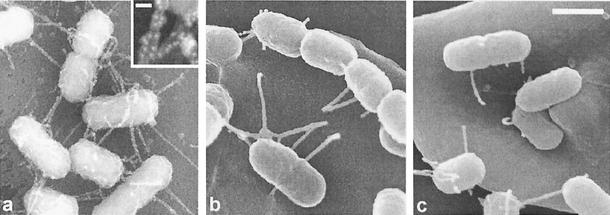
Our group recently demonstrated EspA filament-mediated attachment of EPEC to red blood cell (RBC) membranes (24) and by scanning electron microscopy demonstrated filaments resembling EspA filaments mediating attachment of EHEC O157 to RBCs (24), filaments which we have now confirmed as EspA filaments by immunofluorescence. Although we were unable to demonstrate EspA filaments produced by most EPEC and EHEC serotypes by immunofluorescence (Table 1), scanning electron microscopy of RBCs infected with EPEC and EHEC strains which did not react with either EspAE2348/69 or EspA85-170 antiserum did reveal filaments which mediated bacterial attachment to the RBC membrane and which resembled filaments confirmed as EspA by immunogold labeling with 10-nm gold particles detected with a backscattered electron detector (Fig. 2).
FIG. 2.
Scanning electron micrographs showing attachment of EPEC strains to RBC monolayers. Filamentous structures which did not react with the EPEC or EHEC EspA antisera but which were morphologically identical to EPEC E2348/69 EspA filaments identified by immunogold labeling (a) were seen to promote attachment of EHEC strain O26:H11 (b) and EPEC strain O119:H6 (c) to RBC membranes. Bar, 0.1 μm; inset bar, 0.01 μm.
In this report we have demonstrated polymorphisms between EspA filaments expressed by different EPEC and EHEC isolates. Considering the high level of amino acid sequence identity between different EPEC and EHEC EspA proteins and despite the fact that EspA proteins cross-react in Western blots (23; data not shown) and that EspA genes could be grouped by PCR (2), the lack of cross-reactivity with EspA filaments was unexpected. However, this observation is significant as EspA is considered to be an important component of a veterinary EHEC vaccine. Our results imply that immunization with EHEC O157 EspA would potentially reduce carriage of EHEC O157 but would not protect livestock animals from other EHEC serotypes (i.e., EHEC O26:H11, EHEC O103:H2, and EHEC O111) which, in some countries, are more prevalent than O157 EHEC. Indeed, using the Citrobacter rodentium mouse model of infection, we recently showed that immunization with EspAE2348/69 did not prevent colonization of the mouse gut following oral challenge with C. rodentium (unpublished data). Taken together, the results presented in this report imply that, for a broad-range EspA-based vaccine, a combination of recombinant or secreted EspA preparations would be required.
Acknowledgments
This work was supported by the Wellcome Trust and the CNPq agency, Brazil (BCN).
Bianca Neves and Robert Shaw contributed equally to the study.
Editor: A. D. O'Brien
REFERENCES
- 1.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncaleves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimin α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.China, B., F. Goffaux, V. Pirson, and J. Mainil. 1999. Comparison of eae, tir, espA and espB genes of bovine and human attaching and effacing Escherichia coli by multiplex polymerase chain reaction. FEMS Microbiol. Lett. 178:177-182. [DOI] [PubMed] [Google Scholar]
- 3.Daniell, S. J., N. Takahashi, R. Wilson, D. Friedberg, I. Rosenshine, F. P. Booy, R. K. Shaw, S. Knutton, G. Frankel, and S. Aizawa. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3:865-871. [DOI] [PubMed] [Google Scholar]
- 4.Ebel, F., T. Podzadel, M. Rohde, A. U. Kresse, S. Kramer, C. Deibel, C. A. Guzman, and T. Chakraborty. 1998. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol. 30:147-161. [DOI] [PubMed] [Google Scholar]
- 5.Elliott, S. J., E. O. Krejany, J. L. Mellies, R. M. Robins-Browne, C. Sasakawa, and J. B. Kaper. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 7.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 8.Frankel, G., A. D. Phillips, L. R. Trabulsi, S. Knutton, G. Dougan, and S. E. Matthews. 2001. Intimin and the host cell—is it bound to end in Tir(s)? Trends Microbiol. 9:214-218. [DOI] [PubMed] [Google Scholar]
- 9.Hartland, E. L., M. Batchelor, R. M. Delahay, C. Hale, S. Matthews, G. Dougan, S. Knutton, I. Connerton, and G. Frankel. 1999. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32:151-158. [DOI] [PubMed] [Google Scholar]
- 10.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleating activity and is preceded by additional host modifications. Mol. Microbiol. 31:1229-1241. [DOI] [PubMed] [Google Scholar]
- 12.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 13.Kenny, B., and M. Jepson. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2:579-590. [DOI] [PubMed] [Google Scholar]
- 14.Kenny, B., L. C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313-323. [DOI] [PubMed] [Google Scholar]
- 15.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knutton, S., D. R. Lloyd, and A. S. McNeish. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect. Immun. 55:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine, M. M., E. J. Berquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, S. Stoman, and B. Rowe. 1978. Escherichia coli that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:119-122. [DOI] [PubMed]
- 19.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNamara, B. P., and M. S. Donnenberg. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71-78. [DOI] [PubMed] [Google Scholar]
- 21.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neves, B. C., S. Knutton, L. R. Trabulsi, V. Sperandio, J. B. Kaper, G. Dougan, and G. Frankel. 1998. Molecular and ultrastructural characterisation of EspA from different enteropathogenic Escherichia coli serotypes. FEMS Microbiol. Lett. 169:73-80. [DOI] [PubMed] [Google Scholar]
- 24.Shaw, R. K., S. Daniell, F. Ebel, G. Frankel, and S. Knutton. 2001. EspA filament-mediated protein translocation into red blood cells. Cell. Microbiol. 3:213-222. [DOI] [PubMed] [Google Scholar]
- 25.Tzipori, S., H. Karch, K. I. Wachsmuth, R. M. Robins-Browne, A. D. O'Brien, H. Lior, M. L. Cohen, J. Smithers, and M. M. Levine. 1987. Role of a 60-megadalton plasmid and Shiga-like toxins in the pathogenesis of infection caused by enterohemorrhagic Escherichia coli O157:H7 in gnotobiotic piglets. Infect. Immun. 55:3117-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wachter, C., C. Beinke, M. Mattes, and M. A. Schmidt. 1999. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 31:1695-1707. [DOI] [PubMed] [Google Scholar]
- 27.Whittam, T. S., and E. A. McGraw. 1996. Clonal analysis of EPEC serogroups. Rev. Microbiol. Sao Paulo 27(Suppl. 1):7-16. [Google Scholar]
- 28.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff, C., I. Nisan, E. Hanski, G. Frankel, and I. Rosenshine. 1998. Protein translocation into HeLa cells by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 28:143-155. [DOI] [PubMed] [Google Scholar]