Abstract
Brucella abortus reportedly produces the monocatechol siderophore 2,3-dihydroxybenzoic acid (2,3-DHBA) in response to iron limitation. Nucleotide sequence analysis of the cloned DHBA biosynthesis locus from virulent B. abortus 2308 and genetic complementation of defined Escherichia coli mutants were used to identify the B. abortus genes (designated dhbC, -B, and -A) responsible for synthesis of this siderophore. Reverse transcriptase PCR analysis of total RNA with dhb-specific primers demonstrated that dhbC, -B, and -A are transcribed as components of an operon, together with dhbE, a functional homolog of the Escherichia coli entE gene. Homologs of the E. coli entD and Vibrio cholerae vibH genes were also detected in the flanking regions immediately adjacent to the B. abortus dhbCEBA operon, suggesting that B. abortus has the genetic capacity to produce a more complex 2,3-DHBA-based siderophore. Slot blot hybridization experiments and primer extension analysis showed that transcription of the B. abortus dhbCEBA operon originates from two iron-regulated promoters located upstream of dhbC. Consistent with their iron-dependent regulation, both of the dhbCEBA promoter sequences contain typical consensus Fur-binding motifs. Although previously published studies have shown that 2,3-DHBA production is not required for the establishment and maintenance of chronic spleen infection by B. abortus in mice, experimental infection of pregnant cattle with the B. abortus dhbC mutant BHB1 clearly showed that production of this siderophore is essential for wild-type virulence in the natural ruminant host.
Iron deprivation is an effective host defense that is used by plants and mammals to restrict the growth of microbial pathogens within their tissues (10, 37). Pathogens have developed multiple strategies to overcome iron limitation, and one of the best-known methods is through the synthesis of low-molecular-weight iron chelators known as siderophores. Brucella abortus synthesizes the monocatechol siderophore 2,3-dihydroxybenzoic acid (2,3-DHBA) in response to iron limitation in vitro, and experimental studies have clearly shown that this compound functions efficiently in iron acquisition under in vitro conditions (27, 28). Although 2,3-DHBA production is relatively common among bacteria, this compound is frequently observed only as an intermediate in the synthesis of more complex catechol-based siderophores (9, 30). For example, 2,3-DHBA serves as the precursor for the bacterial siderophores enterochelin (19, 32), vibriobactin (54), and amonabactin (7). Upon initial characterization of the cloned B. abortus 2,3-DHBA biosynthesis operon, the presence of several potential genes that could lead to the production of a more complex DHBA-based siderophore was detected (8). These findings were intriguing, since previous studies have shown that B. abortus does not produce nor can it utilize complex polycatechol-based siderophores (27).
In vitro studies have shown that the addition of 2,3-DHBA to cultured murine macrophages infected with B. abortus results in increased numbers of intracellular bacteria at 48 h postinfection (25). This observation suggested that 2,3-DHBA acts as a virulence determinant by contributing to the intracellular survival of this organism. Subsequent studies employing a genetically defined B. abortus mutant (designated BHB1) that does not produce 2,3-DHBA, however, indicated that production of this siderophore is not required for wild-type virulence in either BALB/c (Brucella sensitive) (8) or C57BL/6 (Brucella resistant) mice (34). Preliminary studies with pregnant goats, on the other hand, suggested that 2,3-DHBA production plays a major role in the virulence of B. abortus in pregnant ruminants (B. Bellaire, C. Baldwin, P. Elzer, and R. Roop, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. B-17, 2000).
The studies described in this report were undertaken to define the genetic composition and organization of the B. abortus 2,3-DHBA biosynthesis operon and to evaluate the contribution of 2,3-DHBA to the virulence of B. abortus in its natural ruminant host. The results of these studies suggest that B. abortus has the capacity to use 2,3-DHBA to produce a more complex catechol-based siderophore under certain environmental conditions, which is consistent with the recent report by González Carreró et al. (18) describing the production of a complex 2,3-DHBA-based siderophore known as brucebactin in response to iron limitation. The studies described in this report also clearly show that 2,3-DHBA production is essential for the wild-type virulence of B. abortus 2308 in pregnant cattle.
MATERIALS AND METHODS
Culture media and growth conditions.
The bacterial strains and vectors used in this study are listed in Table 1. For the routine culture of Escherichia coli strains, Luria-Bertani (LB) broth and agar were used. Brucella abortus strains were routinely grown in brucella broth (Difco) at 37°C or on Schaedler agar (Difco) supplemented with 5% defibrinated bovine blood incubated at 37°C under 5% CO2. Ampicillin and kanamycin were added to these culture media as needed at final concentrations of 100 and 45 μg/ml, respectively. Chrome azurol S (CAS) plates were prepared as previously described (48). Endogenous iron was removed from the CAS medium prior to use by 8-hydroxyquinilone extraction. Low-iron minimal medium was prepared as previously described (27), and 50 μM FeCl3 was added to this medium as a control for growth under iron-replete conditions when appropriate.
TABLE 1.
Bacterial strains and cloning vectors used in this study
| Strain, vector, or plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | General cloning strain | Gibco BRL |
| SAB11 | HB101 entC; 2,3-DHBA-deficient strain | 7 |
| AN90 | AB1515 entD | 11 |
| AN93 | AB1515 entE405 | 53 |
| AN192 | AB1515 entB402 | 53 |
| AB1515.24 | AB1515 entB::Tn5 | 53 |
| B. abortus | ||
| 2308 | Virulent laboratory strain | Laboratory stock |
| BHB1 | 2308 ΔdhbC; Knr | 8 |
| BHB2 | 2308 ΔdhbC | This study |
| Cloning vectors | ||
| pUC9 | High-copy-number plasmid; Ampr | NEB |
| pBluescriptII KS+ | High-copy-number plasmid; Ampr | Stratagene |
| pBC SK | High-copy-number plasmid; Cmr | NEB |
| pBBR1MCS-4 | Broad-host-range plasmid; Ampr | 23 |
| pHC79 | Cosmid; Ampr Knr | 20 |
| pEX100T | sacB-containing counterselection vector; Ampr | 47 |
| Recombinant plasmids | ||
| pPS50 | 4.5-kb Sau3A fragment containing B. abortus dhbC, dhbB, and a portion of dhbE cloned into BamHI site of pUC9; Ampr | 8 |
| p2-9H | 45-kb Sau3A fragment of B. abortus 2308 genomic DNA containing the dhbCEBA operon and surrounding regions cloned into BamHI site of pHC79; Ampr | This study |
| pEC2-M | 2.3-kb EcoRI fragment from p2-9H containing B. abortus dhbC cloned into pBBR1MCS-4; Ampr | This study |
| pEC2-B | 2.3-kb EcoRI fragment from p2-9H containing B. abortus dhbC cloned into pBluescriptII KS+; Ampr | This study |
| pEC6-B | 10-kb EcoRI fragment from p2-9H containing B. abortus dhbB, -A, and -D cloned into pBluescriptII KS+; Ampr | This study |
Genetic techniques.
Standard genetic techniques (6) were employed for the cloning and subcloning of the B. abortus dhb operon. A cosmid library was constructed by ligating BamHI-digested pHC79 with B. abortus 2308 genomic DNA fragments partially digested with Sau3A and size fractionated by centrifugation on a 5 to 25% NaCl gradient. The fractions selected for ligation were those rich in fragments between 35 and 45 kb in length determined by 0.7% agarose gel electrophoresis. The ligation mix was packaged into λ phage by using Packagene (Stratagene). E. coli SAB11 grown overnight in LB broth supplemented with 2% maltose was infected with packaged phage for 20 min. One milliliter of LB broth was added to the mixture containing infected cells, which was then incubated at 37°C for 1 h, and cosmid-containing transformants were selected by plating onto LB agar supplemented with ampicillin. Individual colonies were transferred to 96-well plates containing LB broth supplemented with ampicillin and incubated without shaking for 48 h at 37°C. Total DNA was prepared from these cells in the 96-well format, transferred to a positively charged nylon membrane (Immobilon; Millipore) with a dot blot apparatus (Bio-Rad), and cross-linked to the membrane by UV irradiation (Stratalinker, Stratagene). Immobilized DNA was screened by dot blot Southern hybridization for reactivity with a radiolabeled dhbC-specific probe (a 2.3-kb EcoRI fragment from pPS50) (8), generated by using [γ-32P]dCTP and random priming procedures (43). Nucleotide sequence analysis of cloned DNA fragments was performed by the dideoxy chain termination procedures described by Sanger et al. (46). Computer-assisted analysis of nucleotide sequences was performed with the software program VectorNTI (InforMax), and the Basic Local Alignment Search Tool (BLAST) algorithm (3) was used to compare the sequences with the nucleotide and amino acid sequences currently deposited in GenBank.
Genetic complementation of E. coli ent mutants.
Restriction fragments encompassing selected regions of the cloned B. abortus DHBA biosynthesis operon were subcloned into pBluescript II KS+, and the resulting recombinant plasmids were tested for their ability to restore siderophore production in E. coli strains carrying defined ent mutations (Table 1). Halo production on CAS plates and the catechol-specific Arnow assay (5, 36) were used to measure siderophore production by the recombinant E. coli strains.
RNA isolation and analysis.
Total RNA was isolated from B. abortus cultures grown to an optical density at 600 nm (OD600) of 0.25 to 0.70 in brucella broth (iron replete) or brucella broth supplemented with 600 μM EDDA (iron deplete) by the RNeasy Total RNA isolation method (Qiagen) modified in the following manner. Cells from 10 ml of culture were collected by centrifugation, resuspended in 700 μl of Tris-EDTA (TE) buffer at pH 8.0, and vortexed briefly with an equal volume of chloroform. After phase separation, the aqueous phase was removed, incubated at room temperature with 300 μl of 50 mg of lysozyme per ml for 5 min, and then transferred to the Qiagen RNA isolation column. The remaining steps were carried out according to the manufacturer's instructions. RNA was recovered in 400 μl of nuclease-free water, and RNA preparations were treated with RQ1 RNase-free DNase (Promega) according to the manufacturer's instructions. The absence of contaminating chromosomal DNA was confirmed by failure of the gene amplification reactions to generate a product detectable by agarose gel electrophoresis, in the absence of reverse transcriptase. Concentrations of RNA in these samples were determined by measuring the A260, and these samples were used for both Northern and reverse transcriptase-assisted PCR (RT-PCR) analyses. For Northern analysis by slot blotting, equal amounts of RNA isolated from high- and low-iron cultures were cross-linked to a positively charged membrane (Immobilon; Millipore) by UV irradiation (Stratalinker; Stratagene) and hybridized with a dhbC-specific DNA probe generated by NdeI-EcoRI digestion of pEC-2 (Fig. 1). Determination of the transcriptional start of dhbC and the detection of specific dhb transcripts were performed by primer extension and RT-PCR, respectively, with the oligonucleotides listed in Table 2. RT-PCR analysis was performed with the Access RT-PCR system (Promega) according to the manufacturer's instructions, employing PCR cycling temperatures of 60°C for annealing and 70°C for extension. Amplification products were visualized alongside 100-bp DNA markers in 2% agarose gels after ethidium bromide staining. Primer extension was used to map the 5′ ends of the dhbC transcript, and the methods employed, including [γ-32P]ATP labeling of oligonucleotides by polynucleotide kinase, have been described previously (38).
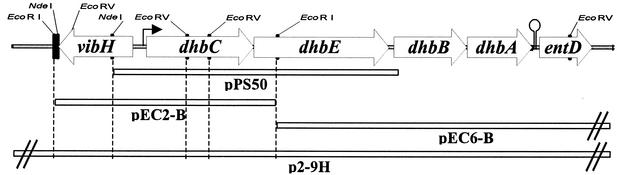
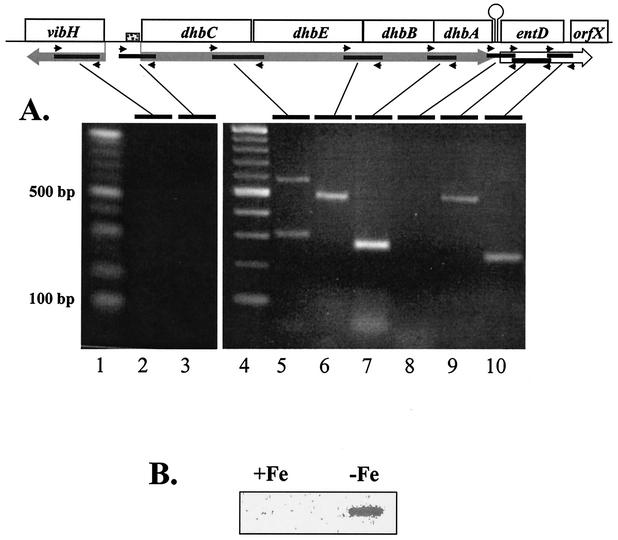
FIG. 1.
Genetic organization of the B. abortus 2308 vihH-dhbCEBA-entD locus. Arrows indicate the direction of transcription and bars represent the cloned DNA fragments present in the plasmids described in Table 1. A functional transcription initiation site upstream of dhbC is depicted by an arrow and a predicted rho-independent transcriptional terminator located downstream of dhbA is represented by a stem-loop structure.
TABLE 2.
Sequences of oligonucleotides used in this study
| Oligonucleotidea | Sequence (5′→3′) |
|---|---|
| Primer extension | |
| Cstrt1 | GGGTATTTCAGGGCGCTCGTTCT |
| Cstrt2 | GGCGTCGCGATCACCTCA |
| RT-PCR analysis | |
| dhbC internal | |
| Up | CGGTAGAACATGCCATCAT |
| Down | CAAGCTGCAAGGCTGTCAT |
| dhbC limiting | |
| Up | CCACATCGCCTTCGATT |
| Down | GGGTATTTCAGGGCGCTCGTTCT |
| dhbC-E | |
| Up | CGCCTGGTGATCGAGGAT |
| Down | GCCAATGGGTAAATTCGAT |
| dhbE-B | |
| Up | TGCCGCCGAGGAGATTGAA |
| Down | GGGCCGTTTGCGTCGTAAA |
| dhbB-A | |
| Up | CTACGGCCTCGATTCGCTA |
| Down | GATGACGGTTGCGCCTTCGG |
| dhbA-entD | |
| Up | GCCGGAACAGTTCAAGCTTG |
| Down | GGCTTGCCCGGCGACAAAT |
| entD internal | |
| Up | CGGCCAGCAAACCCTTG |
| Down | GGAAAAGGCTGCGGCATGG |
| entD-orfX | |
| Up | GGCAGCGGTTCCCGATCC |
| Down | CCGGAAAATCCGCAAGGT |
| vibH | |
| Up | GGCGGAGGTTCTTTCCAT |
| Down | CGTCCTCCTCATGAATG |
Measurement of dhbC promoter activity with a lacZ reporter construct.
Plasmid pPdhbC::lacZ was constructed by cloning a 221-bp region containing the B. abortus dhbC promoter upstream of the promoterless lacZ gene in pMR15 (17). This plasmid was introduced into B. abortus 2308 by electroporation (13), and β-galactosidase production by the plasmid-bearing 2308 cell culture was determined by the methods of Miller (31). pMR15 is a derivative of the low-copy-number, broad-host-range plasmid RK2, and plasmids of this lineage are typically maintained in B. abortus 2308 at 2 to 4 copies per genome equivalent (39).
Genetic complementation of the B. abortus dhbC mutant.
To facilitate the complementation of the dhbC mutation in trans, the dhbC::Knr disruption present in BHB1 was replaced with an in-frame, nonpolar ΔdhbC mutation. The nonpolar dhbC mutation harbors the same 261-bp EcoRV internal deletion used for construction of the polar dhbC::Knr allele; however, the remaining portions of the dhbC open reading frame (ORF) were ligated together without introducing an antibiotic resistance cassette, resulting in the construction of plasmid pBH28. A 2.1-kb fragment containing the dhbC gene harboring the internal in-frame deletion was removed from pBH28 by digestion with EcoRI. This fragment was incubated with the Klenow fragment of DNA polymerase I (Promega) to fill in its protruding 5′ ends and cloned into the SmaI site of pEX100T (47). The resulting plasmid construct was used in a sucrose resistance-based counterselection strategy (47) to introduce the nonpolar dhbC mutation into B. abortus 1. The nonpolar dhbC mutant constructed in this manner was designated BHB2. The genotype of this mutant was confirmed by PCR, and the 2,3-DHBA-negative phenotype of this strain was verified by the Arnow assay. A 2.3-kb EcoRI fragment containing the B. abortus dhbC gene was cloned into pBBR1MCS-4 (23), and the resulting plasmid, pEC2-M (B. H. Bellaire, P. H. Elzer, S. Hagius, J. Walker, C. L. Baldwin, and R. M. Roop II, submitted for publication), was introduced into BHB2 by electroporation (13).
Virulence assessment of B. abortus strains in pregnant cattle.
Virulence studies with pregnant cattle were performed by previously described methods (12). Pregnant mixed-breed heifers were infected with 107 CFU of B. abortus 2308 or BHB1 via the conjunctival route at the beginning of the 3rd trimester of gestation (∼220 days). After abortion or birth, fetuses or calves were examined by necropsy. Samples of the lungs and abomasal fluid were collected and cultured. Heifers that aborted were euthanized and necropsied. For those heifers that bore live calves, milk samples from all four quarters of the mammary gland and intrauterine fluid were collected during the 48-h period following delivery and cultured. Approximately 1 month postdelivery, these heifers were euthanized and necropsied. Portions of the spleen, liver, and mammary glands and the supramammary, inguinal, and parotid lymph nodes were collected for bacteriologic culture. Bacterial isolates were identified as B. abortus by their colony morphology, Gram stain, urease and oxidase reactions, and reaction with B. abortus-specific antiserum. Blood was collected from the heifers prior to challenge and at necropsy. Serum was removed and stored at −20°C. Sera were evaluated for Brucella-specific antibodies by using the Rose Bengal card test (2).
Nucleotide sequence accession numbers.
The nucleotide sequences of the B. abortus dhb operon and surrounding regions have been deposited in GenBank under accession no. AF302798.
RESULTS
Cloning of the complete B. abortus 2,3-DHBA biosynthesis locus.
The cloning and characterization of the B. abortus dhbC and dhbE genes and a portion of the dhbB gene have been previously described (8). These genes were originally given the designations entC, -E, and -B based on their homology to the corresponding enterochelin biosynthesis genes of E. coli (33). Because 2,3-DHBA, and not enterochelin, is the end product of the enzymatic pathway encoded by these genes in B. abortus, we propose changing their designation to dhb to more accurately reflect the activity of their products. Subsequent studies revealed that the cloned fragment containing these genes was a central component of a larger siderophore biosynthesis operon. To obtain the complete 2,3-DHBA biosynthesis operon, B. abortus 2308 genomic DNA fragments approximately 40 kb in length were cloned into the cosmid vector pHC79, and the resulting recombinant cosmids were used to transform the E. coli entC mutant SAB11 (7). Screening the resulting transformants for halo production on CAS plates failed to uncover siderophore-producing colonies, but the presence of the Brucella dhbC in the cosmid library was confirmed by Southern blot analysis of cosmid DNA isolated from a heterogeneous pool of the recombinant SAB11 with a dhbC-specific probe. The same probe was subsequently used in colony blot hybridization to isolate a recombinant E. coli strain carrying the dhbC-containing cosmid. This cosmid was given the designation p2-9H and, consistent with our previous findings, was unable to restore the ability of E. coli SAB11 to produce siderophore activity on CAS plates. To determine if the inability of p2-9H to complement the entC was the result of the low copy number of the base vector in E. coli, a 2.3-kb EcoRI fragment from p2-9H containing the entire dhbC open reading frame was subcloned into the medium-copy-number vector pBBR1MCS-4 (23) and into the high-copy-number vector pBluescript II KS+. The resulting plasmids, pEC2-M and pEC2-B (Fig. 1), respectively, were then introduced into E. coli SAB11, and the resulting transformants were screened for siderophore production on CAS agar plates and by Arnow analysis of supernatants from cultures grown under iron-deprived conditions. SAB11(pEC2-B) produced siderophore levels similar to those observed for SAB11(pPS50) (8) in both assays (data not shown). Strain SAB11(pEC2-M), on the other hand, displayed no detectable siderophore activity on CAS plates and significantly lower activity than SAB11(pEC2-B) in the Arnow assay (data not shown). One possible explanation for the requirement for a high-copy vector for functional complementation of the enterochelin-negative phenotype of E. coli SAB11 by the cloned B. abortus dhbC gene might be inefficient recognition of the B. abortus dhbCEBA promoter sequences by the recombinant E. coli host. This possibility was not further examined experimentally, however.
Genetic complementation of E. coli ent mutants with the corresponding homologs from B. abortus.
Analysis of the nucleotide sequences surrounding dhbC suggested that the B. abortus dhb locus consists of ORFs encoding homologs of five (EntC, -E, -B, -A, and -D) of six of the enzymes involved in enterochelin biosynthesis by E. coli (16, 26, 32, 33). As noted earlier, we have designated the entC, -E, -B, and -A homologs in B. abortus as dhbC, -E, -B, and -A, respectively. Because the function of the B. abortus entD homolog has yet to be defined, however, this gene is presently being referred to as “entD.” An ORF encoding a homolog of VibH, an amide synthase that participates in the synthesis of the 2,3-DHBA-containing siderophore vibriobactin in V. cholerae (21, 22, 55), was also detected. To assess the biochemical activity of these predicted gene products, selected restriction fragments from p2-9H were subcloned into pBluescript II KS+ (Fig. 1), and the capacity of the resulting plasmids to complement well-characterized E. coli enterochelin biosynthesis mutants was evaluated. Introduction of pPS50 and pEC6-B into E. coli SAB11(entC) and AN192(entB), respectively, restored the capacity of these strains to produce enterochelin. The latter finding is notable in that pEC6-B lacks the promoter sequences from the B. abortus dhbCEBA operon, and, consequently, expression of the cloned B. abortus dhbB in AN192 is likely dependent upon the activity of the lac promoter resident in the pBluescript II KS+ cloning vector. With respect to the B. abortus genes encoding homologs of enzymes involved in the conversion of 2,3-DHBA into enterochelin, introduction of pPS50 into E. coli AN193 (entE) restored enterochelin production by this strain, but the introduction of pEC6-B failed to restore enterochelin biosynthesis in the E. coli entD mutant AN190. Interestingly, introduction of pEC6-B failed to restore enterochelin production in the E. coli entB mutant AB1515.24 (53). EntB is a bifunctional enzyme: the isochorismate lyase activity of this enzyme is involved in the biosynthesis of 2,3-DHBA, while the serine phosphopantetheinylation domain of this enzyme participates in the conversion of 2,3-DHBA to enterochelin (16). The E. coli entB mutant AN192 lacks the isochorismate lyase activity of EntB due to a nonpolar point mutation in the corresponding gene, but retains the serine phosphopantetheinylation domain and corresponding enterochelin synthetase activity (formerly known as EntG activity) of this enzyme (53). The E. coli entB mutant AB1515.24, on the other hand, lacks both isochorismate lyase and enterochelin synthase activities (53) due to a polar mutation in the 5′ end of entB created by the insertion of Tn5. Consequently, the ability of the B. abortus dhbB to complement AN192 but not AB1515.24 suggests that the B. abortus DhbB has lost its capacity to participate in the conversion of 2,3-DHBA to enterochelin. Loss of enterochelin synthetase activity by the B. abortus DhbB was not verified biochemically, however.
Genetic organization and iron-responsive expression of the B. abortus 2,3-DHBA biosynthesis operon.
Computer-assisted analysis of the nucleotide sequences of the B. abortus dhbC, -E, -B, and -A genes suggested that these genes are transcribed as a polycistronic message (Fig. 1). The polycistronic nature of the dhbCEBA mRNA and termination of the dhbCEBA transcript in the dhbA-entD intergenic region were confirmed by RT-PCR analysis employing primers designed to detect transcripts arising from contiguous genes (Fig. 2A). These studies also indicated that the B. abortus entD homolog is transcribed as part of a separate operon that includes an unidentified downstream gene. Transcripts arising from the B. abortus vibH were not detected. Consistent with dhbC, -E, -B, and -A being transcribed as a discrete operon, a region capable of forming a stem-loop structure (ΔG° = −13.3 kcal/mol) is present within the dhbA-entD intergenic region (Fig. 1).
FIG. 2.
Polycistronic nature (A) and iron-regulated production (B) of the B. abortus dhbCEBA transcript. (A) A schematic of the dhbCEBA operon is shown above the agarose gel analysis of the RT-PCR products generated with the primers listed in Table 2. Large rectangular arrows indicate the predicted transcript direction, small arrows represent the relative location of primers and solid bars depict regions targeted for amplification by RT-PCR. The small shaded box represents a putative Fur-binding sequence within the dhbC promoter region. Lanes 1 and 4 contain a 100-bp DNA ladder (NEB). The sample shown in lane 3 illustrates the 5′ end of the transcript as well as the absence of contaminating DNA. Lane order and the predicted size of amplified products are as follows: lane 2, vibH internal (365 bp); lane 3, dhbC limiting (303 bp); lane 5, dhbC-E intergenic (585 bp); lane 6, dhbE-B intergenic (479 bp); lane 7, dhbB-A intergenic (321 bp); lane 8, dhbA-entD intergenic (334 bp); lane 9, entD internal (454 bp); and lane 10, entD-orfX (214 bp). The same DNase-treated RNA samples were used for all reactions. (B) Slot blot Northern analysis of total RNA isolated from B. abortus 2308 cultures grown under iron-replete (+) and iron-depleted (−) conditions with a dhbC-specific probe.
B. abortus 2308 produces 2,3-DHBA in response to iron deprivation, but not during growth under iron-replete conditions (27), suggesting that expression of the dhbCEBA operon is regulated in an iron-repressible manner. Consistent with such a relationship, dhbCEBA-specific transcripts were readily detected in RT-PCR (Fig. 2A) and Northern blot (Fig. 2B) analyses of total RNA preparations from B. abortus 2308 cultures grown under iron-restricted conditions. On the other hand, dhbCEBA-specific transcripts were not detectable in Northern blot (Fig. 2B) and RT-PCR (data not shown) analyses of total RNA preparations from B. abortus 2308 cultures grown under iron-replete conditions. Initiation of transcription of the dhbCEBA operon could potentially occur at either of the two predicted promoter sequences depicted in Fig. 3, both of which contain putative ferric iron uptake regulator (Fur)-binding sites (15). Surprisingly, primer extension analysis showed that both of these promoters are functional in B. abortus when this organism is cultivated under low-iron conditions (Fig. 4). In contrast, transcription initiation from neither promoter was detected when the primer extension analysis was performed on RNA preparations from B. abortus 2308 grown under iron-replete conditions (data not shown).
FIG. 3.
Relevant features of the B. abortus dhbCEBA promoter region. Transcript start sites are marked by +1, and the corresponding −35 and −10 regions are underlined. Putative Shine-Dalgarno (S.D.) sequences are also indicated. Oligonucleotides CStrt1 (P1) and CStrt2 (P2) used for primer extension analysis are indicated by arrows. Shaded boxes highlight predicted start codons, and large arrows indicate translated regions with amino acid sequence. Predicted Fur binding sites (15) are designated by boxed areas.
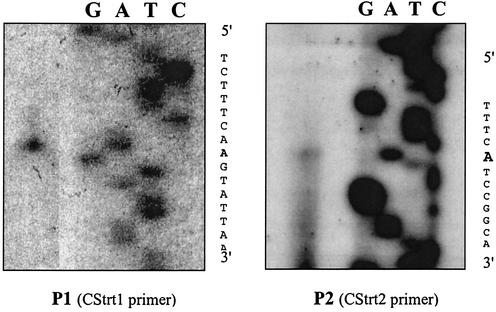
FIG. 4.
Primer extension analysis of the B. abortus dhbC promoter region. Primer extension products from Cstrt1 (P1) and Cstr2 (P2) reactions were loaded alongside end-labeled DNA sequencing reaction products to map the 5′ ends of the dhbCEBA transcript (boldface). Lane designations of sequencing products are presented as being complementary to that of the transcript to allow for 5′-to-3′ reading to coincide with the sequence presented in Fig. 3.
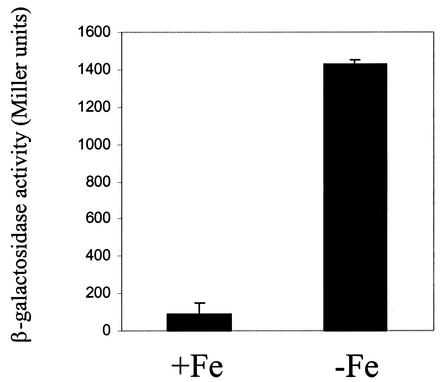
To further evaluate the capacity of iron to repress expression from the dhbCEBA promoter, we evaluated β-galactosidase production by a B. abortus 2308 derivative carrying a plasmid-borne dhbC::lacZ fusion following growth under iron-restricted or iron-replete conditions. Significant levels of β-galactosidase production were only observed when this strain was grown under iron-restricted conditions (Fig. 5), further supporting the notion that expression of the B. abortus dhbCEBA operon is repressed in response to increasing levels of iron in the environment.
FIG. 5.
Repression of dhbC::lacZ expression in response to iron supplementation in B. abortus 2308. β-Galactosidase activity was measured after culture of both strains for 48 h in either low-iron minimal medium alone (−Fe) or the same medium supplemented with 50 μM FeCl3 (+Fe) as described in Materials and Methods. The graph shown is representative of multiple experiments performed. β-Galactosidase activity is presented on the y axis in Miller units (31).
The B. abortus dhbC mutant BHB1 displays significant attenuation in pregnant cattle.
To evaluate the contribution of 2,3-DHBA production to the virulence of B. abortus in its natural ruminant host, pregnant heifers were experimentally infected with virulent strain 2308 and the isogenic dhbC mutant BHB1. The construction of this mutant has been described previously (8). Briefly, BHB1 was constructed by replacing a 261-bp EcoRV fragment internal to the dhbC coding region with the kanamycin resistance gene aphA3. Because dhbC is the first gene in the B. abortus 2,3-DHBA biosynthesis operon (Fig. 1), the placement and orientation of the antibiotic resistance cassette in BHB1 prevent the expression of the remaining downstream genes in this locus, dhbE, -B, and A. BHB1 is unable to produce 2,3-DHBA as measured by the Arnow and liquid CAS siderophore assays (8).
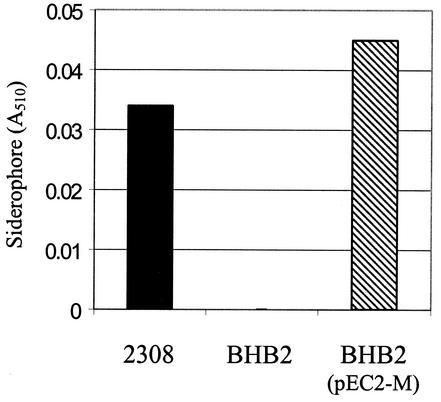
A two-step approach was used to confirm the link between the dhbC mutation in BHB1 and the inability of this strain to produce 2,3-DHBA in response to iron limitation. First, the polar dhbC mutation in BHB1 was replaced with a nonpolar dhbC mutation, resulting in the construction of BHB2. These two mutants carry the same 219-bp deletion in the dhbC coding region, the only difference being that this deletion was replaced with the aph3A gene in BHB1 (8), rendering this a polar mutation in this strain. Like BHB1, BHB2 is unable to produce 2,3-DHBA in response to iron limitation (Fig. 6). Introduction of a cloned copy of the B. abortus dhbC on a pBBR1MCS-based plasmid restored the capacity of BHB2 to produce 2,3-DHBA in response to iron limitation, confirming the participation of the dhbC gene product in 2,3-DHBA biosynthesis.
FIG. 6.
Genetic complementation of the siderophore-negative phenotype of the B. abortus nonpolar dhbC mutant BHB2 by a plasmid-borne cloned copy of dhbC. Catechol-type siderophore production by the B. abortus strains was determined with the Arnow assay (5) after 48 h of culture in low-iron medium. The results presented are from a representative experiment.
Eight out of 10 pregnant heifers inoculated with B. abortus 2308 terminated gestation by either abortion or by giving birth to a weak calf (Table 3). All 10 of the animals in this experimental group were culture positive for B. abortus at necropsy, as were their offspring, including 2 healthy calves. All of the adult cattle in this group also displayed positive serological responses in the Rose Bengal card test for Brucella-specific antibodies (data not shown). In contrast, only one of the pregnant heifers inoculated with the dhbC mutant BHB1 experienced abortion (Table 3). B. abortus was not recovered from this animal at necropsy, nor was it recovered from its aborted fetus. Brucellae were recovered, however, from another adult animal in the BHB1 experimental group as well as from its full-term, healthy calf. Only a single pregnant heifer inoculated with strain BHB1 developed a positive serological response, and this animal neither aborted nor was colonized by BHB1 at the time of necropsy (data not shown).
TABLE 3.
Virulence of B. abortus 2308 and BHB1 (ΔdhbC) in pregnant cattle
| Strain | No. of heifers with result/no. tested
|
||
|---|---|---|---|
| Maternal colonizationa,b | Fetal colonizationb,c | Fetal pathologyd | |
| 2308 | 10/10 | 10/10 | 8/10 |
| BHB1 | 1/10e | 1/10e | 1/10e |
B. abortus (≥ 1 CFU) isolated from a vaginal swab and/or one or more of the following tissues from the heifer: parotid, internal iliac, or supramammary lymph node, mammary gland, or milk.
B. abortus isolates were identified by standard biochemical reactions and their reactivity with B. abortus antiserum.
B. abortus (≥1 CFU) isolated from the lungs and/or abomasal fluid of the fetus or calf.
Fetus aborted or birth of a weak calf (e.g., a calf that cannot stand alone and cannot or will not nurse).
P ≤ 0.005 versus 2308 by Fisher's exact test (41).
Although we have successfully used a pBBR1MCS-based vector in genetic complementation experiments in vitro to confirm the role of the B. abortus dhbCEBA operon in 2,3-DHBA biosynthesis, these plasmids have proven to be unreliable for this purpose in the ruminant host (40). Consequently, we were unable to apply a similar strategy to verify the link between the dhbC mutation in BHB1 and the attenuation of this strain in goats or cattle from a genetic standpoint.
DISCUSSION
Brucella spp. produce the catechol siderophore 2,3-DHBA in response to iron limitation (27, 28). The biosynthesis of 2,3-DHBA from chorismate in bacteria requires the products of the entC, entB, and entA genes or their homologs (9). Therefore, it was not surprising that analysis of the genes downstream of the B. abortus dhbC revealed the presence of functional entB and entA homologs (which we have designated dhbB and dhbA). The presence of a functional entE homolog positioned between dhbC and dhbB is also consistent with the genetic organization of other 2,3-DHBA biosynthesis operons, as is the transcription of the B. abortus dhbC, dhbE, dhbB, and dhbA genes into a polycistronic mRNA (30, 32, 33, 54). Like other prokaryotic 2,3-DHBA biosynthesis operons, transcription of the B. abortus dhbCEBA operon is induced in response to iron limitation and is repressed when sufficient iron is available. Consistent with this finding, both of the promoters identified for this operon are overlapped by consensus Fur-binding sites (15). The relevance of the presence of two functional promoters upstream of the B. abortus dhbCEBA operon, if any, is unknown. One intriguing, potentially biologically significant possibility is that the dual iron-responsive promoters and Fur-recognition sites upstream of the dhbCEBA operon allow this set of genes to be regulated both in concert with and independently from the genes immediately surrounding this operon.
In E. coli, the products of the entB, -D, -E, and F genes work together to assemble 2,3-DHBA and serine into enterochelin (19, 32). Previous studies have shown that B. abortus does not produce enterochelin, nor can it utilize this complex catechol as a siderophore (27). Consequently, although it was not surprising to find a functional entE homolog (dhbE) embedded in the B. abortus 2,3-DHBA biosynthesis operon, it was initially intriguing to find entD and vibH homologs flanking this locus. The recent description of brucebactin, a complex 2,3-DHBA-containing siderophore of unknown structure produced by B. abortus (18), however, offers a possible explanation for the presence of these genes as well as their close proximity to the dhbCEBA operon. This novel siderophore was identified through the failure of a B. abortus 2308 derivative carrying a Tn5 insertion in the vibH homolog to produce this catechol. VibH is a stand-alone nonribosomal peptide synthetase that utilizes a condensation domain (22) to fuse 2,3-DHBA to the polyamine norspermidine during the biosynthesis of the catechol-based siderophore vibriobactin in V. cholerae. Prior to this reaction in V. cholerae, 2,3-DHBA is tethered to the phosphopantetheinyl domain of VibB (a homolog of the B. abortus DhbB) through the activity of VibE (a homolog of the B. abortus DhbE) (29). VibD, a homolog of the B. abortus EntD, adds the phosphopantetheinyl domain to VibB. The functional domains required for the amide synthase activity of the V. cholerae VibH are highly conserved in the B. abortus VibH homolog (Table 4), and it is easy to envision a biochemical pathway in which B. abortus DhbB, EntD, DhbE, and VibH fuse 2,3-DHBA with a small nucleophilic molecule, possibly a polyamine or an amino acid, giving rise to brucebactin.
TABLE 4.
Conservation of siderophore biosynthesis domains in B. abortus DhbB, EntD, and VibHa
| Domain (reference) | Sequence |
|---|---|
| Serine phosphopantetheinylation domain of EntB (14) | |
| B. abortus DhbB (aa 242-248) | LYGLDSL |
| E. coli EntB (aa 240-246) | DYGLDSL |
| Phosphopantetheinyl transferase domains of EntD (21) | |
| B. abortus EntD (aa 89-94) | IGIDIE |
| E. coli EntD (aa 108-113) | GGIDIE |
| B. abortus EntD (aa 130-140) | FSAKESLFKAL |
| E. coli EntD (aa 151-161) | FSAKESAFKAS |
| Amide synthase active site of VibH (19) | |
| B. abortus VibH (aa 166-172) | HHIIIDG |
| V. cholerae VibH (aa 125-131) | HHIVLDG |
Amino acids (aa) that are underlined are considered critical residues in these motifs based on their highly conserved nature and/or the results of mutational analysis.
Failure of the B. abortus dhbB and entD to restore the enterochelin synthetase activities of the E. coli entB mutant AB1515.24 and entD mutant AN90 suggests that the products of these genes may not be functional, at least with regard to their capacity to participate in the conversion of 2,3-DHBA into enterochelin. However, the enterochelin synthase of E. coli is a multienzyme complex consisting of EntB, -E, -D, and -F (19), and it is possible that the inability of B. abortus EntD and DhbB to function in enterochelin synthetase activity results from their failure to properly interact with the other components in complex formation, and not their lack of aryl carrier protein and phosphopantetheinyl transferase activities, respectively. Indeed, the proposed serine phosphopantetheinylation domain of EntB (16) is highly conserved in the B. abortus DhbB, and DhbD contains two highly conserved phosphopantetheinylation domains (24) (Table 4). Our inability to detect transcription of the B. abortus vibH homolog in response to iron limitation raises another question with regard to the potential participation of these genes in brucebactin synthesis. However, this siderophore appears to be produced later in growth than 2,3-DHBA and at much lower levels (18). This would be consistent with expression of the B. abortus vibH homolog being regulated in a different manner from the dhbCEBA operon in response to environmental iron levels, and this may explain our inability to detect the vibH transcript under the conditions employed in our study. Obviously, a definitive evaluation of the participation of the B. abortus dhbB, dhbE and entD homologs in brucebactin synthesis will require mutational analysis of these genes.
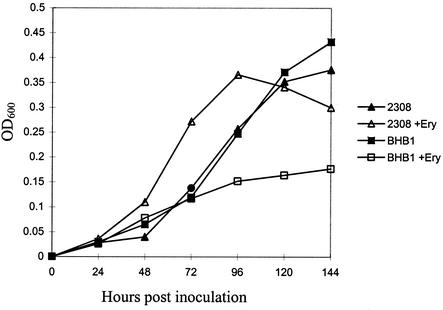
2,3-DHBA production is not required for the survival and replication of B. abortus in cultured murine macrophages or its virulence in BALB/c or C57BL/6 mice (8, 34). Our results with pregnant cattle, on the other hand, indicate that 2,3-DHBA is essential for wild-type virulence in the natural host. These findings also corroborate those of an earlier pilot study showing that BHB1 is attenuated in pregnant goats (Bellaire et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000). The reason for the differential behavior of the B. abortus dhbC mutant in murine and ruminant hosts is unknown, but one particular phenotypic characteristic of this strain offers a possible explanation. Unlike its parental strain, the B. abortus dhbC mutant BHB1 displays considerable growth restriction in vitro when grown under low-iron conditions in the presence of erythritol (Fig. 7) (Bellaire et al., submitted). This defect can be alleviated by adding 2,3-DHBA or FeCl3 to the growth medium, or by genetic complementation of the dhbC mutation, and is not observed when BHB1 is cultivated in low-iron medium with other carbon and energy sources or when this strain is grown in iron-replete medium. At present, our experimental results suggest that utilization of erythritol by B. abortus imposes an unusually high physiologic demand for iron on this bacterium and that the biosynthesis of 2,3-DHBA is required to meet this demand (Bellaire et al., submitted). Erythritol is the preferred carbon and energy source of B. abortus (4, 51), and this four-carbon sugar alcohol is produced in large quantities by ruminant placental trophoblasts during the latter stages of pregnancy (14, 35, 49). Massive intracellular replication of B. abortus in these cells is associated with abortion (1, 14, 35, 42, 50), the cardinal clinical presentation observed with bovine brucellosis. It has long been postulated that the capacity of B. abortus to efficiently metabolize erythritol in the reproductive tract of ruminants plays a key role in virulence (49). Indeed, the attenuation of the bovine vaccine strain B. abortus S19 has been ascribed at least in part to its inability to catabolize erythritol (44, 45, 52). In contrast, erythritol is not a major component of murine tissues, and an isogenic derivative of B. abortus 2308 carrying Tn5 inserted in the erythritol catabolism operon retains its virulence in the mouse model (44). Combined, these findings strongly suggest that the basis for the attenuation of the B. abortus dhbC mutant BHB1 is its inability to efficiently utilize erythritol in the ruminant reproductive tract. Comparison of the phenotype of a B. abortus dhbC mutant with that of an isogenic strain carrying a defined lesion in its erythritol catabolism genes (e.g., eryA) (45) in pregnant ruminants will be important for better defining the relationship between 2,3-DHBA production, efficient erythritol utilization, and virulence in the natural host. Should subsequent studies verify the participation of dhbE, vibH, and entD in the conversion of 2,3-DHBA into brucebactin, B. abortus strains carrying defined lesions in these loci will be useful for separating the contributions of 2,3-DHBA and brucebactin to efficient erythritol metabolism and virulence in the bovine reproductive tract. Developing a genetic complementation strategy for B. abortus that can be reliably used in ruminants will be critical for these types of studies.
FIG. 7.
Growth of B. abortus 2308 and BHB1 in low-iron minimal medium and low-iron minimal medium supplemented with 0.5% meso-erythritol. The culture conditions are detailed in Materials and Methods, and the results presented are from a representative experiment.
Acknowledgments
Plasmid pMR15 was a gift from Lucy Shapiro, Department of Developmental Biology, Stanford University School of Medicine, and the E. coli ent mutants were graciously provided by Shelley Payne, Department of Microbiology, University of Texas at Austin.
This work was supported by grants (95-09195 and 98-02620) from the United State Department of Agriculture's National Research Initiative Competitive Grants Program.
Editor: J. T. Barbieri
REFERENCES
- 1.Alexander, B., P. R. Schnurrenberger, and R. R. Brown. 1981. Numbers of Brucella abortus in the placenta, umbilicus and fetal fluid of two naturally infected cows. Vet. Rec. 108:500.. [DOI] [PubMed] [Google Scholar]
- 2.Alton, G. G., L. M. Jones, R. D. Angus, and J. M. Verger. 1988. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique, Paris, France.
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J. D., and H. Smith. 1965. The metabolism of erythritol by Brucella abortus. J. Gen. Microbiol. 38:109-124. [DOI] [PubMed] [Google Scholar]
- 5.Arnow, L. E. 1937. Colorimetric determination of the components of 3,4-dihydroxyphenyalanine-tyrosine mixtures. J. Biol. Chem. 118:531-537. [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2000. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 7.Barghouthi, S., S. M. Payne, J. E. L. Arceneaux, and B. R. Byers. 1991. Cloning, mutagenesis, and nucleotide sequence of a siderophore biosynthetic gene (amoA) from Aeromonas hydrophila. J. Bacteriol. 173:5121-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellaire, B. H., P. H. Elzer, C. L. Baldwin, and R. M. Roop II. 1999. The siderophore 2,3-dihydroxybenzoic acid is not required for virulence of Brucella abortus in BALB/c mice. Infect. Immun. 67:2615-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, V., K. Hantke, and W. Koster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation. Met. Ions Biol. Syst. 35:67-145. [PubMed] [Google Scholar]
- 10.Byers, B. R., and J. E. Arceneaux. 1998. Microbial iron transport: iron acquisition by pathogenic microorganisms. Met. Ions Biol. Syst. 35:37-66. [PubMed] [Google Scholar]
- 11.Cox, G. B., F. Gibson, R. K. J. Luke, N. A. Newton, I. G. O'Brien, and H. Rosenberg. 1970. Mutations affecting iron transport in Escherichia coli. J. Bacteriol. 104:219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmonds, M., N. Booth, S. Hagius, J. Walker, F. Enright, R. M. Roop II, and P. Elzer. 2000. Attenuation and immunogenicity of a Brucella abortus htrA cycL double mutant in cattle. Vet. Microbiol. 76:81-90. [DOI] [PubMed] [Google Scholar]
- 13.Elzer, P. H., R. W. Phillips, M. E. Kovach, K. M. Peterson, and R. M. Roop II. 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62:4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, F. M. 1990. The pathogenesis and pathobiology of Brucella infections in domestic animals, p. 301-320. In K. H. Nielsen and J. R. Duncan (ed.), Animal brucellosis. CRC Press, Boca Raton, Fla.
- 15.Escolar, L., J. Pérez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring, A. M., K. A. Bradley, and C. T. Walsh. 1997. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry 36:8495-8503. [DOI] [PubMed] [Google Scholar]
- 17.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González Carreró, M. I., F. J. Sangari, J. Agüero, and J. M. García Lobo. 2002. Brucella abortus strain 2308 produces brucebactin, a highly efficient catecholic siderophore. Microbiology 148:353-360. [DOI] [PubMed] [Google Scholar]
- 19.Greenwood, K. T., and R. J. Luke. 1976. Studies on the enzymatic synthesis of enterochelin in Escherichia coli K-12. Four polypeptides involved in the conversion of 2,3-dihydroxybenzoate to enterochelin. Biochim. Biophys. Acta 454:285-297. [DOI] [PubMed] [Google Scholar]
- 20.Hohn, B., and J. Collins. 1980. A small cosmid for efficient cloning of large DNA fragments. Gene 11:291-298. [DOI] [PubMed] [Google Scholar]
- 21.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513-15521. [DOI] [PubMed] [Google Scholar]
- 22.Keating, T. A., G. C. Marshall, C. T. Walsh, and A. E. Keating. 2002. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat. Struct. Biol. 9:522-526. [DOI] [PubMed] [Google Scholar]
- 23.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 24.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 25.Leonard, B. A., I. López-Goñi, and C. L. Baldwin. 1997. Brucella abortus siderophore 2,3-dihydroxybenzoic acid protects brucellae from killing by macrophages. Vet. Res. 28:87-92. [PubMed] [Google Scholar]
- 26.Liu, J., K. Duncan, and C. T. Walsh. 1989. Nucleotide sequence of a cluster of Escherichia coli enterobactin biosynthesis genes: identification of entA and purification of its product 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase. J. Bacteriol. 171:791-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Goñi, I., I. Moriyón, and J. B. Neilands. 1992. Identification of 2,3-dihydroxybenzoic acid as a Brucella abortus siderophore. Infect. Immun. 60:4496-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Goñi, I., and I. Moriyón. 1995. Production of 2,3-dihydroxybenzoic acid by Brucella species. Curr. Microbiol. 31:291-293. [Google Scholar]
- 29.Marshall, C. G., M. D. Burkhart, T. A. Keating, and C. T. Walsh. 2001. Heterocycle formation in vibriobactin biosynthesis: alternative substrate utilization and identification of a condensed intermediate. Biochemistry 40:10655-10663. [DOI] [PubMed] [Google Scholar]
- 30.Massad, G., J. E. Arceneaux, and B. R. Byers. 1994. Diversity of siderophore genes encoding biosynthesis of 2,3-dihydroxybenzoic acid in Aeromonas spp. Biometals 7:227-236. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Nahlik, M. S., T. P. Fleming, and M. A. McIntosh. 1987. Cluster of genes controlling synthesis and activation of 2,3-dihydroxybenzoic acid in production of enterobactin in Escherichia coli. J. Bacteriol. 169:4163-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nahlik, M. S., T. J. Brickman, B. A. Ozenberger, and M. A. McIntosh. 1989. Nucleotide sequence and transcriptional organization of the Escherichia coli enterobactin biosynthesis cistrons entB and entA. J. Bacteriol. 171:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parent, M. A., B. H. Bellaire, E. A. Murphy, R. M. Roop II, P. H. Elzer, and C. L. Baldwin. 2002. Brucella abortus siderophore 2,3-dihydroxybenzoic acid (DHBA) facilitates intracellular survival of the bacteria. Microb. Pathog. 32:239-248. [DOI] [PubMed] [Google Scholar]
- 35.Payne, J. M. 1959. The pathogenesis of experimental brucellosis in the pregnant cow. J. Pathol. Bacteriol. 78:447-463. [DOI] [PubMed] [Google Scholar]
- 36.Payne, S. M. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 37.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 38.Robertson, G. T., M. E. Kovach, C. A. Allen, T. A. Ficht, and R. M. Roop II. 2000. The Brucella abortus Lon functions as a generalized stress response protease and is required for wild-type virulence in BALB/c mice. Mol. Microbiol. 35:577-588. [DOI] [PubMed] [Google Scholar]
- 39.Robertson, G. T., A. Reisenauer, R. Wright, R. B. Jensen, A. Jensen, L. Shapiro, and R. M. Roop II. 2000. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J. Bacteriol. 182:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roop, R. M., II, R. W. Phillips, S. Hagius, J. V. Walker, N. J. Booth, W. T. Fulton, M. D. Edmonds, and P. H. Elzer. 2001. Reexamination of the role of the Brucella melitensis HtrA stress response protease in virulence in pregnant goats. Vet. Microbiol. 82:91-95. [DOI] [PubMed] [Google Scholar]
- 41.Rosner, B. 2000. Fundamentals of biostatistics, 5th ed. Duxbury, Pacific Grove, Calif.
- 42.Samartino, L. E., and F. M. Enright. 1993. Pathogenesis of abortion of bovine brucellosis. Comp. Immunol. Microbiol. Infect. Dis. 16:95-101. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Sangari, F. J., M. J. Grilló, M. P. Jiménez de Bagües, M. I. González-Carreró, J. M. García-Lobo, J. M. Blasco, and J. Agüero. 1998. The defect in the metabolism of erythritol of the Brucella abortus B19 vaccine strain is unrelated with its attenuated virulence in mice. Vaccine 16:1640-1645. [DOI] [PubMed] [Google Scholar]
- 45.Sangari, F. J., J. Agüero, and J. M. García-Lobo. 2000. The genes for erythritol catabolism are organized as an inducible operon in Brucella abortus. Microbiology 146:487-495. [DOI] [PubMed] [Google Scholar]
- 46.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schweitzer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 48.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 49.Smith, H., A. E. Williams, J. H. Pearce, J. Keppie, P. W. Harris-Smith, R. B. Fitzgeorge, and K. Witt. 1962. Foetal erythritol: a cause of the localization of Brucella abortus in bovine contagious abortion. Nature 193:47-49. [DOI] [PubMed] [Google Scholar]
- 50.Smith, T. 1919. A characteristic localization of Bacillus abortus in the bovine fetal membranes. J. Exp. Med. 29:451-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sperry, J. F., and D. C. Robertson. 1975. Erythritol catabolism by Brucella abortus. J. Bacteriol. 121:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sperry, J. F., and D. C. Robertson. 1975. Inhibition of growth by erythritol catabolism in Brucella abortus. J. Bacteriol. 124:391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staab, J. F., and C. F. Earhart. 1990. EntG activity of Escherichia coli enterobactin synthetase. J. Bacteriol. 172:6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyckoff, E. E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyckoff, E. E., S. L. Smith, and S. M. Payne. 2001. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J. Bacteriol. 183:1830-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]