Abstract
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli are extracellular pathogens that employ a type III secretion system to export translocator and effector proteins, proteins which facilitates colonization of the mucosal surface of the intestine via formation of attaching and effacing (A/E) lesions. The genes encoding the proteins for A/E lesion formation are located on a pathogenicity island, termed the locus of enterocyte effacement (LEE), which contains eae encoding intimin as well as the type III secretion system and effector genes. Many type III secreted proteins are stabilized and maintained in a secretion-competent conformation in the bacterial cytosol by specific chaperone proteins. Three type III chaperones have been described thus far within the EPEC LEE region: CesD, for the translocator proteins EspB and EspD; CesT, for the effector proteins Tir and Map; and CesF, for EspF. In this study we report the characterization of CesD2 (previously Orf27), a second LEE-encoded chaperone for EspD. We show specific CesD2-EspD protein interaction which appears to be necessary for proper EspD secretion in vitro and pathogenesis in vivo as demonstrated in the A/E-lesion-forming mouse pathogen Citrobacter rodentium.
Enteropathogenic Escherichia coli (EPEC) is a common cause of infant diarrhea, particularly in developing countries (42). EPEC infection is associated with formation of a typical microscopic lesion on intestinal epithelial cells, the attaching and effacing (A/E) lesion (41). A/E lesions are characterized by destruction of cellular microvilli and intimate attachment of bacteria to cup-like pedestals at the apical enterocyte cell membrane (31), triggered by activation of a number of signal transduction pathways and rearrangement of cytoskeletal proteins (46). A/E lesions are induced by other enteric pathogens, such as enterohemorrhagic E. coli (EHEC) (54). EHEC infection, which is frequently associated with outbreaks in the developed world, can cause severe diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome (42). EPEC and EHEC are the most prominent members among the A/E-lesion-causing pathogens, which are also represented by animal pathogens, including rabbit EPEC (45) and Citrobacter rodentium, which causes transmissible murine colonic hyperplasia (47).
The A/E lesion phenotype is encoded by a chromosomal pathogenicity island termed the locus of enterocyte effacement (LEE) region (37). The LEE consists of 41 open reading frames (11, 16), encoding a transcriptional regulator (Ler) (40); type III secretion system (TTSS) proteins (Esc and Sep) (16, 25); type III translocator (EspA, EspB, EspD) (12, 30, 34) and effector (EspF, EspG, Map, Tir) (14, 28, 29, 38, 39) proteins; an outer membrane adhesin, intimin (26); and a number of proteins of unknown function (16).
TTSSs deliver effector proteins through the bacterial inner and outer membranes, with no periplasmic intermediate, and through the plasma membrane into the eukaryotic cell cytosol (23). The structural TTSS proteins are highly conserved among pathogenic bacteria, and many show an intriguing similarity to proteins involved in flagellar biosynthesis (23). The TTSS has been purified from Salmonella enterica serovar Typhimurium, Shigella flexneri (33, 51) and most recently from EPEC (8, 49). It consists of a multiple ring structure and a needle projection. In contrast to the high degree of sequence similarity between the structural TTSS proteins, the effector, secreted, proteins show a high level of variation from one system to another. These proteins vary greatly in size, structure, and function and account for the species-specific pathogenicity phenotypes associated with different bacterial infections (23).
In EPEC there are seven LEE-encoded type III secreted proteins—EspA, EspB, EspD, EspF, EspG, Map, and Tir (12, 14, 28-30, 34, 38, 39). EspA, EspB, and EspD are translocator proteins, necessary for A/E lesion formation and signaling within host cells (19). EspA is the main or only component of a transiently expressed filamentous surface organelle (32), which is required for the translocation of other translocator and effector proteins (28, 32). In contrast, immunofluorescence assays and a calmodulin-dependent adenylate cyclase reporter system have demonstrated that EspB is translocated into the host cell, where it is distributed between the cytosol and plasma membrane compartments (61). The presence of EspB in the cytosolic fraction suggests that it may also have, in addition to its translocator activities, an effector function (52). Indeed, it has been shown that EspB causes changes in host cell morphology and reorganization of stress fibers, acting possibly as a cytoskeletal toxin (52).
The third translocator protein, EspD, is inserted into a trypsin-sensitive location in the host cell plasma membrane, at sites of bacterial contact, but is not translocated into the cytoplasm (55). Based on its homology to YopB, it is believed that EspD is the main component of the TTSS translocation pore in the plasma membrane (19). Indeed, we have recently shown that EspD exhibits intermolecular interaction which involved its carboxy-terminal coiled-coil domain (7). A radical disruption of this region, although did not affect EspA filament production, caused reduced EspA filament-mediated cell attachment and EspD-mediated A/E lesion formation (7). The fact that an espD mutant secretes low levels of EspA and produces truncated EspA filaments (32) suggests that EspD might be a minor component of the filaments. Indeed, the current model of the EPEC TTSS predicts interaction between EspA filaments and the EspD translocation pores (19), interaction which allows protein translocation.
Many of the type III secreted proteins are dependent on specific chaperones for stabilization in the bacterial cytosol prior to secretion and prevention of premature interactions with secreted proteins and/or with parts of the secretion and translocation machinery (4, 58). In addition, chaperones have been shown to be required for the exportation of translocator and effector proteins. Three chaperones encoded within the LEE region have been described thus far. The first one described was CesD, a chaperone for EspB and EspD (56). CesD is a 17.5-kDa protein which was shown to interact specifically with EspD. In a CesD-deficient mutant, EspD secretion was abolished and the amount of EspB secreted was reduced, but little effect on secretion of EspA was observed (56). Despite sharing features that are common to other type III secretion chaperones, CesD is distinct due to its membrane localization (56). CesT was the second reported LEE-encoded chaperone (1, 13). CesT, 15 kDa, is also a bivalent chaperone that binds to, and is required for translocation of, both Tir (1, 13) and Map (6). CesF, the most recently reported EPEC TTSS chaperone, is a 14-kDa protein which specifically interacts with EspF (15). In this study we characterize the gene product of the LEE region orf27, a protein with unknown function which exhibits features common to TTSS chaperones (16). We demonstrate that Orf27 is a second secretion partner for EspD and consequently have renamed it CesD2.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains used in this study are listed in Table 1. Plasmids used and constructs generated in this study are listed in Table 2. Unless otherwise stated, bacterial strains were grown in Luria-Bertani (LB) broth at 37°C. Growth media were supplemented with ampicillin (50 to 100 μg/ml), chloramphenicol (30 μg/ml), kanamycin (30 μg/ml), or nalidixic acid (100 μg/ml) as required.
TABLE 1.
Bacterial strains used in this study
| Strain | Description or genotype | Source or reference |
|---|---|---|
| E. coli TG1 | supE hsd Δ5thi Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | Stratagene |
| E. coli BL21(DE3)pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3) pLysS (Cmr) | Novagen |
| E. coli JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) | New England Biolabs |
| E. coli E2348/69 | EPEC prototype strain | 36 |
| C. rodentium | Wild-type, nalidixic acid-resistant strain (formerly C. freundii biotype 4280) | 47 |
| C. rodentium ICC172 | E2348/69 ΔcesD2::aphT | This study |
| C. rodentium ICC173 | C. rodentium cesD2::aphT | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Description or genotype | Source or reference |
|---|---|---|
| pET 28a | PT7lac N-terminal His6 expression vector | Novagen |
| pGEM-T-Easy | ColE1-based vector for cloning of PCR products | Promega |
| pET3d | PT7 expression vector | Novagen |
| pMal-c2X | PtacmalE N-terminal fusion expression vector | New England Biolabs |
| pACYC184 | Origin of replication from p15A; low copy number; Cmr Tcr | New England Biolabs |
| pICC232 | pET3d-CesD2, unmodified CesD2 | This study |
| pICC233 | pMAL-c2X-CesD2; full-length CesD2 fused to MBP in N-terminal end | This study |
| pICC234 | pET28a-CesD2; full-length CesD2 fused to His6 in the N-terminal end | This study |
| pICC240 | PCR-amplified 4.071-kbp DNA fragment containing cesD2; cloned into pGEM-T-Easy | This study |
| pICC241 | In-frame deletion subclone of pICC240; missing bp 97 to 351 of cesD2 | This study |
| pICC260 | ΔcesD2::aphT in pGEM-T-Easy | This study |
| pKD46 | Low-copy-number plasmid encoding the phage λ Red recombinase, expressing γ, β, and exo from the arabinose-inducible ParaB | 9 |
| pSB315 | Plasmid containing cloned aphT kanamycin cassette | 21 |
| pICC243 | PCR-amplified 1.208-kbp DNA fragment containing cesD2CR and cloned in pGEM-T-Easy | This study |
| pICC244 | Insertion subclone of pICC243; cesD2CR::aphT | This study |
| pICC245 | pACYC184-CesD2CR | This study |
| pICC246 | pACYC184-HisCesD2EPEC | This study |
Molecular techniques.
Cloning which required PCR amplification was performed with the proofreading DNA polymerase Deep Vent (New England Biolabs), unless stated otherwise, and all the clones were verified by sequencing. The synthetic oligonucleotides used as primers in the PCR and DNA sequencing procedures were obtained from Pharmacia (United Kingdom) and are listed in Table 3. Automated sequencing was performed at the Advanced Biotechnology Centre, Imperial College of Science, Technology, and Medicine, Faculty of Medicine. DNA analysis was performed with Gene Jockey II and programs available at http://www.expasy.ch/.
TABLE 3.
Oligonucleotide primers used in this study
| Primer name (restriction site) | Sequencea |
|---|---|
| CesD2-F (NdeI) | 5′ TATCATATGGTCGATACGTTTAATGATG 3′ |
| CesD2-R (BamHI) | 5′ TATGGATCCTTAACTATTTACGTTCATTACGAAC 3′ |
| LEE-4K-F | 5′ GGTGAAGTTACGAGTCGAACAGAGG 3′ |
| LEE-4K-R | 5′ GGCTATACAGGAGCGGTTTGGTCTG 3′ |
| CesD2-INV-F (SmaI) | 5′CCGCCACCCGGGAAAAAAGGCACTGCCACAAAGAAACTCC 3′ |
| CesD2-INV-R (SmaI) | 5′AAGGGTCCCGGGCAGCAAGAAACAGAAGACGGAATTTGGTTC 3′ |
| CesD2CR(+1)-F (NcoI) | 5′ TATCCATGGTCGATACGTTTAATGACG 3′ |
| CesD2CR(+408)-R (SalI) | 5′ TATGTCGACCTAATTATTTACTTTCATTACATACC 3′ |
| CesD2CR(−400)-F | 5′ AATCGTACATCAGGTATCGCTGATG 3′ |
| CesD2CR(+808)-R | 5′ TCTGATAAGATTTGCGGCAATACATTCA 3′ |
| CesD2CR(−45)-F (BamHI) | 5′ TATGGATCCATTAATCGTATGGGGCAATCGGC 3′ |
| AphT-R | 5′ GCTTGATGGTCGGAAGACG 3′ |
| pACYC-F | 5′ TCACAGTTAAATTGCTAACGCAGTCAGGCA 3′ |
Restriction sites are underlined.
Cloning, expression, and purification of histidine-tagged CesD2.
The 408-bp DNA fragment encoding orf27 (cesD2) was amplified by PCR, using the primer pair cesD2-F (NdeI) and cesD2-R (BamHI) and EPEC genomic DNA from the prototype strain E2348/69 (36) as the DNA template. The PCR product was digested with NdeI and BamHI and cloned into NdeI/BamHI-digested pET28-a, generating plasmid pICC234 (Table 2). A His6-CesD2 fusion was expressed in BL21(DE3)pLysS(pICC234). An overnight culture was diluted 1:100 in 100 ml of LB broth, supplemented with kanamycin (30 μg/ml), chloramphenicol (30 μg/ml), and 0.2% glucose. The culture was grown to an optical density at 600 nm (OD600) of 0.4 to 0.8, at 37°C with shaking, and was induced with the addition of 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Induction was performed for 4 h at 30°C, with shaking. Bacterial cells were pelleted by centrifugation at 8,000 × g for 20 min and resuspended in cold binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]). Bacterial suspensions were sonicated and cell debris was removed by centrifugation at 45,000 × g for 30 min. The cell extracts were filtered through a 0.45-μm-pore-size filter device prior to the following purification procedures. His6-CesD2 was purified as recommended by the manufacturer (Novagen). Briefly, the bacterial lysate obtained as described above was loaded onto a 2.5-ml nickel-charged column, previously equilibrated with binding buffer. The column was then washed with 25 ml of binding buffer, 7.5 ml of wash buffer 1 (30 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]), and 7.5 ml of wash buffer 2 (60 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]). The bound protein was eluted with 15 ml of elution buffer (500 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]). Fractions (1 ml) were collected and monitored by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE).
Column capture assay.
His-tag columns containing bound His6-CesD2 or His-T7 tag (negative control), were used to test the ability of recombinant CesD2 to bind the secreted forms of EspA, EspB, and EspD. The procedure was performed as described for the purification steps, involving two additional stages which consisted of loading 25 ml of culture supernatant from E2348/69, grown in Dulbecco's modified Eagle medium (DMEM), onto the columns and an extra washing step followed by elution as for the purification procedures. Fractions (1 ml) were collected, separated by SDS-12% PAGE, and analyzed by Western blotting in order to verify copurification of any of the secreted proteins with the recombinant CesD2, or the control column His6-T7.
Generation of a nonpolar cesD2 E2348/69 mutant.
PCR was performed using primers LEE-4K-F and LEE-4K-R to obtain a 4,071-bp DNA fragment of the LEE region containing cesD2, from strain E2348/69, which was ligated into pGEM-T-Easy vector (Promega), creating plasmid pICC240 (Table 2). Inverse PCR was used to construct an in-frame deletion within cesD2 using primers CesD2-INV-F (SmaI) and CesD2-INV-R (SmaI) and pICC240 as the template. Digestion with SmaI and self-ligation excised nucleotides 97 to 351 and added a SmaI site to the deletion locus (Fig. 1B), creating plasmid pICC241 (Table 2). For DNA amplification, the Expand High Fidelity PCR System (Roche, Mannheim, Germany) was employed, which included a hot start of 2 min at 94°C, and the preparation of two master mixes. For a 100-μl reaction mixture, mix 1 (50 μl) contained 400 μM concentrations of the deoxynucleoside triphosphates, 600 nM concentrations of upstream and downstream primers, and 10 ng of template (genomic DNA from strain E2348/69), while mix 2 (50 μl) contained the Expand HF buffer with 15 mM MgCl2 and 2.6 U of Expand High Fidelity PCR System enzyme mix. For primers LEE-4K-F and LEE-4K-R, cycling was performed as follows: 94°C for 2 min (hot start); 10 cycles of 94°C for 30 s, 65°C for 30 s, and 68°C for 6 min; 20 cycles of 94°C for 30 s, 65°C for 30 s, and 68°C for 6 min with 15-s increase; and a final extension at 68°C for 7 min. The same PCR cycling conditions were used for the inverse PCR, using primers CesD2-INV-F (SmaI) and CesD2-INV-R (SmaI), except for the annealing temperature, which, in this case, was 60°C. An aphT cassette (21) with a HincII site in the correct reading frame was digested out of plasmid pSB315 and cloned into the SmaI-digested plasmid pICC241. The orientation of the cassette was verified by PCR, using primers CesD2-F (NdeI) and AphT-R, which tested one of the junction fragments. Primers LEE-4K-F and LEE-4K-R were used to amplify a fragment of approximately 4.8 kbp from plasmid pICC260, which contained the disrupted cesD2 gene and its flanking regions. One microgram of the amplified fragment was treated with DpnI, purified, and transformed by electroporation into E2348/69 carrying a Red helper plasmid, pKD46 (Table 2) (9). Transformants were first selected for Kmr and then tested for ampicillin sensitivity to confirm loss of the helper plasmid. The ΔcesD2::aphT EPEC derivative was named ICC172 (Table 1). In order to trans-complement ICC172, pICC234 was double digested with XbaI/BamHI and a 509-bp DNA fragment, which contained the entire cesD2 and a sequence coding for an N-terminal His6 tag, was ligated into the BamHI site of pACYC184, generating plasmid pICC246. Prior to ligation, both vector and insert were previously filled in with Klenow DNA polymerase I (New England Biolabs), according to the manufacturer's instructions. Fragment orientation was verified by PCR, using primers pACYC-F and CesD2-R (BamHI).
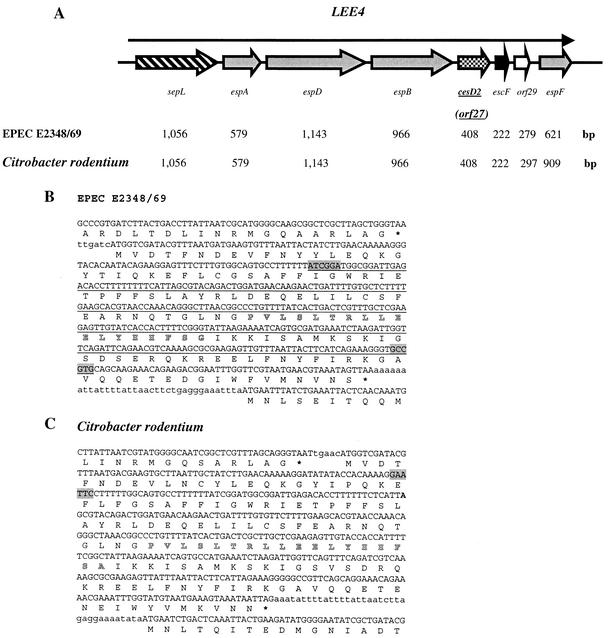
FIG. 1.
Schematic representation of LEE4, DNA sequence, and deduced amino acid sequence of cesD2 (formerly orf27). (A) Overview (not to scale) of the organization of the LEE4 operon. Arrows represent each of the open reading frames, with their length indicated below. (B and C) Nucleotide and amino acid sequences of espB (carboxyl-terminal region), cesD2 (orf27), and escF (amino terminal region) from EPEC E2348/69 and C. rodentium. Outlined amino acid sequences indicate a potential amphipathic α-helix in CesD2. (B) The underlined DNA sequence indicates the deletion in EPEC ΔcesD2 and the insertion point of the aphT cassette. Highlighted sequences indicate the points wherein the SmaI sites were introduced for construction of EPEC ΔcesD2::aphT. (C) Highlighted sequence indicates the EcoRI site where the aphT cassette was inserted, creating a C. rodentium strain that is cesD2::aphT.
Generation of a nonpolar cesD2 C. rodentium mutant.
PCR was used to generate a 1,208-bp fragment containing the C. rodentium cesD2 gene (11), using primers CesD2CR(−400)-F and CesD2CR(+808)-R (Table 3) and genomic DNA from a wild-type C. rodentium strain (47). The amplified fragment was inserted into pGEM-T Easy vector, creating plasmid pICC243. aphT cassette (21) was digested out of pSB315 with EcoRI and ligated into EcoRI-cut pICC243, generating an insertion at position 76 of cesD2 (Fig. 1C) and creating pICC244. The orientation of the cassette was verified by PCR with primers CesD2CR(+1)-F (NcoI) and AphT-R (Table 3). A 2,200-bp fragment containing the disrupted cesD2 was generated by PCR using plasmid pICC244 as the template, with primers CesD2CR(−400)-F and CesD2CR(+808)-R, and treated with DpnI. One microgram of the PCR product was electrotransformed into the C. rodentium strain carrying the Red helper plasmid pKD46 (Table 2), and this was followed by screening for recombinants as described above. The generated cesD2::aphT C. rodentium strain was named ICC173 (Table 1). The cesD2 (CR27) (11) gene was also cloned into plasmid pACYC184 (Table 2), to be used in the trans-complementation experiments. A fragment of 453 bp containing the full-length cesD2 and 45 bp of upstream sequence was amplified from C. rodentium genomic DNA, with primers CesD2CR(−45)-F (BamHI) and CesD2CR(+408)-R (SalI) and was ligated into BamHI/SalI-double-digested pACYC184, generating pICC245.
Preparation of EPEC-secreted proteins.
Analysis of culture supernatants of EPEC-secreted proteins was performed as described previously (32). Bacteria were grown in 20 ml of DMEM at 37°C with shaking to an OD600 of 1.0. The bacterial cells were pelleted by centrifugation at 8,000 × g for 10 min, and supernatants were passed through filters with a pore size of 0.45 μm. Phenylmethylsulfonyl fluoride (PMSF) (50 μg/ml; Sigma), aprotinin (0.5 μg/ml; Sigma), and EDTA (0.5 mM) were added (all final concentrations), and the Esp proteins were concentrated 100-fold with centrifugal filter devices (Millipore Corporation, Bedford, Mass.). A volume of concentrated samples was loaded and separated by SDS-12% PAGE followed by Western blotting.
Bacterial cell fractionation. (i) Periplasm extraction.
All the EPEC cultures (100 ml) were grown to an OD600 of 1.0 in DMEM. The periplasmic contents were collected following osmotic shock as follows. Bacterial cells were pelleted by centrifugation at 4,000 × g for 20 min, at 4°C. The supernatant was removed, the pellets were washed twice and resuspended in 2.5 ml of ice-cold 20 mM Tris-HCl (pH 7.5). Two ml of ice-cold 40% sucrose and 150 μl of 500 mM EDTA were subsequently added, followed by gentle agitation at 4°C for 20 min. Cells were once more spun down as before, the supernatant was discarded, and pellets were immediately resuspended in 1 ml of ice-cold deionized water, following gentle agitation at 4°C for 20 min. The suspension was centrifuged at 8,000 × g for 20 min at 4°C. The resulting supernatant containing periplasmic proteins was then collected (P fraction).
(ii) Preparation of cytoplasmic and membrane fractions.
The pellet from the periplasmic extraction was then fractionated as described previously (24), with modifications. The pellet was initially resuspended in 8 ml of lysis buffer (10 mM Tris-HCl [pH 7.5], 0.5 mM PMSF, aprotinin [0.5 μg/ml]), freeze-thawed, and passed twice through a cell disrupter (Constant Cell Disruption System). Cell envelopes and unbroken bacteria were removed by centrifuging twice (5,000 × g for 10 min at 4°C). The supernatant, containing soluble proteins (cytosolic) and insoluble proteins (inner and outer membranes), was removed and ultracentrifuged for 1 h at 50,0000 × g, at 4°C, to pellet the membranes. The supernatant containing cytoplasmic proteins (C fraction) was collected and concentrated to 0.4 ml with centrifugal filter devices (Millipore Corporation). The membrane pellet was washed once with lysis buffer and resuspended in 0.4 ml of Sarkosyl buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.5 mM PMSF, aprotinin [0.5 μg/ml], 0.5% N-lauroylsarcosine [Sigma]). Under this condition the inner membrane is dissolved but not the outer membrane. Following centrifugation at 50,000 × g for 1 h, at 4°C, the supernatant containing the inner membrane proteins (IM fraction) was collected. The remaining pellet was washed once with Sarkosyl buffer, under the same conditions as described above, and dissolved in 0.4 ml of 1× SDS loading buffer (OM fraction). Equivalent amount of proteins were loaded after normalization in relation to the volume of the original bacterial cultures.
Enrichment and purity of each fraction were estimated by detection of proteins known to be localized within each of the compartments. Enrichment of the cytoplasm was determined by immunoblotting using a mouse polyclonal antiserum against β-galactosidase (1:2,000; Sigma), a well-characterized cytoplasmic enzyme (35). Enrichment of the IM fraction was determined by detection of Etk (24) with a specific rabbit polyclonal antiserum (1:2,000). Maltose binding protein (antiserum diluted 1:2,000, New England Biolabs) was used as a marker for periplasmic material (27), while intimin (26), an outer membrane adhesin of EPEC (antiserum diluted 1:2,000 [2]), was used as a marker for the OM fraction.
Immunoblot analysis.
Proteins separated by SDS-PAGE were transferred electrophoretically onto nitrocellulose membranes (0.45-μm pore size; Bio-Rad) and immunoblotted according to the method of Towbin et al. (53). Proteins were blotted using a Bio-Rad Wet Blot apparatus, the membranes were blocked overnight in 3% bovine serum albumin (Sigma) and washed thoroughly with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST). Primary antisera were diluted in PBST, and the concentrations used are indicated for each experiment. Secondary antibodies, goat anti-rabbit or anti-mouse (1:5,000 dilution, Sigma), were conjugated with horseradish peroxidase. An ECL Western blotting analysis system (Amersham Life Science) was used according to the manufacturer's instructions. The immunoblots were then exposed to a high-performance chemiluminescence film, Hyperfilm ECL (Amersham Life Science), and the was film developed using a Fuji X-ray film developer.
Fluorescence actin staining (FAS).
Subconfluent cultures of HEP-2 cells on glass coverslips were placed in wells of a 24-well tissue culture plate containing 1 ml of HEPES-buffered minimal essential medium containing 2% fetal calf serum. Ten microliters of an overnight bacterial broth culture was added to each well, and the cells were incubated for 1, 3, and 6 h at 37°C; in 6-h assays fresh medium was added after 3 h. After three washes in PBS to remove nonadhering bacteria, cells were fixed in 4% buffered formalin for 30 min. Washed cells were permeabilized by treating coverslips with 0.1% Triton X-100 in PBS for 5 min. After three PBS washes, coverslips were incubated with a solution of fluorescein isothiocyanate-phalloidin (5 μg/ml; Sigma) in PBS for 20 min to stain filamentous cell actin. Bacteria were immunostained using an antibody to whole E2348/69 bacteria and a secondary Alexa-594 antibody (Molecular Probes). Coverslips were washed a further three times in PBS and mounted in Citifluor mountant (agar). Specimens were examined using a Leica TCS SP2 spectral confocal microscope.
Challenge of mice with C. rodentium.
Male, specific-pathogen-free C3H/Hej mice (4 to 5 weeks old) were purchased from Harlan Olac (Bicester, United Kingdom). All mice were housed in individual ventilated cages with free access to food and water. Bacterial inocula were prepared by culturing bacteria overnight at 37°C in LB broth containing nalidixic acid (100 μg/ml) plus chloramphenicol (30 μg/ml) (wild-type C. rodentium carrying pACYC184), LB broth containing kanamycin (30 μg/ml) (C. rodentium ICC173), or LB broth containing kanamycin plus chloramphenicol (30 μg/ml) (C. rodentium ICC173 carrying pICC245 or pICC246). After incubation, bacteria were harvested by centrifugation and resuspended in equal volumes of PBS. Mice were then orally inoculated with 0.2 ml of a bacterial suspension using a gavage needle (50). The viable count of each inoculum was determined by retrospective plating on LB agar containing appropriate antibiotics.
The murine infection assay was performed twice. In the first set of experiments, five mice were used per strain and two mice were used as uninfected controls. In the second set of experiments, three mice were used per group and again two mice were used as uninfected controls. There was one case of fatality among the mice infected with the wild-type strain.
Mesurement of pathogen burden.
Mice were killed 13 days postinfection, and the distal 8 cm of colon was aseptically removed and weighed, after removal of fecal pellets. Colons were then homogenized mechanically using a Seward 80 stomacher (Seward Medical, London, England), and the numbers of viable bacteria in colonic homogenates were determined by viable count on LB agar containing appropriate antibiotics as described previously (50).
Statistical analysis.
All statistical analysis was carried out using the two-tailed Student's t test.
Histological and immunofluorescence analysis.
Distal colons of noninfected mice and those of mice infected with wild-type, ICC173, or complemented stains were snap-frozen and stored at −70°C until cutting. Cryosections (6 μm thick) were cut, air dried for 1 h, fixed with acetone for 10 min, and processed for histological analysis or immunofluorescence analysis. For histological analysis cryosections were stained with hematoxylin and eosin and analyzed using a Nikon Eclipse E600 microscope.
For immunofluorescence analysis cryosections were blocked using 1% bovine serum albumin followed by the addition of a rabbit universal intimin sera (3) and were incubated for 1 h at room temperature. After extensive washing, cryosections were incubated for 45 min with goat anti-rabbit immunoglobulin G labeled with fluorescein isothiocyanate (Sigma). Host cell F-actin was stained with TRITC (tetramethyl rhodamine isothiocyanate)-conjugated phalloidin (Sigma). Specimens were mounted in Mowiol (Aldrich) and viewed through an inverted Zeiss LSM 510 Meta confocal microscope.
RESULTS
Binding of native secreted EspD to His6-Orf27.
Type III secretion chaperones form a heterogeneous family of proteins that have little or no similarity at the amino acid level but are generally small (14- to 19-kDa), cytoplasmic, acidic proteins with predicted carboxyl-terminal amphipathic α-helix and often adjacent to or in the proximity of the gene coding for the target protein (4, 23, 58). Although no function has been determined for orf27 of the LEE region, its deduced gene product has all the characteristics of a putative type III secretion chaperone (16). Orf27 is encoded within the polycistronic LEE4 operon (16, 40), which encodes the TTSS translocator (EspA, EspD, EspB, and EscF) and effector (EspF) proteins (Fig. 1A). Orf27 has a calculated molecular mass of 15.8 kDa and pI 5.3. Although it does not show primary sequence similarity to any protein currently in the databases, its carboxyl-terminal region contains a putative amphipathic α-helix, proposed to mediate interactions with target proteins (Fig. 1B). Based on these observations, we investigated whether Orf27 had the ability to directly interact with any of the translocator proteins encoded upstream on the LEE4, i.e., EspA, EspD, and EspB.
Preliminary results obtained from microarray-based transcriptional analyses in our laboratory have demonstrated that orf27 is expressed in the early exponential growth phase (M. Batchelor, unpublished data). To examine whether the orf27 gene produces a detectable protein product, the full-length orf27 gene was cloned into pET3-d (pICC232) and expressed in BL21(DE3)pLysS, producing an unmodified recombinant Orf27 peptide. A protein band with an apparent molecular mass of 13 kDa, slightly smaller than that predicted from the amino acid sequence (15.8 kDa), was visualized by SDS-PAGE (data not shown). In addition, the entire Orf27 peptide was expressed as a histidine-tagged N terminus fusion protein (His6-Orf27) in E. coli BL21(DE3)pLysS from plasmid pET28a (pICC234). To investigate whether recombinant Orf27 binds any of the native translocator proteins from EPEC, columns containing bound His6-Orf27 or His6-T7 were overlaid with 25 ml of filtered culture supernatant from E2348/69, grown in DMEM under conditions favorable for LEE gene expression. Following elution of His6-T7 or His6-Orf27 with the imidazole buffer, 1-ml fractions were collected and probed with either His6, EspA, EspB, or EspD antisera by Western blotting. The results showed that EspD was coeluted with His6-Orf27 (Fig. 2). Neither EspA nor EspB bound to the column, as they were not detected within the eluted His6-Orf27 fractions (data not shown). The control, His6-T7 column, did not bind any of the tested proteins from the culture supernatant (Fig. 2). These results showed specific Orf27-EspD protein interaction and suggested that Orf27, like CesD, might display an EspD chaperone activity. Consequently, we renamed Orf27 CesD2.
FIG. 2.
Immunoblot analysis of fractions eluted from the His-CesD2 capture column. (a) Fractions 2, 3, and 4 (lanes 1, 2, and 3, respectively) probed with polyclonal anti-His sera. (c) The same fractions, probed with monoclonal anti-EspD antibody, demonstrating that the secreted form of EspD copurifies with His-CesD2. (b and d) Control, His-T7 column fractions 2, 3, and 4, probed with anti-His polyclonal sera and monoclonal anti-EspD antibody, respectively, showing no copurification.
A nonpolar mutation in EPEC cesD2 affects the level of intracellular and secreted EspD but not A/E lesion formation in vitro.
In order to determine the role of CesD2 during infection in vitro, we disrupted its gene in the prototype EPEC strain E2348/69 and subjected the mutant to a variety of phenotypic characterizations. For the mutagenesis, a 4-kbp DNA fragment containing cesD2 was cloned. Inverse PCR was then performed to generate an in-frame internal deletion (Fig. 1B) in which a kanamycin resistance cassette aphT (21) was inserted (pICC260). The disrupted cesD2 was amplified, and the PCR product was transformed into E2348/69 carrying plasmid pKD46 (encoding the Red recombinase, which enhances the rate of recombination of linear DNA) (9). Allelic exchange was selected, generating a ΔcesD2::aphT EPEC, strain ICC172. The nonpolar nature of the mutation was confirmed by the detection of wild-type levels of EspF, which is encoded downstream cesD2 (Fig. 1; data not shown), in Western blots.
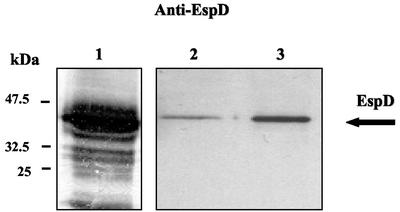
Initially, the effect of the cesD2 mutation on protein secretion was examined. Supernatants from E2348/69 and ICC172 were collected and concentrated. Proteins were separated by SDS-PAGE, electrotransferred, and probed with antisera directed against EspA, EspB, or EspD (Fig. 3). This revealed that mutation in cesD2 resulted in a substantially reduced level of EspD in the culture supernatant, compared to that observed with the wild-type EPEC. A slight reduction in secretion of EspB and no effect on secretion of EspA were observed in the culture supernatant of ICC172 (data not shown). Complementation of the cesD2 mutation with pACYC-HisCesD2 (pICC246) restored secretion of EspD, but not to the wild-type levels (Fig. 3).
FIG. 3.
Immunoblot analysis of culture supernatants showing the EspD secretion profile of EPEC strains. Secretion of EspD is greatly reduced in ICC172 (lane 2) compared with the level of secreted EspD in the supernatant of the wild-type E2348/69 strain (lane 1). Complementation of ICC173 with pICC246 resulted in an increased level of secreted EspD, although not to the wild-type level. Blots were probed with monoclonal EspD antibodies and developed by alkaline phosphates (lane 1) or chemiluminescence (lanes 2 and 3).
In order to determine if the reduced levels of secreted EspD were due to reduced stability of the protein in the absence of CesD2, we determined the levels of EspD within the intracellular compartments of wild-type, ICC172, and complemented strains. We observed that EspD could be detected predominantly in membrane fractions of both E2348/69 and ICC172 (Fig. 4B). However, and similarly to the secretion profiles, lower levels of intracellular EspD were detected in the ICC172 cesD2 mutant strain. Interestingly, complementation of the cesD2 mutation with pICC246 (pACYC-HisCesD2) restored the intracellular levels of EspD to nearly wild-type levels, with EspD localization predominantly in the Sarkosyl-soluble membrane fraction, corresponding to the inner membrane cellular compartment (Fig. 4B).
FIG. 4.
Localization of CesD2 and EspD. (A) Whole ICC172(pICC246) bacterial cells (WC) and culture supernatant (S) were probed with polyclonal anti-His antiserum, demonstrating that CesD2 is intracellular.(B) EPEC strains E2348/69, ICC172, and ICC172(pICC246) were fractionated into C, IM, OM, and P fractions. Samples were analyzed by Western blotting and probed with polyclonal anti-His antiserum (ICC172-pICC246) and monoclonal anti-EspD antibody (strains E2348/69, ICC172, and ICC172-pICC246). His6-CesD2 was detected mostly in the inner membrane, with small amount detected in the cytoplasm. EspD was detected in IM and OM fractions from the three strains. Samples were also probed with anti-β-galactosidase, anti-Etk, anti-intimin, and anti-MBP to verify fraction enrichment. Anti-β-galactosidase, anti-Etk, anti-intimin, and anti-MBP reacted with bands of the size expected for β-galactosidase (116 kDa), Etk (81.2 kDa), intimin (94 kDa), and MBP (43 kDa), respectively. As predicted, β-galactosidase and Etk were detected only in C and IM fractions, respectively. Intimin, which is an outer membrane protein, was also detected in the IM fraction, probably representing the unprocessed form of the molecule. MBP was detected in the periplasmic preparation, while it was also present in some of the C fractions. The upper band that reacted with the MBP antiserum is likely to represent a cross-reactivity with another cytosolic protein. Molecular mass markers are shown at right, and fractionation markers are identified on the left side of the panel.
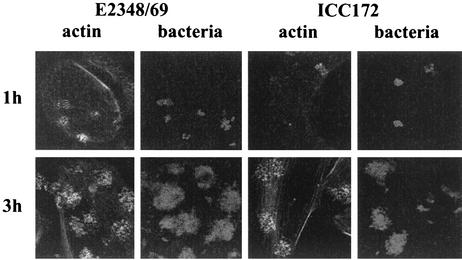
To determine if the mutation in cesD2 affected EPEC virulence properties, the FAS test, which is commonly used as an in vitro measure for A/E lesion formation, was performed. Rearrangement of filamentous actin was examined after 1, 3, and 6 h by staining infected HEp-2 cell monolayers with fluorescein-tagged phalloidin and subsequently using confocal microscopy. After 1 h the wild-type strain adhered to HEp-2 cells in small microcolonies and induced significant actin accumulation, whereas strain ICC172 showed reduced adhesion with little actin accumulation (Fig. 5). By 3 h, however, and after 6 h (data not shown) there was little difference in levels of adhesion and both strains produced discrete foci of actin accumulation beneath adherent bacteria, i.e., a positive FAS reaction (Fig. 5). These results show that despite reduced secretion of EspD, CesD2 is not essential for A/E lesion formation in vitro.
FIG. 5.
FAS tests. After 1 h the wild-type strain adhered to HEp-2 cells in small microcolonies and induced disorganized actin accumulation, whereas strain ICC172 showed reduced adhesion with little actin accumulation. By 3 h however, both strains showed good adhesion and both strains produced discrete foci of actin accumulation beneath adherent bacteria, i.e., produced a positive FAS reaction.
CesD2 is localized to IM and C cellular fractions.
Based on its features and the lack of any obvious signal peptide sequence, as predicted by SignalP V1.1 and iPSORT prediction (http://hypothesiscreator.net/iPSORT/predict.cgi), we hypothesized that CesD2, like other type III secretion chaperones, would be a cytosolic protein. In order to confirm this experimentally, we took advantage of the fact that pICC246 (pACYC-HisCesD2), expressing a tagged CesD2, is biologically active in vivo (Fig. 4 and below). We employed a polyclonal anti-His antiserum to localize His-CesD2 in different EPEC cellular compartments.
Whole-cell lysates and concentrated culture supernatants were first examined to determine whether the His6-CesD2 was either intracellular or secreted to the external milieu. Once it was observed that CesD2 is a bacterial-associated protein (Fig. 4A), cells were fractionated into C, IM, OM, and P fractions. The fractions were assayed for enrichment by using protein markers specific for each of the tested compartments (Fig. 4). β-Galactosidase, the cytoplasmic marker, is an abundant E. coli cytosolic enzyme of 116 kDa (35). Etk, the inner membrane marker, is an 81-kDa protein tyrosine kinase associated with the inner membrane of E. coli (24). Maltose-binding protein (MBP), the periplasmic marker, is a 43-kDa periplasmic E. coli protein involved in the active transport of maltose (27). Intimin, the outer membrane marker, is a 94-kDa EPEC adhesin (26), which is also subjected to N-terminal processing during secretion and, as shown before (24), can also be detected in the inner membrane. The banding pattern of intimin is likely to represent degradation products that are commonly seen on Western blots (2). Equal amounts of subcellular fractions were separated by SDS-PAGE, electrotransferred, and probed with anti-His antiserum. The results demonstrated that CesD2 predominantly localized to the inner membrane with some present in the cytosolic compartment (Fig. 4B). These are the same cellular distributions previously reported for CesD (56). These data are consistent with the intracellular pool of EspD being membrane associated (Fig. 4B).
The effect of cesD2 mutagenesis on virulence in vivo.
EPEC and EHEC have a narrow host specificity range and are restricted human pathogens. Therefore, infection of mice with C. rodentium has been used as a surrogate model to study A/E lesion formation and colonization of A/E-lesion-causing pathogens (18, 22, 43, 47, 48). Importantly, C. rodentium possesses an equivalent LEE region (11), and the A/E lesion induced by C. rodentium is ultrastructurally identical to those formed by EHEC and EPEC in animals and humans (47).
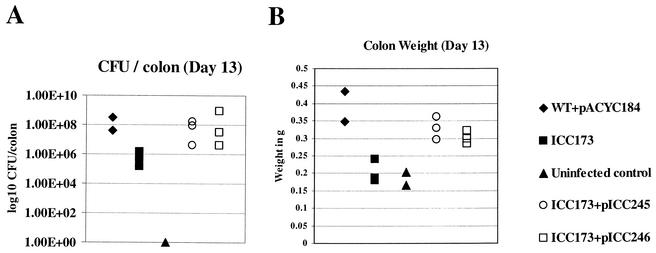
To determine if CesD2 is required for virulence in vivo, an insertional, nonpolar mutation in cesD2 was generated in a wild-type C. rodentium strain. A 1.208-kbp PCR fragment containing cesD2 was cloned into pGEM-T-Easy vector (pICC243), and an aphT cassette (21) was inserted into an EcoRI site within the cloned cesD2 gene (Fig. 1C), resulting in pICC244. The disrupted cesD2 was introduced into the chromosome of wild-type C. rodentium carrying pKD46, by allelic exchange, generating strain ICC173. ICC173 was complemented by either pACYC-CesDCR (pICC245) or pACYC-HisCesDEPEC (pICC246). Wild-type C. rodentium, ICC173 and its complemented derivatives were then used to orally infect mice. Pathogen burden was assessed 13 days postchallenge. This revealed that mice infected with the wild-type strain had ca. 108 CFU/colon (Fig. 6A) and an extensive colonic hyperplasia, as indicated by increased colon weight (Fig. 6B) and microscopic visualization of crypt length in frozen colon sections (Fig. 7). In contrast, mice infected with ICC173 had only ca. 106 CFU/colon (Fig. 6A), and no hyperplasia was observed, either by colon weight, which was similar to that in the uninfected control, or microscopic examination of crypt length (Fig. 6B and 7). Importantly, in ICC173 complemented with either the C. rodentium or EPEC cesD2 genes, the virulence properties were restored to wild-type levels, showing high numbers of challenged bacteria in the colons and hyperplasia (Fig. 6 and 7).
FIG. 6.
Virulence of C. rodentium strains. (A) Data depict the number of C. rodentium CFU recovered from colonic tissues of individual mice, on day 13 postinfection. Mice infected with the wild-type strain had a high pathogen burden. In contrast, mice infected with the cesD2-mutant strain (ICC173) had lower bacterial counts, although the difference was not statistically significant (P > 0.05). In mutant strains complemented with either pICC245 or pICC246, the CFU level was restored to wild-type level. (B) The distal 8 cm of the colons were weighed 13 days postchallenge. The weights of colons from mice infected with the wild-type (WT) strain or complemented mutant strains, carrying either pICC245 or pICC246, were equivalent and significantly greater than those of colons from mice infected with the mutant strain and uninfected controls (P < 0.1). There was no significant difference between the colon weights of uninfected mice and mice infected with ICC173 (P > 0.05).
FIG. 7.
Hematoxylin and eosin-stained colonic frozen sections from C3H/Hej mice experimentally infected with C. rodentium. (A) Colonic sections from an uninfected mouse, showing normal colonic architecture. (B) Crypt hyperplasia and inflammation in a mouse infected with wild-type C. rodentium. (C) Infection with cesD2-minus derivative (ICC173), similarly to the uninfected control, revealed neither hyperplasia nor inflammatory response. (D and E) Complementation of ICC173 with pICC245 (D) or pICC246 (E) restored both hyperplastic and inflammatory responses. Original magnification, ×200.
Challenged bacteria in the frozen colon sections were also stained for intimin using immunofluorescence. Actin staining was used to counterstain the tissue. High numbers of challenged bacteria were observed along the epithelial surface of mice infected with the wild-type (Fig. 8) or complemented (not shown) strains. In contrast, and in agreement with the viable counts, a very low level of challenged bacteria, mainly in the lumen, was observed in mice infected with ICC173 (Fig. 8).
FIG. 8.
Actin and intimin staining of C. rodentium-infected mice. Colonic sections were stained with anti-intimin antiserum to directly label adherent bacteria and counterstained with phalloidin to stain tissue actin. Large numbers of wild-type challenged bacteria can be seen in association with the surface epithelium (A), while only small numbers of ICC173, mainly luminal, were observed (B). (C) No intimin staining was seen in uninfected colon sections. Original magnification, ×200.
DISCUSSION
The hallmark of EPEC infection is the A/E lesion histopathology. A/E lesion formation and intimate bacterial attachment are mediated by close interaction between the outer membrane adhesin intimin and the translocated intimin receptor, Tir, which is believed to be delivered to the host cell plasma membrane via the TTSS needle complex and through EspA filaments and an EspD-associated translocation pore (19). Similar to translocator proteins in other TTSSs, EspD was reported to require a chaperone, CesD, for its proper secretion (56). Here we report that EspD is unique among the TTSS translocator proteins, in that it requires a second chaperone, CesD2, for stabilization and secretion.
TTSS chaperones form a disparate group of proteins with little or no sequence similarity, but which commonly share a predicted α-helical propensity, small size (14 to 19 kDa), acidic nature (pH 4 to 7), and invariably a carboxyl-terminal amphipathic α-helical segment (4, 58). The recognition that not all chaperones have single substrate specificity, but rather are able to bind and are required for the proper secretion of more than one substrate, suggests that the chaperones may not form a single homogeneous family but may rather be segregated into two subfamilies. The first is typified by the Yersinia SycE chaperone (57) that serves a single substrate, YopE. The archetype of the second family, the Yersinia SycD protein, is required for the proper secretion of two proteins, YopB and YopD (44). Interestingly, CesD2 belongs to the SycE family, since it binds only EspD, while CesD belongs to the SycD family as it has chaperone activity for both EspB and EspD (56). Importantly, CesD and CesD2 are unique not only because they chaperone and are required for the stability and secretion of a single translocator protein, EspD, but also because they are located mainly within the bacterial inner membrane.
CesD2 shows all the characteristics of a TTSS chaperone. Indeed, direct and specific EspD-CesD2 protein interaction was demonstrated using the column pull down methods. Interestingly, the phenotypes of the cesD2 mutant (ICC172) are similar to those described previously for the cesD mutant (56). Specifically, mutagenesis of cesD2, like that of CesD (56), substantially affected the stability of EspD and consequently the level of its secretion. However, unlike the cesD mutant, which also affected secretion of EspB but not of EspA (56), mutation in cesD2 had no major effect on secretion of either translocator protein EspA or EspB. However, evaluating the virulence potential of the cesD2 mutant by the FAS test revealed that while after 1 h of infection the wild-type strain adhered to HEp-2 cells in small microcolonies and induced significant actin accumulation, ICC172 showed reduced adhesion with little actin accumulation. Nevertheless, by 3 h there was little difference in the levels of adhesion, and both strains produced a positive FAS reaction. Of note is the fact that the cesD mutant also required a longer incubation time to produce a typically positive FAS test (56). These results suggest that either chaperone enables EPEC to maintain and secrete sufficient amounts of EspD for interaction with epithelial cells in vitro.
The contribution of CesD2 for pathogenesis in vivo was assessed using the C. rodentium mouse model. This revealed that in vivo CesD, on its own, is insufficient for colonization and disease as, in comparison with the wild-type strain, 100-fold fewer challenged bacteria were recovered following oral infection with ICC173, with no sign of colonic hyperplasia. Interestingly, the fact that a mutation of cesD2 caused significant reduction virulence in vivo without affecting FAS and hence A/E lesion formation in vitro suggests that actin polymerization and pedestal formation might not be directly associated with colonization and virulence. The trans complementation of cesD2 mutation with either EPEC or C. rodentium cesD2 restored the virulence potential to wild-type levels. These results demonstrate that CesD2 plays an important role during infection in vivo.
Like EspD, YopN, a secreted protein in Y. pestis, requires two “cochaperones” for its secretion, SycN and YscB (10). SycN and YscB form a complex, a heterodimer, that functions as a specific chaperone for YopN. The present study is the first report of a TTSS translocator protein (EspD) having two chaperones (CesD and CesD2).
The precise role of chaperones has not been determined. They have been described as antiassociation factors or “bodyguards,” whereby chaperone binding to interactive surfaces prevents premature and nonproductive homo- or hetero-oligomeric cytosolic substrate interactions, which would otherwise target the substrate for degradation (4, 20, 44, 60). Chaperones were also implicated in the hierarchical release of effector proteins from the bacterium organizing trafficking through the secretion apparatus (5) and in control of virulence gene expression, by establishing a regulatory hierarchy of protein synthesis during infection (17, 59).
EspD, with its two putative transmembrane domains (55), is completely insoluble (7). Indeed, large EspD aggregates were seen in EPEC supernatants (7). Moreover, as EspD is essential for EspA filament biosynthesis (possibly by having a cap-like activity), it is likely to be the first protein to be secreted once the TTSS needle complex had been assembled. The reason why EspD requires two chaperones is not clear. However, this might be needed to keep EspD in a soluble state, to prevent EspD-EspD (7) and possible EspD-EspA protein interactions from occurring prematurely, while giving EspD an advantage in secretion over other translocator and effector proteins. We are currently testing these hypotheses experimentally.
Acknowledgments
This work was supported by the Wellcome Trust and the BBSRC. B.C.N. was supported by a fellowship from CNPy Brazil.
Editor: J. T. Barbieri
REFERENCES
- 1.Abe, A., M. de Grado, R. A. Pfuetzner, C. Sanchez-Sanmartin, R. Devinney, J. L. Puente, N. C. Strynadka, and B. B. Finlay. 1999. Enteropathogenic Escherichia coli translocated intimin receptor, Tir, requires a specific chaperone for stable secretion. Mol. Microbiol. 33:1162-1175. [DOI] [PubMed] [Google Scholar]
- 2.Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncaleves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimin α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor, M., S. Knutton, V. Huter, M. Zanial, G. Dougan, and G. Frankel. 1999. Development of a universal intimin antiserum and PCR primers. J. Clin. Microbiol. 37:3822-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, J. C., and C. Hughes. 2000. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 8:202-204. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, A. P., I. Lambermont, and G. R. Cornelis. 2000. Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182:4811-4821. [DOI] [PMC free article] [PubMed]
- 6.Creasey, E. A., R. M. Delahay, A. A. Bishop, R. K. Shaw, B. Kenny, S. Knutton, and G. Frankel. 2002. CesT is a bivalent enteropathogenic Escherichia coli chaperone required for translocation of both Tir and Map. Mol. Microbiol. 47:209-221. [DOI] [PubMed] [Google Scholar]
- 7.Daniell, S. J., R. M. Delahay, R. K. Shaw, E. L. Hartland, M. J. Pallen, F. Booy, F. Ebel, S. Knutton, and G. Frankel. 2001. The coiled coil domain of enteropathogenic Escherichia coli type III secreted protein EspD is involved in EspA filament-mediated cell attachment and hemolysis. Infect. Immun. 69:4055-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniell, S. J., N. Takahashi, R. Wilson, D. Friedberg, I. Rosenshine, F. P. Booy, R. K. Shaw, S. Knutton, G. Frankel, and S. Aizawa. 2001. The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell. Microbiol. 3:865-871. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day, J. B., and G. V. Plano. 1998. A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis. Mol. Microbiol. 30:777-788. [DOI] [PubMed] [Google Scholar]
- 11.Deng. W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg, M. S., J. Yu, and J. B. Kaper. 1993. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J. Bacteriol. 175:4670-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, S. J., M. S. Dubois, S. W. Hutcheson, L. A. Wainwright, M. Batchelor, G. Frankel, S. S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in Enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176-1189. [DOI] [PubMed] [Google Scholar]
- 14.Elliott, S. J., E. O. Krejany, J. L. Mellies, R. M. Robins-Browne, C. Sasakawa, and J. B. Kaper. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott, S. J., C. B. O'Connell, A. Koutsouris, C. Brinkley, M. S. Donnenberg, G. Hecht, and J. B. Kaper. 2002. A gene from the locus of enterocyte effacement that is required for enteropathogenic Escherichia coli to increase tight-junction permeability encodes a chaperone for EspF. Infect. Immun. 70:2271-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol 28:1-4. [DOI] [PubMed] [Google Scholar]
- 17.Francis, M. S., S. A. Lloyd, and H. Wolf-Watz. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42:1075-1093. [DOI] [PubMed] [Google Scholar]
- 18.Frankel, G., A. D. Phillips, M. Novakova, H. Field, D. C. Candy, D. B. Schauer, G. Douce, and G. Dougan. 1996. Intimin from enteropathogenic Escherichia coli restores murine virulence to a Citrobacter rodentium eaeA mutant: induction of an immunoglobulin A response to intimin and EspB. Infect. Immun. 64:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 20.Frithz-Lindsten, E., R. Rosqvist, L. Johansson, and A. Forsberg. 1995. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensable for targeting to the secretion loci. Mol. Microbiol. 16:635-647. [DOI] [PubMed] [Google Scholar]
- 21.Galan, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 174:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghaem-Maghami, M., C. P. Simmons, S. Daniell, M. Pizza, D. Lewis, G. Frankel, and G. Dougan. 2001. Intimin-specific immune responses prevent bacterial colonization by the attaching-effacing pathogen Citrobacter rodentium. Infect. Immun. 69:5597-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilan, O., Y. Bloch, G. Frankel, H. Ullrich, K. Geider, and I. Rosenshine. 1999. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J. 18:3241-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellermann, O., and S. Szmelcman. 1974. Active transport of maltose in Escherichia coli K12. Involvement of a “periplasmic” maltose binding protein. Eur. J. Biochem. 47:139-149. [DOI] [PubMed] [Google Scholar]
- 28.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 29.Kenny, B., and M. Jepson. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell. Microbiol. 2:579-590. [DOI] [PubMed] [Google Scholar]
- 30.Kenny, B., L. C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313-323. [DOI] [PubMed] [Google Scholar]
- 31.Knutton, S., D. R. Lloyd, and A. S. McNeish. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect. Immun. 55:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 34.Lai, L. C., L. A. Wainwright, K. D. Stone, and M. S. Donnenberg. 1997. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect. Immun. 65:2211-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, C., P. Li, H. Inouye, E. R. Brickman, and J. Beckwith. 1989. Genetic studies on the inability of β-galactosidase to be translocated across the Escherichia coli cytoplasmic membrane. J. Bacteriol. 171:4609-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine, M. M., E. J. Berquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, S. Stoman, and B. Rowe. 1978. Escherichia coli that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:119-122. [DOI] [PubMed]
- 37.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNamara, B. P., and M. S. Donnenberg. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71-78. [DOI] [PubMed] [Google Scholar]
- 39.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. P. Nougayrede, M. S. Donnenberg, and G. Hecht. 2000. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed]
- 41.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman, J. V., B. A. Zabel, S. S. Jha, and D. B. Schauer. 1999. Citrobacter rodentium espB is necessary for signal transduction and for infection of laboratory mice. Infect. Immun. 67:6019-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neyt, C., and G. R. Cornelis. 1999. Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol. Microbiol. 31:143-156. [DOI] [PubMed] [Google Scholar]
- 45.Robins-Browne, R. M., A. M. Tokhi, L. M. Adams, V. Bennett-Wood, A. V. Moisidis, E. O. Krejany, and L. E. O'Gorman. 1994. Adherence characteristics of attaching and effacing strains of Escherichia coli from rabbits. Infect. Immun. 62:1584-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenshine, I., S. Ruschkowski, M. Stein, D. J. Reinscheid, S. D. Mills, and B. B. Finlay. 1996. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 15:2613-2624. [PMC free article] [PubMed] [Google Scholar]
- 47.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype 4280 that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schauer, D. B., and S. Falkow. 1993. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect. Immun. 61:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sekiya, K., M. Ohishi, T. Ogino, K. Tamano, C. Sasakawa, and A. Abe. 2001. Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. USA 98:11638-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simmons, C. P., N. S. Goncalves, M. Ghaem-Maghami, M. Bajaj-Elliott, S. Clare, B. Neves, G. Frankel, G. Dougan, and T. T. MacDonald. 2002. Impaired resistance and enhanced pathology during infection with a noninvasive, attaching-effacing enteric bacterial pathogen. Citrobacter rodentium, in mice lacking IL-12 or IFN-gamma. J. Immunol. 15:1804-1812. [DOI] [PubMed] [Google Scholar]
- 51.Tamano, K., S. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. 19 EMBO J. 3876-3887. [DOI] [PMC free article] [PubMed]
- 52.Taylor, K. A., P. W. Luther, and M. S. Donnenberg. 1999. Expression of the EspB protein of enteropathogenic Escherichia coli within HeLa cells affects stress fibers and cellular morphology. Infect. Immun. 67:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzipori, S., I. K. Wachsmuth, C. Chapman, R. Birden, J. Brittingham, C. Jackson, and J. Hogg. 1986. The pathogenesis of hemorrhagic colitis caused by Escherichia coli O157:H7 in gnotobiotic piglets. J. Infect. Dis. 154:712-714. [DOI] [PubMed] [Google Scholar]
- 55.Wachter, C., C. Beinke, M. Mattes, and M. A. Schmidt. 1999. Insertion of EspD into epithelial target cell membranes by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 31:1695-1707. [DOI] [PubMed] [Google Scholar]
- 56.Wainwright, L. A., and J. B. Kaper. 1998. EspB and EspD require a specific chaperone for proper secretion from enteropathogenic Escherichia coli. Mol. Microbiol. 27:1247-1260. [DOI] [PubMed] [Google Scholar]
- 57.Wattiau, P., and G. R. Cornelis. 1993. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol. Microbiol. 8:123-131. [DOI] [PubMed] [Google Scholar]
- 58.Wattiau, P., S. Woestyn, and G. R. Cornelis. 1996. Customized secretion chaperones in pathogenic bacteria. Mol. Microbiol. 20:255-262. [DOI] [PubMed] [Google Scholar]
- 59.Williams, A. W., and S. C. Straley. 1998. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J. Bacteriol. 180:350-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woestyn, S., M. P. Sory, A. Boland, O. Lequenne, and G. R. Cornelis. 1996. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol. Microbiol. 20:1261-1271. [DOI] [PubMed] [Google Scholar]
- 61.Wolff, C., I. Nisan, E. Hanski, G. Frankel, and I. Rosenshine. 1998. Protein translocation into HeLa cells by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 28:143-155. [DOI] [PubMed] [Google Scholar]