Abstract
The ability of E. coli strains to colonize the mouse large intestine has been correlated with their ability to grow in cecal and colonic mucus. In the present study, an E. coli MG1655 strain was mutagenized with a mini-Tn5 Km (kanamycin) transposon, and mutants were tested for the ability to grow on agar plates with mouse cecal mucus as the sole source of carbon and nitrogen. One mutant, designated MD42 (for mucus defective), grew poorly on cecal-mucus agar plates but grew well on Luria agar plates and on glucose minimal-agar plates. Sequencing revealed that the insertion in MD42 was in the waaQ gene, which is involved in lipopolysaccharide (LPS) core biosynthesis. Like “deep-rough” E. coli mutants, MD42 was hypersensitive to sodium dodecyl sulfate (SDS), bile salts, and the hydrophobic antibiotic novobiocin. Furthermore, its LPS core oligosaccharide was truncated, like that of a deep-rough mutant. MD42 initially grew in the large intestines of streptomycin-treated mice but then failed to colonize (<102 CFU per g of feces), whereas its parent colonized at levels between 107 and 108 CFU per g of feces. When mouse cecal mucosal sections were hybridized with an E. coli-specific rRNA probe, MD42 was observed in cecal mucus as clumps 24 h postfeeding, whereas its parent was present almost exclusively as single cells, suggesting that clumping may play a role in preventing MD42 colonization. Surprisingly, MD42 grew nearly as well as its parent during growth in undiluted, highly viscous cecal mucus isolated directly from the mouse cecum and, like its parent, survived well after reaching stationary phase, suggesting that there are no antimicrobials in mucus that prevent MD42 colonization. After mini-mariner transposon mutagenesis, an SDS-resistant suppressor mutant of MD42 was isolated. The mini-mariner insertion was shown to be in the bipA gene, a known regulator of E. coli surface components. When grown in Luria broth, the LPS core of the suppressor mutant remained truncated; however, the LPS core was not truncated when the suppressor mutant was grown in the presence of SDS. Moreover, when the suppressor mutant was grown in the presence of SDS and fed to mice, it colonized the mouse large intestine. Collectively, the data presented here suggest that BipA may play a role in E. coli MG1655 LPS core biosynthesis and that because MD42 forms clumps in intestinal mucus, it is unable to colonize the mouse large intestine.
The large intestine of the mouse consists of the cecum and the colon, each of which contains the mucosa and the luminal contents. Two components of the mucosa are the layer of epithelial cells on the intestinal wall and the mucus layer, which covers them. The relatively thick (up to 400 μm) mucus layer consists of mucin, a 2-MDa gel-forming glycoprotein, and a large number of smaller glycoproteins, proteins, glycolipids, lipids, and sugars (1, 12, 18, 36, 39, 43, 45). Presumably, shed epithelial cells are a source of many of the smaller mucus components (36, 39). The mucus layer itself is in a dynamic state, constantly being synthesized and secreted by specialized goblet cells and degraded to a large extent by the indigenous intestinal microbes (17, 32). Degraded mucus components are shed into the intestinal lumen, forming a part of the luminal contents which are excreted in feces (17).
The prevalent theory as to how bacteria colonize the mammalian gut is that the ∼500 indigenous species (31) can coexist as long as each member of the flora is able to utilize one or a few limiting nutrients better than all the others and its rate of growth during the colonization process is at least equal to the washout rate from the intestine (13, 14). The rate of growth of a particular bacterium in the intestine is presumably determined by the nature of the limiting nutrient it utilizes, and the density to which it grows is determined by the available concentration of that nutrient. It is also possible for a species that does not compete well for limiting nutrients to avoid washout and to colonize if it is able to adhere to the intestinal wall (14).
In recent studies, the location of a commensal strain, Escherichia coli BJ4, was examined in the mouse large intestine using in situ hybridization with species-specific rRNA probes (37). E. coli BJ4 was found dispersed in intestinal mucus and in luminal contents but was not found associated with epithelial cells (21, 37). While E. coli is present in both mucus and intestinal contents, a large body of experimental evidence shows that it grows rapidly in intestinal mucus both in vitro and in vivo but does not grow or grows poorly in luminal contents (23, 33, 45, 46, 49). Furthermore, E. coli leuX mutants have difficulty surviving in intestinal mucus and eda mutants have difficulty growing in intestinal mucus in vitro, and both are unable to colonize the mouse large intestine (33, 46). It is therefore highly likely that the ability of an E. coli strain to grow and survive in intestinal mucus plays a critical role in its ability to colonize the intestine. With this in mind, genes critical to the ability of E. coli to colonize the mouse large intestine were identified by screening mutants unable to grow well on agar plates containing mouse cecal mucus as the sole source of carbon and nitrogen. These mutants were tested for the ability to colonize the mouse large intestine. Surprisingly, the first mutant we isolated was a lipopolysaccharide (LPS) core mutant. In the present study, we characterize that mutant with respect to its ability to grow and survive in cecal mucus and its ability to colonize the mouse large intestine.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| MG1655 | Wild type (CGSC no. 7740) | E. coli Genetic Stock Culture Collection; Yale University |
| MG1655 Strr | Spontaneous streptomycin-resistant mutant of MG1655 | This study |
| MG1655 Strr Nalr | Spontaneous nalidixic acid-resistant mutant of MG1655 Strr | This study |
| MD42 Strr Nalr | Mini-Tn5 Km::waaQ mutant of MG1655 Strr Nalr | This study |
| MD42 | P1 mini-Tn5 Km::waaQ transductant of MD42 Strr | This study |
| MD42mm5 | Mini-mariner Gm::bipA mutant of MD42 | This study |
| SM10 λ pir (pFAC) | Contains mini-mariner Gm in the pFAC suicide vector | 50 |
| ATM161 | Contains mini-Tn5 Km in the pUT suicide vector | 10 |
| F470 | Rough LPS derivative of an O8:K27 strain | 51 |
| CWG296 | waaP::aacC1 mutant of F470 | 51 |
| CWG297 | waaQ::aacC1 mutant of F470 | 51 |
| CWG303 | waaG::aacC1 mutant of F470 | 52 |
| Plasmids | ||
| pLD55 | Suicide vector for allelic exchange; bla tetAR | 28 |
| pBR322 | bla tet | 4 |
Bacterial growth media.
Luria broth was made as described by Revel (41). Luria agar is Luria broth containing 12 g of Bacto Agar (Difco Laboratories, Detroit, Mich.) per liter. MacConkey agar (Difco) was prepared according to the package instructions. Glucose minimal M9 agar plates (Difco Agar Noble; 15 g/liter) were made with M9 minimal medium (30) supplemented with glucose (0.2% [wt/vol]). Cecal-mucus agar plates (Difco Agar Noble; 15 g/liter) were made using UV light-sterilized mouse cecal mucus (34) at a concentration of 2 mg/ml with respect to protein in HEPES-Hanks buffer (pH 7.4).
Insertional mutagenesis.
E. coli MG1655 Strr Nalr mini-Tn5 Km (kanamycin) mutants were constructed by insertional mutagenesis. Briefly, E. coli ATM161 carrying the suicide vector pUT, which contains the mini-Tn5 Km transposon (10), was conjugated with the recipient E. coli MG1655 Strr Nalr in the following manner. The donor and recipient strains were grown overnight, with shaking, in Luria broth at 30°C. Aliquots of 100 liters of each culture were mixed together in 5 ml of 10 mM MgSO4 and filtered through a Millipore 0.45-μm-pore-size membrane filter. The filter was placed on the surface of a Luria agar plate and incubated for 5 h at 37°C. Following incubation, the bacteria on the filter were suspended in 5 ml of 10 mM MgSO4, and 100-liter aliquots of the suspension were plated on Luria agar (Difco) containing streptomycin sulfate (100 g/ml) and kanamycin sulfate (40 g/ml) and incubated at 37°C overnight. The colonies were then screened for the ability to grow on glucose (0.2% [wt/vol]) minimal M9 agar plates containing streptomycin sulfate (100 g/ml) and kanamycin sulfate (40 g/ml) by toothpicking them and incubating them overnight at 37°C. Mutants that grew on Luria agar and glucose minimal M9 agar were toothpicked to cecal-mucus agar plates. The cecal-mucus agar plates were incubated at 37°C for 18 h.
Isolation of suppressor mutants.
E. coli MD42 recipients, sensitive to <0.1 mg of sodium dodecyl sulfate (SDS)/ml, were mated with E. coli SM10 λ pir (pFAC) as described above. pFAC houses the Himar1-based mini-mariner transposon containing the aacC1 gentamicin resistance gene (50). Five hours after mating, mutants (≈106) were plated on Luria agar plates containing streptomycin (100 μg/ml), kanamycin sulfate (40 μg/ml), gentamicin (15 μg/ml), and SDS (1 mg/ml). Following incubation at 37°C overnight, five MD42 SDS-resistant colonies were isolated. Further characterization of the isolates is presented in Results.
Bacteriophage P1 transduction.
Bacteriophage P1 transduction and curing of transductants of bacteriophage P1 were performed as previously described (24). The transductants were constructed by infecting E. coli MG1655 Strr with the P1 lysate from an induced Luria broth culture of lysogenized E. coli MD42 Strr Nalr and selection on Luria agar containing streptomycin sulfate (100 g/ml) and kanamycin sulfate (40 g/ml).
SDS, novobiocin, and bile salts sensitivity testing.
SDS, novobiocin, and bile salts sensitivity testing were performed essentially as described by Yethon et al. (51). Briefly, twofold serial dilutions were made of SDS (200 to 0.1 mg/ml), novobiocin (200 to 1.6 μg/ml), and Bacto Bile Salts no. 3 (Difco) (6.0 to 0.18 mg/ml). Each series of tubes was inoculated with ∼105 CFU/ml from an overnight culture of the strain to be tested and then incubated standing at 37°C. Growth was scored as positive if, after 8 h of incubation, the absorbancy at 600 nm (A600) was >0.2.
In vitro growth in mouse cecal mucus.
Mouse cecal mucus was isolated as previously described (8). Briefly, 18 mice (35 to 42 days old) were fed Charles River Valley Rat, Mouse, and Hamster Formula for 5 days. The drinking water was then replaced with sterile distilled water containing streptomycin sulfate (5 g/liter). Twenty-four hours later, the mice were sacrificed by CO2 asphyxiation and their ceca were removed. The cecal contents were collected for use in growth experiments (see below), and any remaining cecal contents were washed out of each cecum with sterile HEPES-Hanks buffer (pH 7.4). Viscous cecal mucus was prepared from the washed ceca by scraping them with a rubber spatula as described previously (8). The cecal mucus obtained from the mice was pooled and mixed to obtain a homogeneous, highly viscous sample, which is undiluted cecal mucus.
The ceca from each set of 18 mice yielded ∼2 ml of undiluted cecal mucus. One-milliliter aliquots of undiluted cecal mucus or undiluted cecal contents were separately inoculated with 105 CFU of either E. coli MG1655 Strr or E. coli MD42. E. coli strains used for the inocula were grown in Luria broth (37°C; shaking; 18 h), centrifuged at 8,000 × g, washed three times in sterile HEPES-Hanks buffer (pH 7.4), and diluted in HEPES-Hanks buffer (pH 7.4). Cecal-mucus samples were incubated standing at 37°C. Samples were withdrawn at the times indicated in the figures, diluted in HEPES-Hanks buffer (pH 7.4), and plated on Luria agar plates containing either streptomycin sulfate (100 μg/ml) for MG1655 Strr or streptomycin sulfate (100 μg/ml) and kanamycin sulfate (40 μg/ml) for MD42. The plates were incubated at 37°C for 18 to 24 h prior to bacteria being counted. Growth experiments utilizing cecal contents were performed identically to those utilizing cecal mucus. Cecal mucus was also scraped into HEPES-Hanks buffer (pH 7.4) and centrifuged and sterilized by UV irradiation as described previously (8, 33) and was adjusted to a concentration of 2 mg/ml with respect to protein using sterile HEPES-Hanks buffer (pH 7.4) as described previously (33). The cecal mucus isolated from five mice yielded ∼30 mg of protein. The growth of E. coli MG1655 Strr and E. coli MD42 in cecal mucus (2 mg/ml with respect to protein) was determined as described above for undiluted cecal mucus. Alternatively, 1.0-ml aliquots were read at 600 nm in a Pharmacia Biotech (Ultrospec 2000) UV-visible-light spectrophotometer.
Mouse colonization experiments.
The method used to compare the large-intestine-colonizing abilities of E. coli strains in mice has been described previously (24, 26, 33, 45, 46, 49). Briefly, three male CD-1 mice (5 to 8 weeks old) were given drinking water containing streptomycin sulfate (5 g/liter) for 24 h to eliminate resident facultative bacteria (28). Following 18 h of starvation, the mice were fed 1.0 ml of 20% (wt/vol) sucrose containing either 5 × 105, 1 × 108, or 1 × 1010 CFU of Luria broth-grown E. coli MG1655 strains, depending on the experiment. After the bacterial suspension was ingested, both the food (Charles River Valley Rat, Mouse, and Hamster Formula) and streptomycin-water were returned to the mice, and 1 g of feces was collected after 5 and 24 h and, on odd-numbered days, at the times indicated in the figures. The mice were housed individually in cages without bedding and were placed in clean cages daily. One-gram fecal samples (no older than 24 h) were homogenized in 1% Bacto Tryptone (10 ml) and diluted in the same medium, and 100-μl samples were plated on Luria agar plates containing streptomycin sulfate (100 μg/ml) and kanamycin sulfate (40 μg/ml) for determining numbers of MD42, streptomycin sulfate (100 μg/ml) for determining numbers of MG1655 Strr, kanamycin sulfate (40 μg/ml) and gentamicin (15 μg/ml) for determining numbers of MD42 mm5 in feces of mice fed Luria broth-grown MD42 mm5, and kanamycin sulfate (40 μg/ml) and SDS (1.0 mg/ml) for determining numbers of MD42 mm5 in feces of mice fed MD42 mm5 grown in Luria broth containing SDS (1 mg/ml). All plates were incubated for 18 to 24 h at 37°C prior to counting of bacteria. Each colonization experiment was repeated at least once to confirm the initial colonization results. The log10 mean number of CFU per gram of feces and the standard error of the log10 mean number of CFU per gram of feces for each set of three mice was calculated for each time point. The limit of detection of CFU per gram of feces is 102. In one experiment, when the concentration of an E. coli strain in mouse feces was below the detection limit on day 7, three mice were sacrificed and their cecal mucus was isolated as described above. The mucus from the cecum of each mouse (∼100 μl) was plated onto Luria agar plates containing the appropriate antibiotics to determine if the E. coli strain could be detected in mouse cecal mucus.
DNA procedures and sequencing.
The gene interrupted in MD42 was determined by sequencing out in one direction from the mini-Tn5 Km insertion. The gene interrupted by the mini-mariner transposon in MD42 mm5 was determined by sequencing out in both directions from the mini-mariner transposon. E. coli MD42 and MD42 mm5 chromosomal DNAs were purified using Qiagen Genomic tip-100. The 40-μl sequencing reaction mixture contained 2 μg of chromosomal DNA, 13 pmol of primer (30-mer), and 16 μl of BigdyeTM Terminator Cycle Sequencing Ready Reaction mix. The PCR sequencing program consisted of 5 min at 95°C; 10 cycles of 30 s at 95°C and 4 min 20 s at 65°C with a 0.5°C decrease for each subsequent cycle; 10 cycles of 30 s at 95°C, 20 s at 60°C with a 0.5°C decrease for each subsequent cycle, and 4 min 20 s at 60°C; 80 cycles of 30 s at 95°C, 20 s at 55°C, and 4 min 20 s at 60°C; and cool down to 4°C.
Reaction products were purified three times over a Centriflex gel filtration cartridge (Edge BioSystems). Samples were analyzed on the ABI 310 genetic analyzer (Perkin-Elmer, Foster City, Cal.) by injection for 100 s. Up to 500 bases could be read from a single run.
Sequence analysis was performed and the results were collected using ABI Prism 310 collection software version 1.04 (Perkin-Elmer) and analyzed with Sequence Analysis version 3.0 (Perkin-Elmer). Database searches were done using BLAST (http://www.ncbi.nlm.nih.gov/).
The primers used to amplify part of the the pir gene (602 bp) were as follows: forward, 5′-ATG AGA CTC AAG GTC ATG ATG G-3′, and reverse, 5′-CTC TTC CTT TAA CTC ATC AAC GG-3′. The primers upstream and downstream of waaQ, used to amplify a 4,000-bp fragment containing the mini-Tn5 Km transposon in waaQ and flanking sequences, were as follows: forward, 5′-CTA GCT GCA GGC TGA CTT ATG GAT GTG CTG GG-3′, and reverse, 5′-CTA GCT GCA GCC CAC GAC TGT GTA TAT ACC CG-3′.
Allelic exchange.
The bipA::mini-mariner transposon was inserted into a fresh MD42 background by a modification of the system described by Datsenko and Wanner (9). The forward primer, including a 5′ BamHI site, was 5′-CTA GGG ATC CCA TCG TCA GTA CGT GTT CAG CG-3′, and the reverse primer, including a 5′ BamHI site, was 5′-CTA GGG ATC CAT GAC TTT GCA GAT AAT TGT GAC-3′. The primers were used to PCR amplify the bipA::mini-mariner insertion flanked by a 399-bp region upstream of the bipA ATG start codon and a 188-base region downstream of the bipA TGA stop codon. The PCR product was used to transfect MD42 (pKD46) as described by Datsenko and Wanner (9), and recombinants were selected at 37°C on Luria agar plates containing kanamycin sulfate (40 μg/ml) and gentamicin (15 μg/ml). The recombinants were tested for loss of pKD46 by growth at 37°C and testing for loss of ampicillin resistance as described by Datsenko and Wanner (9). Recombinants were confirmed by PCR to have the bipA::mini-mariner transposon inserted correctly in the chromosome.
Isolation of LPS and gel electrophoresis.
Luria broth-grown cells (3 ml) were centrifuged, resuspended in 1.0 ml of phosphate-buffered saline, vortexed, and then incubated at 60°C for 30 min. The suspension was centrifuged at 11,750 × g for 30 min. One hundred microliters of the supernatant was added to 100 μl of Tricine sample buffer (Bio-Rad Laboratories, Hercules, Calif.) containing 2-β-mercaptoethanol (2%), and the mixture was boiled for 10 min. Fifty microliters of the boiled sample was added to 10 μl of a protease K solution (2.5 mg of protease K in 1.0 ml of sample buffer), and the sample was then incubated at 60°C for 60 min and centrifuged at 16,000 × g for 30 min. LPS in supernatants from samples prepared in this manner was separated on 18% Tricine sodium SDS-polyacrylamide gel electrophoresis (PAGE) gels as described by Pradel and Schnaitman (38) and visualized by silver staining (48).
Motility.
Motility assays were performed by inoculating 10 μl of overnight Luria broth cultures on the tops of 10-cm Luria soft-agar tubes (3.5 g of Bacto Agar/liter). Migration toward the bottoms of the tubes was recorded after 24 h of incubation at 37°C. In this assay, nonmotile mutants do not migrate.
Serotyping.
Serotyping (O and H) was performed using specific antisera produced by the World Health Organization International Escherichia and Klebsiella Centre, Statens Seruminstitut, Copenhagen, Denmark.
Assays for type 1 fimbriae.
Type 1 fimbriation was assayed by slide agglutination using fresh guinea pig erythrocytes (5% suspension) in the presence and absence of mannose (100 mM). Strains that have type 1 fimbriae agglutinate guinea pig erythrocytes in the absence of mannose but not in its presence. Type 1 fimbriation was confirmed by agglutination with specific antibodies against type 1 fimbriae (20).
Preparation of histological sections for hybridization.
After the mice were sacrificed, the cecum was removed from each mouse and was cut in two halves. One half was immediately transferred to a phosphate-buffered 4% (vol/vol) formalin solution. The other half was used for determination of the number of CFU per gram of cecum. Each cecum was dehydrated and embedded in paraffin prior to preparation of 5-μm-thick cross sections. The sections were placed on glass microscope slides. Prior to hybridization, the sections were deparaffinated by treatment (three times; 10 min each) with xylene (Bie & Berntsen, Rødovre, Denmark) and dehydrated for 10 min in 96% ethanol. Before the hybridization solution was applied, the intestinal sections were circumscribed with a hydrophobic PAP-pen (Daido Sangyo Co. Ltd., Tokyo, Japan).
Oligonucleotide probes.
A probe specific to E. coli 23S rRNA (EC1531; 5′-CAC CGT AGT GCC TCG TCA TCA-3′) was used (37). The probe was labeled at the 5′ end with CY3 red fluorescent dye (cyanine dye CY3.29-OSu; Biological Detection Systems, Pittsburg, Pa.). In addition, probe EU338 (44), which is specific to the eubacterial 23S rRNA domain, was used. This probe (5′-GCT GCC TCC CGT AGG AGT-3′) was labeled at the 5′ end with the green fluorescent compound fluorescein (Peninsula Laboratories, Inc., Belmont, Calif.).
Hybridization and microscopy.
For visualization of bacteria in the intestinal sections, the eubacterial probe labeled with fluorescein was used in combination with the CY3-labeled E. coli-specific probe. Hybridization was carried out as described by Poulsen et al. (37) with the following modifications. The hybridization solution contained 10% formamide and 2.5 ng of each probe and was at pH 7.2. Washing solution I contained 10% formamide and was at pH 7.2. Washing solution II was at pH 7.2. Following hybridization, the sections were viewed by confocal microscopy.
RESULTS
Isolation and biochemical characterization of an MG1655 Strr Nalr mutant that grows poorly on cecal-mucus agar plates.
Three hundred mini-Tn5 Km mutants that grew on both Luria agar plates and glucose M9 plates were toothpicked from Luria agar to cecal-mucus agar plates (2 mg/ml with respect to protein) containing streptomycin and kanamycin. One mutant, designated MD42 Strr Nalr (for mucus defective), grew poorly on the cecal-mucus agar plates, i.e., the colony diameter was less than half that of the other mutants and growth was relatively sparse. MD42 Strr Nalr was also ampicillin sensitive, ruling out the possibility that pUT, the suicide vector containing the mini-Tn5 Km transposon, had integrated into the chromosome. Moreover, the pir gene did not amplify from MD42 Strr Nalr DNA using pir-specific primers (see Materials and Methods) but did amplify from DNA isolated from E. coli ATM161, the mini-Tn5 Km donor strain. In addition, MD42 Strr Nalr was transformable with pBR322 but was not transformable with the pir-dependent suicide vector pLD55 (28). These results effectively rule out the possibility that MD42 Strr Nalr became lysogenized with bacteriophage λ pir during mating, a reported possible pitfall (10). To be sure that the growth defect of MD42 Strr Nalr on cecal-mucus agar plates was due to the mini-Tn5 Km insertion, kanamycin resistance was transduced from MD42 into MG1655 Strr. The transductants had the same defective growth characteristics as MD42 Strr Nalr on cecal-mucus agar plates, which proves that the mini-Tn5 Km insertion in MD42 Strr Nalr was indeed responsible for the growth defect. E. coli MG1655 Strr and one of the MG1655 Strr transductants, which for simplicity will hereafter be referred to as MD42, were chosen for further study.
In addition to growing poorly on cecal-mucus agar plates, MD42 grew poorly on MacConkey agar plates, suggesting that it might be sensitive to bile salts. E. coli MG1655 Strr and MD42 behaved identically in 36 of the 41 biochemical tests listed in Table 5.3 of the 1984 edition of Bergey's Manual of Systematic Bacteriology (6) (the tests not performed were for production of lipase, DNase, and phenylalanine deaminase and for utilization of α-methyl-d-glucoside and d-arabitol). In addition, MG1655 Strr and MD42 were tested for motility and production of type 1 fimbriae. MG1655 Strr and MD42 were found to be identical with respect to all biochemical characteristics. The serotype of both strains was OR:H48, i.e., both were rough and both contained the H48 flagellar antigen, typical of K-12 strains. Furthermore, the strains contained approximately equal amounts of type 1 fimbriae. MD42 was motile, but less so than MG1655 Strr, i.e., MG1655 Strr traveled 6.5 ± 0.29 cm (n = 3) down Luria soft-agar tubes in 24 h (see Materials and Methods), whereas MD42 traveled only 3.8 ± 0.03 cm (n = 3) in the same time.
The gene interrupted in MD42 is waaQ.
DNA sequencing (449 bp from the point of insertion) showed that the mini-Tn5 Km inserted 192 bp upstream of the 3′ end of the waaQ coding sequence. The waaQ gene (1,035 bp) is the most upstream of the 10 genes in the E. coli K-12 waa operon involved in assembly of the LPS core (15). The mini-Tn5 Km transposon was the only segment of the pUT suicide vector inserted, which was confirmed by PCR using waaQ-specific upstream and downstream primers (see Materials and Methods). Using these primers, the expected 4.0-kb fragment was amplified.
Sensitivity of MD42 to SDS, bile salts, and novobiocin.
As stated above, MD42 grew poorly on MacConkey agar plates, which contain a mixture of bile salts. E. coli strains with severe defects in the LPS core (“deep-rough” mutants) are known to be hypersensitive to SDS, bile salts, and hydrophobic antibiotics, such as novobiocin (51). Therefore, MD42 and MG1655 Strr were tested for sensitivity to these agents (see Materials and Methods). As shown in Table 2, MD42 had a >1,000-fold increase in sensitivity to SDS, a >8-fold increase in sensitivity to bile salts, and an ∼33-fold increase in sensitivity to novobiocin compared to MG1655 Strr, suggesting that MD42 is a deep-rough mutant.
TABLE 2.
MICs of SDS, novobiocin, and bile salts for MG1655 Strr and MD42a
| Strain | MIC
|
||
|---|---|---|---|
| SDS (mg/ml) | Novobiocin (μg/ml) | Bile salts (mg/ml) | |
| MG1655 Strr | 100 | 50 | >6.0 |
| MD42 | <0.1 | 1.56 | 1.5 |
| MD42mm5 (Luria broth) | <0.1 | 6.25 | 1.5 |
| MD42mm5 (Luria broth + SDS) | 25 | 50 | 3.0 |
MICs were determined as described in Materials and Methods.
LPS profiles of MG1655 Strr and MD42.
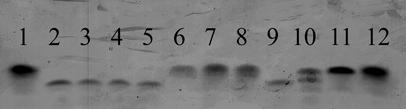
LPS core mutants defective only in waaQ are not deep-rough mutants, i.e., they are not hypersensitive to bile salts, novobiocin, or SDS and make an essentially full-length LPS core oligosaccharide (51, 52). However, the mini-Tn5 Km transposon used in the present study has identical strong transcription termination sequences flanking both ends of the kanamycin resistance gene (10). It was therefore likely that the entire waa operon was inactive in MD42 due to downstream polarity. Mutants in which the only defective waa gene is waaG make a truncated LPS core oligosaccharide (52), and mutants in which the only defective waa gene is waaP make both truncated and full-length LPS core oligosaccharides (53). Therefore, to determine whether the entire waa operon in MD42 was inactivated, the mobilities of the LPS cores of MD42, a known waaQ mutant (51), a known waaG mutant (51), and a known waaP mutant (51) were compared on SDS-PAGE gels. As shown in Fig. 1, MD42 LPS core migration was essentially identical to that of the truncated LPS core of the waaG mutant and to that of the lower band of the LPS core isolated from the known deep-rough waaP mutant, and its migration was more rapid than those of the LPS cores of the known waaQ mutant and MG1655 Strr. These data suggest that the mini-Tn5 Km insertion inactivated the entire MD42 waa operon.
FIG. 1.
E. coli LPS profiles. Purified LPS on SDS-PAGE gels was silver stained as described in Materials and Methods. Lane 1, MG1655 Strr; lane 2, MD42; lanes 3 to 5, SDS-sensitive revertants isolated from SDS-resistant MD42 mm5; lanes 6 to 8, individual SDS-resistant MD42 mm clones; lane 9, waaG::aacC1 mutant of F470; lane 10, waaP::aacC1 mutant of F470; lane 11, waaQ::aacC1 mutant of F470; lane 12, wild-type F470.
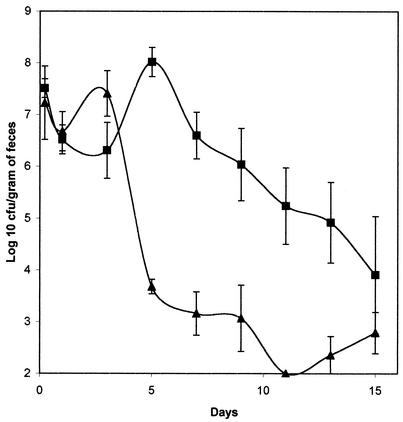
Mouse large-intestine-colonizing ability of MD42.
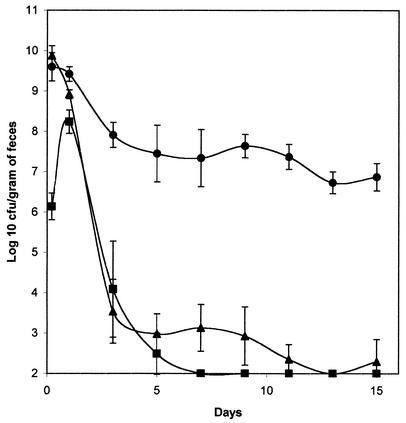
The mouse large-intestine colonization model requires that the mice be fed streptomycin in their drinking water. This treatment eliminates the facultative flora and creates a niche for E. coli but leaves the obligate-anaerobe population largely intact (29). The mini-Tn5 Km insertion in MD42 had no effect on its resistance to streptomycin (MIC > 5.0 mg/ml). MG1655 Strr and MD42 were each fed individually to mice (1010 CFU per mouse). At 5 h postfeeding, MD42 was present in feces at 3.6 × 109 CFU per g, and MG1655 Strr was present at 7.0 × 109 CFU per g. Therefore, the strains survived passage through the intestine equally well, despite the fact that bile salts are present in the small intestines of mammals at levels as high as 20 mM (2). However, MG1655 Strr colonized at a level between 107 and 108 CFU per g of feces (Fig. 2), whereas MD42 decreased in numbers continuously through day 5 and was observed only sporadically (between 102 and 103 CFU per g of feces) thereafter (Fig. 2). Furthermore, in one experiment in which MD42 was undetectable in feces 7 days postfeeding in three mice fed the strain, it was undetectable (<1 CFU) in cecal mucus isolated from the same mice sacrificed 8 days postfeeding, whereas MG1655 Strr was present in cecal mucus isolated from three mice sacrificed on the same day at an average level of 4.5 × 106 CFU per cecum (data not shown).
FIG. 2.
E. coli MG1655 Strr and MD42 colonization of the mouse large intestine. Sets of three mice were fed either 1010 CFU of MG1655 Strr (•), 1010 CFU of MD42 (▴), or 5 × 105 CFU of MD42 (▪). At the indicated times, fecal samples were homogenized, diluted, and plated as described in Materials and Methods. Bars representing the standard error of the log10 mean number of CFU per gram of feces for each set of three mice are presented for each time point.
When MD42 was fed to mice at lower levels (5 × 105 CFU per mouse), it grew from a level of about 2.5 × 106 CFU per g of feces 5 h postfeeding to ∼2.2 × 108 CFU per g of feces 24 h postfeeding (Fig. 2) but was eliminated thereafter, so that by day 7 and beyond (up to day 15), MD42 was no longer detectable in feces (<102 CFU per g) (Fig. 2). It therefore appears that MD42 grew in the intestine for 1 day in vivo and then failed to grow at a rate sufficient to prevent washout from the intestine. As a control, MG1655 Strr was fed to mice at 5 × 105 CFU per mouse. It grew to a level of ∼5 × 108 CFU per g of feces within 24 h postfeeding and colonized at a level of between 107 and 108 CFU per g of feces until the experiment was terminated at day 15 (data not shown).
Survival of MD42 in mouse feces.
It might be argued that if MD42 dies rapidly in mouse feces relative to MG1655 Strr, fecal viable counts would minimize its real colonizing ability. To address this question, samples of feces taken 24 h postfeeding from a mouse fed 5 × 105 CFU of MD42 and samples of feces taken 24 h postfeeding from a mouse fed 5 × 105 CFU of MG1655 Strr were homogenized in 1% Bacto Tryptone immediately after collection and plated, 24 h postcollection and plated, and 48 h postcollection and plated. Both MD42 and MG1655 Strr Nalr died logarithmically in mouse feces so that every 24 h 5% of the live MD42 in feces remained viable, whereas 35% of the live MG1655 Strr remained viable. Since feces no older than 24 h are assayed for viable counts, at most MD42 CFU relative to MG1655 Strr CFU are sevenfold underestimated, which would not change the fact that MD42 is a far worse colonizer of the mouse intestine than MG1655 Strr.
Growth of MG1655 Strr and MD42 in dilute cecal mucus.
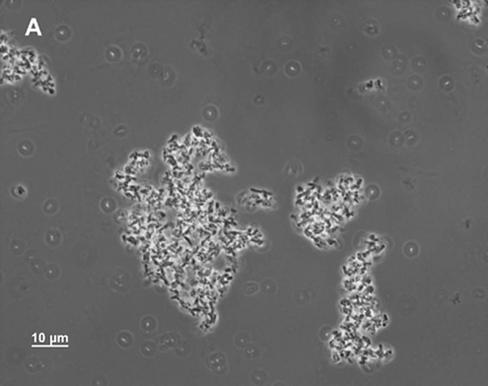
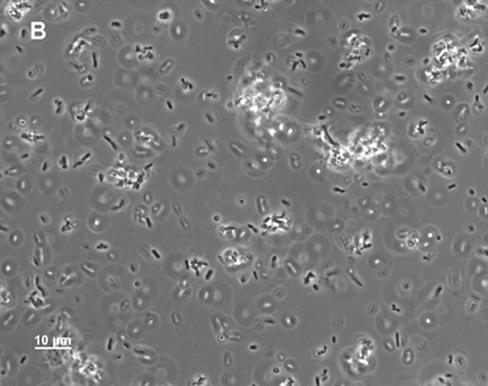
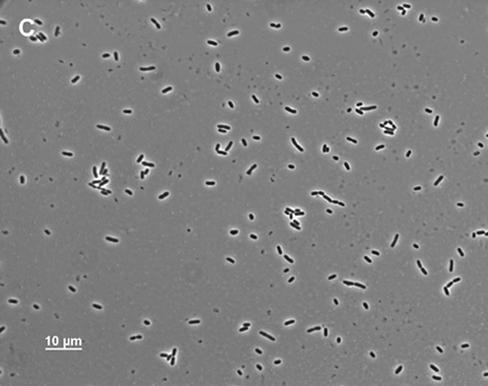
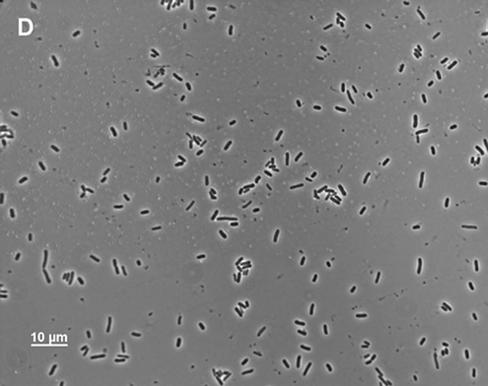
Although MD42 grew poorly on cecal-mucus agar plates (2 mg/ml with respect to protein), when grown standing in liquid cecal mucus at the same protein concentration, it grew as well to stationary phase as MG1655 Strr (55-min generation times); however, after overnight incubation, it settled to the bottom of the culture tubes. As measured by A600, only 22.0% ± 2.2% (n = 3) of the MD42 cells in the overnight culture remained suspended. In contrast, 59.5% ± 1.5% (n = 3) of the MG1655 Strr cells in the culture remained suspended overnight. Microscopic analysis of the MD42 cecal-mucus cultures revealed that by the time the cells settled to the bottom of the culture tubes, they had agglutinated, forming massive clumps (Fig. 3A). When overnight MD42 cecal-mucus cultures were vortexed, the cells in the clumps dispersed and the viable counts were found to be ∼4.0 × 108 CFU/ml, suggesting that all the cells in the clumps were viable. In contrast to MD42, overnight MG1655 Strr cultures contained many small clumps but also contained many single cells (Fig. 3B). MD42 grew with essentially the same doubling time as MG1655 Strr in standing Luria broth cultures (111 min for MG1655 Strr and 116 min for MD42) and to nearly the same extent at 24 h of incubation (4 × 108 to 8 × 108 CFU/ml). Neither strain formed clumps when grown in Luria broth (Fig. 3C and D), and in each case, ∼60% of the cells remained suspended overnight. In standing glucose minimal-medium cultures, MG1655 Strr and MD42 grew with identical generation times (110 min) and to nearly identical extents (4 × 108 to 8 × 108 CFU/ml). MD42 did not agglutinate in glucose minimal medium.
FIG. 3.
Phase-contrast microscopy of MD42 and MG1655 Strr cultures. The cultures were grown standing either in Luria broth or in mouse cecal mucus (2.0 mg/ml with respect to protein). After 24 h of incubation, 10-μl samples were taken from the bottoms of the unvortexed culture tubes and were examined by phase-contrast microscopy. (A) MD42 grown in cecal mucus; (B) MG1655 Strr grown in cecal mucus; (C) MD42 grown in Luria broth; (D) MG1655 Strr grown in Luria broth.
The NaCl concentration in HEPES-Hanks buffer is 137 mM, which mimics the NaCl concentration in jejunal and ileal intestinal fluid (5). The NaCl concentration in Luria broth is 85.5 mM. Since deep-rough mutants have a more hydrophobic surface than their wild-type counterparts, it was possible that the increased NaCl concentration in HEPES-Hanks buffer was responsible for MD42 agglutination. Therefore, cecal mucus (2 mg/ml with respect to protein) was prepared in HEPES-Hanks buffer containing 85.5 and 137 mM NaCl. After overnight growth in cecal mucus with 85.5 mM NaCl, 60.2% ± 0.8% (n = 3) of the MD42 cells remained suspended, whereas in the presence of 137 mM NaCl, only 15.1% ± 1.8% (n = 3) of the MD42 cells remained suspended. It therefore appears that the NaCl concentration in HEPES-Hanks buffer, representative of that in intestinal juice (5), is responsible for agglutination of MD42 after growth in diluted cecal mucus.
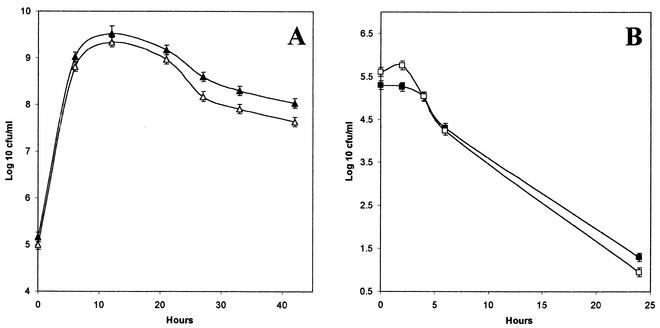
Growth of MG1655 Strr and MD42 in undiluted cecal mucus and in cecal contents.
MG1655 Strr and MD42 were also tested separately for growth during a 42-h period in cecal mucus scraped directly into culture tubes. Each strain was inoculated at a concentration of ∼5 × 105 CFU/ml. Both MG1655 Strr and MD42 grew rapidly in undiluted cecal mucus to viable counts of 2.0 × 109 and 7.0 × 108 CFU/ml, respectively, at 24 h postinoculation, and each stabilized at a level of ∼108 CFU/ml (Fig. 4A). It therefore appears that there is no antimicrobial substance, e.g., bile salts, present in mouse cecal mucus at high enough concentrations to preferentially kill MD42.
FIG. 4.
Growth of MD42 and MG1655 Strr in undiluted viscous mouse cecal mucus and cecal contents. E. coli MG1655 Strr and MD42 were inoculated separately at ∼5 × 105 CFU/ml into 2 ml of mouse cecal mucus scraped directly into 14- by 110-mm culture tubes and into 2 ml of cecal contents in 14- by 110-mm culture tubes. The cultures were incubated standing at 37°C. At the indicated times, the cultures were vortexed gently and 50-μl samples were diluted and plated. Bars representing the standard error of the log10 mean of duplicate cultures for each point are presented. (A) ▴, MG1655 Strr in cecal mucus; ▵, MD42 in cecal mucus. (B) ▪, MG1655 Strr in cecal contents; □, MD42 in cecal contents.
E. coli MG1655 Strr and MD42 were also each inoculated at ∼5 × 105 CFU/ml into cecal luminal contents isolated directly from mouse intestines (Fig. 4B). Both MD42 and MG1655 Strr remained relatively constant in number in cecal contents for 6 h postinoculation. After 6 h, both strains began to die at approximately the same rate (Fig. 4B). Therefore, both MD42 and MG1655 Strr failed to grow in cecal luminal contents, and each had about the same sensitivity to antimicrobials present in cecal contents. The failure of various E. coli strains to grow in cecal luminal contents has been reported previously (25, 33, 45, 46, 49).
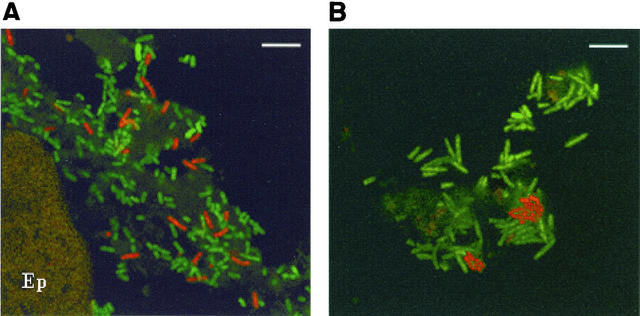
Visualization of MD42 in the mouse cecal-mucus layer.
Experiments were performed to determine whether MD42 agglutinated in cecal mucus in vivo as it did in diluted mouse cecal mucus in vitro. MD42 and MG1655 Strr were grown in Luria broth, conditions under which the majority of MD42 and MG1655 Strr appear as single cells (Fig. 3C and D), and fed to mice individually (1010 CFU per mouse). The colonized bacteria were visualized in cecal mucus in situ 24 h postfeeding by hybridization with an E. coli-specific oligonucleotide probe (see Materials and Methods). MD42 was present in cecal mucus almost exclusively in small clumps ranging between 4 and 20 cells far from epithelial cells (Fig. 5B), whereas MG1655 Strr was present in cecal mucus almost exclusively as single cells, frequently close to epithelial cells (Fig. 5A). These results suggest that MD42 may have difficulty penetrating the mucus layer. In addition, the MD42 clumps were frequently associated with clumps of other eubacteria (Fig. 5B), suggesting the possibility that MD42 is attracted to other hydrophobic bacteria in vivo that may also have trouble penetrating the mucus layer. In summary, these data show that in contrast to MG1655 Strr, MD42 forms clumps when grown in cecal mucus in vivo as well as in cecal mucus in vitro, thus suggesting that clumping may play a role in its elimination from the intestine.
FIG. 5.
In situ hybridization with fluorescence-labeled oligonucleotide probes. Cecal-mucosal sections from mice fed MG1655 Strr (A) or MD42 (B) were hybridized 24 h postfeeding with an E. coli-specific oligonucleotide probe (red) and a eubacterium-specific oligonucoeotide probe (green). E. coli cells appear red, while all other bacteria appear green. Ep, epithelial cell. Bars, 10 μm.
Isolation of an MD42 suppressor mutant.
MD42 was mutagenized with a mini-mariner transposon containing the gentamicin resistance gene (50). Following mutagenesis and plating (see Materials and Methods), five MD42 colonies grew on Luria agar plates containing streptomycin, kanamycin, gentamicin, and SDS (1 mg/ml). Four of the isolates were far more resistant to SDS (50 mg/ml) and novobiocin (50 μg/ml) than MD42. However, as shown by PCR, in each of these isolates the mini-Tn5 Km transposon had moved out of the waaQ gene, presumably because of the transposase introduced with the mini-mariner transposon (not shown). These four isolates were therefore not suppressor mutants and were not studied further. The fifth isolate, designated MD42 mm5 (for mini-mariner), retained the mini-Tn5 Km transposon in waaQ and was used for further study.
Sequencing from both ends of the mini-mariner transposon revealed that the transposon had inserted within the bipA (yihK) gene, just 35 bp upstream of the 3′ end. bipA mutants have been reported to grow poorly at 20°C (35) and to be more motile than their parents (11); however, we were unable to find any difference in doubling times at 20°C or in motility between MD42 and MD42 mm5, suggesting that the bipA gene in MD42 mm5 was not completely inactivated.
The LPS core mobility of MD42 mm5 on SDS-PAGE gels remained like that of a deep-rough mutant, i.e., indistinguishable from that of MD42 and faster than that of MG1655 Strr (Fig. 1). Furthermore, MD42 mm5 grew at the same rate and almost to the same extent as MG1655 Strr in Luria broth, and like MD42, MD42 mm5 agglutinated when grown in cecal mucus (2 mg/ml with respect to protein) containing 137 mM NaCl. These results suggest that MD42 mm5 grown in Luria broth retained the defect in its core LPS.
Further characterization of MD42 mm5.
The defective bipA gene was introduced into a fresh MD42 background by allelic exchange (see Materials and Methods), and three recombinants were selected on Luria agar plates containing kanamycin and gentamicin. All three recombinants were confirmed by PCR as having the mini-mariner insertion in bipA. MD42 mm5 and the three recombinants grew well in Luria broth containing SDS (1.0 mg/ml), whereas MD42 did not. Therefore, the mini-mariner insertion in bipA appears to be responsible for increased MD42 mm5 resistance to SDS.
Although the resistance of MD42 mm5 to novobiocin (6.25 μg/ml) was greater than that of MD42 (1.56 μg/ml), surprisingly, the MIC of SDS for MD42 mm5 did not differ significantly from that for MD42 (Table 2). However, the increased resistance of MD42 mm5 to SDS was found to manifest itself as an increased number of resistant cells in the population (∼1 in 104 CFU) relative to MD42 (<1 in 108 CFU) rather than an increase in resistance of the entire population; MD42 mm5 had a 10−4 plating efficiency on Luria agar containing SDS (1.0 mg/ml) relative to Luria agar, whereas MD42 had a plating efficiency of <10−8 under the same conditions. Moreover, when MD42 mm5 was grown in Luria broth containing SDS, its resistance to SDS increased >250-fold, its resistance to novobiocin increased 8-fold, and its resistance to bile salts increased 2-fold (Table 2).
To further investigate SDS sensitivity and resistance in MD42 mm5, a single MD42 mm5 colony from a Luria agar plate containing SDS (1 mg/ml) was grown overnight in Luria broth containing SDS (1 mg/ml) and then plated on Luria agar without SDS. After overnight incubation, 200 colonies were toothpicked to two Luria agar plates containing SDS (1 mg/ml) and then to a Luria agar plate without SDS. One hundred eighty-nine colonies grew on all three plates, and 11 colonies failed to grow on either of the two plates containing SDS but grew well on the plate lacking SDS. Therefore, SDS-resistant MD42 mm5 cells can revert to the SDS-sensitive state at the relatively high frequency of ∼1 in 20. Thus, MD42 mm5 appears to be capable of a reversible switch between SDS sensitivity and resistance: when grown under nonselective conditions (without SDS), only 1 in 104 cells is resistant; conversely, after being grown under selective conditions in which all cells are SDS resistant, 1 in 20 cells reverts to being SDS sensitive under nonselective conditions.
MD42 mm5 cells (1 in 104 resistant to SDS) grown in Luria broth and in Luria broth containing SDS (1.0 mg/ml) were diluted into cecal mucus (2.0 mg/ml with respect to protein) to a level of ∼1.0 × 105 CFU/ml. The cultures were incubated standing for 24 h at 37°C. After overnight growth in cecal mucus, 66.3% ± 3.6% (n = 3) of the MD42 mm5 cells that had been grown in the presence of SDS remained suspended, whereas only 25.8% ± 1.8% (n = 3) of the MD42 cells grown in Luria broth remained suspended. It therefore appears that growth in the presence of SDS prevents excessive MD42 mm5 clumping in cecal mucus.
Since growth of MD42 mm5 in SDS prevented clumping in cecal mucus, experiments were performed to compare the LPS core structures of MD42 mm5 grown in Luria broth and that grown in Luria broth containing SDS (1 mg/ml). Three SDS-resistant clones and three SDS-sensitive clones that had reverted from SDS resistance were tested. The MD42 mm5 LPS cores derived from the three SDS-resistant clones were the same size as that of the wild type (Fig. 1); however, the LPS cores derived from the three SDS-sensitive clones that had reverted from SDS resistance were truncated (Fig. 1). Since the SDS-resistant MD42 mm5 clones still contained the mini-Tn5 Km insertion in waaQ, these results suggest that in these clones, the mini-mariner insertion in bipA somehow restored expression of the waa operon downstream of waaQ or induced an alternative pathway for core synthesis.
Mouse large-intestine-colonizing ability of MD42 mm5.
Three mice were fed 108 CFU of MD42 mm5 that had been grown in Luria broth (1 in 104 CFU resistant to SDS), and three mice were fed 108 CFU of MD42 mm5 that had been grown in Luria broth containing 1.0 mg of SDS/ml. MD42 mm5 grown in Luria broth acted identically to MD42, i.e., it initially grew but then was rapidly eliminated from the intestine (Fig. 6). In contrast, after an initial period of growth in the intestine, MD42 mm5 grown in the presence of SDS remained at relatively high levels in the intestine and was only slowly eliminated (Fig. 6). Therefore, making the entire MD42 mm5 population resistant to SDS by growing it in the presence of SDS resulted in partial suppression of poor colonizing ability. PCR analysis of colonies from fecal plate counts indicated that the transposon insertions in waaQ and bipA remained intact, ruling out the possibility that the mutants reverted during the experiment. It therefore appears that the better colonizing ability of MD42 mm5 grown in the presence of SDS is due to suppression of the deep-rough phenotype by the transposon insertion in bipA.
FIG. 6.
E. coli MD42 mm5 colonization of the mouse large intestine. Sets of three mice were fed either 108 CFU of MD42 mm5 grown in Luria broth (▴) or 108 CFU of MD42 mm5 grown in Luria broth containing SDS (1 mg/ml) (▪). At the indicated times, fecal samples were homogenized, diluted, and plated on Luria agar plates containing kanamycin sulfate (40 μg/ml) and gentamicin (15 μg/ml) for determining numbers of MD42 mm5 in feces of mice fed Luria broth-grown MD42 mm5 and on Luria agar plates containing kanamycin sulfate (40 μg/ml) and SDS (1.0 mg/ml) for determining numbers of MD42 mm5 in feces of mice fed Luria broth plus SDS-grown MD42 mm5. Bars representing the standard error of the log10 mean number of CFU per gram of feces for each set of three mice are presented for each time point.
DISCUSSION
A large body of experimental evidence shows that E. coli grows rapidly in intestinal mucus both in vivo and in vitro but does not grow, or grows poorly, in luminal contents (23, 33, 45, 46, 49). This suggests that the ability of an E. coli strain to grow in intestinal mucus plays a critical role in its ability to colonize the intestine. In the present study, we isolated MD42, a mutant in which the mini-Tn5 Km transposon had inserted in the waaQ gene. MD42 grew poorly on cecal-mucus agar plates and was unable to colonize the mouse large intestine.
The waaQ gene is the promoter-proximal gene in a 10-gene operon involved in LPS core biosynthesis (15). WaaQ appears to be a HepIII (Hep, l-glycero-d-manno-heptose) transferase that transfers HepIII to the HepII residue in the LPS core (51). However, since the mini-Tn5 Km transposon used here has identical strong transcription termination sequences flanking both ends of the kanamycin resistance gene (10), the entire waa operon would likely be inactivated due to downstream polarity. In support of this view, the LPS mobility on SDS-PAGE gels was like that of a waaG mutant with a truncated LPS core oligosaccharide and not like that of a nonpolar waaQ mutant, which has a more complete LPS core oligosaccharide (Fig. 1) (15, 51). waaG is immediately downstream of waaQ (15). Inactivation of the entire waa operon results in a truncated LPS core oligosaccharide, no HepIII, and lack of phosphorylation of HepI and HepII (51, 52, 53). Lack of phosphorylation of HepI and HepII prevents neighboring LPS molecules in the outer membrane from being cross-linked by divalent cations (15, 51, 52, 53), resulting in extreme destabilization of the outer membrane and a phenotype called deep-rough (51, 52, 53). Deep-rough mutants exhibit hypersensitivity to bile salts, SDS, and the hydrophobic antibiotic novobiocin (51). MD42 is also hypersensitive to all three agents (Table 2), whereas a nonpolar E. coli waaQ mutant is almost as resistant to SDS and novobiocin as its parent (51). Therefore, it appears highly likely that in MD42, the insertion in waaQ has a polar effect on the entire waa operon, thereby explaining its deep-rough phenotype and its truncated core oligosaccharide.
To our knowledge, this is the first time that an LPS deep-rough core mutant has been shown to be unable to colonize the intestine when fed to mice alone; however, various LPS mutants have been shown to have a reduced mouse intestine-colonizing ability when fed simultaneously with their wild-type parents (7, 22, 26, 27). It should be noted, however, that not all LPS mutants are poor colonizers when fed to mice alone, e.g., Salmonella enterica serovar Typhimurium O side chain mutants and Vibrio cholerae O side chain mutants have been shown to colonize the mouse large intestine well under these conditions (3, 26). In fact, E. coli MG1655 Strr, the parent of MD42, has no O side chain (34) yet, as shown here, is an excellent colonizer when fed to mice alone.
Despite MD42 being hypersensitive to SDS and bile salts, it is unlikely that antimicrobials present in cecal mucus are responsible for the inability of MD42 to colonize the mouse large intestine, since MD42 and MG1655 Strr grew and survived equally well in undiluted cecal mucus in vitro (Fig. 4). It might be argued that MD42 is unable to colonize the mouse large intestine because it is less motile than MG1655 Strr and may therefore have trouble maintaining itself in the mucus layer. This hypothesis is also unlikely, however, since it was shown previously that nonmotile and nonchemotactic mutants of E. coli F-18, a human commensal fecal isolate, are excellent colonizers of the mouse large intestine when fed to mice alone or together with their parent (25).
The propensity of MD42 to form clumps may be a major factor in its poor colonizing ability. That is, MD42 formed clumps when grown in cecal mucus in vitro (Fig. 3) and in vivo (Fig. 5) and failed to colonize the mouse large intestine (Fig. 2). Similarly, MD42 mm5 grown in Luria broth formed clumps when grown in mouse cecal mucus in vitro and also failed to colonize the mouse large intestine (Fig. 6). In contrast, when grown in the presence of SDS, MD42 mm5 did not clump when subsequently grown in cecal mucus in vitro and colonized the mouse large intestine far better than either MD42 mm5 grown in Luria broth (Fig. 6) or MD42 (Fig. 2). It was shown previously that the ability of S. enterica serovar Typhimurium to traverse a layer of mucus in vitro and in vivo correlates well with its ability to colonize the mouse large intestine (22, 26). It is likely that, relative to single cells, clumps of cells have difficulty penetrating a layer of mucus. It may be that clumps of MD42 cells are more readily sloughed into the cecal contents as the mucus layer normally turns over and is excreted in the feces. In summary, the results show a correlation between clumping of the waaQ mutant and poor colonization; suppression of the clumping phenotype by mutation of bipA partially restored colonization.
MD42 mm5 is a bipA mutant of MD42. Most of the properties ascribed to the E. coli BipA protein stem from studies with enteropathogenic E. coli (EPEC) strains. BipA appears to be important for several aspects of EPEC infection, including resistance to the host bactericidal-permeability-increasing protein, for triggering characteristic rearrangements of the cytoskeletons of EPEC-infected host cells, for regulating flagellum-mediated cell motility (11), and for expression of K-5 capsular genes (42). In addition, the E. coli K-12 BipA protein appears to be important for achieving maximum growth rate at low temperature (35). BipA is a member of the GTPase superfamily, which includes the elongation factors EF-Tu and EF-G and the TetM/TetO tetracycline resistance proteins (40). BipA may also bind to ribosomes (40). The EPEC BipA protein is phosporylated on a tyrosine residue, but the K-12 BipA protein is not phosphorylated (11). BipA regulates E. coli target gene expression by an as-yet-unknown mechanism.
Surprisingly, the LPS core of SDS-resistant MD42 mm5 was not truncated (Fig. 1). This result suggests that although SDS-resistant MD42 mm5 still contains a kanamycin resistance gene with transcription termination sequences inserted in waaQ, the downstream genes of the operon are somehow transcribed. This would explain the normal-size LPS core; resistance to SDS, bile salts, and novobiocin; and improved colonizing ability of MD42 mm5. At the present time, it is unclear how the mini-mariner insertion in bipA would allow transcription of the entire waa operon in SDS-resistant MD42 mm5, but it appears that the mutant BipA protein prevents transcription termination caused by the mini-Tn5 Km insertion in waaQ or that the mini-mariner insertion results in activation of a promoter immediately upstream of waaG. It is also interesting that the mini-mariner insertion in bipA eliminates only 11 amino acids at the carboxyl-terminal end of the protein and that 5 of the last 6 are basic amino acids. The short (11-amino-acid) truncation of BipA in the MD42 background did not confer the reported phenotype of a bipA null mutant: increased motility and poor growth at 20°C. The bipA mutation in MD42 mm5 instead conferred restored synthesis of the LPS core (and related sensitivity phenotypes), but with a pattern reminiscent of phase variation (16, 19, 47), in this case between SDS sensitivity and resistance. Thus, it appears that the truncated bipA gene has altered regulation of genes that are important for colonization. It will be interesting to determine if BipA is either directly or indirectly involved in a phase variation mechanism that regulates E. coli MG1655 LPS core synthesis.
Acknowledgments
This research was supported by Public Health Service grant AI48945 to T.C. and P.S.C. and by grant H-401 from the University of Rhode Island Agricultural Experiment Station to P.S.C.
We thank C. Whitfield of the University of Guelph, Guelph, Ontario, Canada, for bacterial strains; J. J. Mekalanos of Harvard Medical School, Boston, Mass. for the mini-mariner transposon; Paul Johnson of the University of Rhode Island for his skill in photographing MD42 and MG1655; and April Anderson of the University of Oklahoma for critical reading of the manuscript. We also thank our Danish colleagues in Copenhagen, Carsten Struve of the Statens Seruminstitut for help with in situ hybridization, Anni Ravn of the Statens Veterinaere Seruminstitut for preparing cecal sections, and Bent Roldgaard of the Foedevaredirektoratet for help with confocal microscopy.
Editor: A. D. O'Brien
REFERENCES
- 1.Allan, A. 1981. Structure and function of gastrointestinal mucus, p. 637-639. In L. R. Johnson (ed.), Physiology of the gastrointestinal tract. Raven Press, New York, N.Y.
- 2.Altman, P. L., and D. S. Dittman. 1968. Metabolism, p. 262. Federation of American Societies for Experimental Biology, Bethesda, Md.
- 3.Angelichio, M. J., J. Spector, M. K. Waldor, and A. Camilli. 1999. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 67:3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balbas, P., X. Soberon, E. Merino, M. Zurita, H. Lomeli, F. Valle, N. Flores, and F. Bolivar. 1986. Plasmid vector pBR322 and its special derivatives—a review. Gene 50:3-40. [DOI] [PubMed] [Google Scholar]
- 5.Banwell, J. G., S. L. Gorbach, N. F. Pierce, R. Mitra, and A. Mondal. 1971. Acute undifferentiated human diarrhea in the tropics. II. Alterations in intestinal fluid and electrolyte movements. J. Clin. Investig. 50:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner, D. J. 1984. Facultatively anaerobic Gram-negative rods, p. 408-516. In N. R. Krieg (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 7.Chiang, S. L., and J. J. Mekalanos. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 27:797-805. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, P. S., and D. C. Laux. 1995. Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods Enzymol. 253:309-315. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farris, M., A. Grant, T. B. Richardson, and C. D. O'Connor. 1998. BipA: a tyrosine-phosphorylated GTPase that mediates interactions between enteropathogenic Escherichia coli (EPEC) and epithelial cells. Mol. Microbiol. 28:265-279. [DOI] [PubMed] [Google Scholar]
- 12.Forstner, G. G. 1970. [1-14C]glucosamine incorporation by subcellular fractions of small intestine mucosa. J. Biol. Chem. 245:3584-3592. [PubMed] [Google Scholar]
- 13.Freter, R., H. Brickner, M. Botney, D. Cleven, and A. Aranki. 1983. Mechanisms that control bacterial populations in continuous-flow culture models or mouse large intestinal flora. Infect. Immun. 39:676-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freter, R., H. Brickner, J. Fekete, M. M. Vickerman, and K. E. Carey. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 39:686-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinrichs, D. E., J. A. Yethon, and E. Whitfield. 1998. Molecular basis of structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella typhimurium. Mol. Microbiol. 30:221-231. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the on and off of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 17.Hoskins, L. 1984. Mucin degradation by enteric bacteria: ecological aspects and implications for bacterial attachment to gut mucosa, p. 51-65. In E. C. Boedecker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, Fla.
- 18.Kim, Y. S., A. Morita, S. Miura, and B. Siddiqul. 1984. Structure of glycoconjugates of intestinal mucosal membranes. Role of bacterial adherence, p. 99-109. In E. C. Boedecker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, Fla.
- 19.Klemm, P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 5:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogfelt, K. A., and P. Klemm. 1988. Investigation of minor components of Escherichia coli Type 1 fimbriae: Protein chemical and immunological aspects. Microb. Pathog. 4:231-238. [DOI] [PubMed] [Google Scholar]
- 21.Krogfelt, K. A., L. K. Poulsen, and S. Molin. 1993. Identification of coccoid Escherichia coli BJ4 cells in the large intestine of streptomycin-treated mice. Infect. Immun. 61:5029-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Licht, T. R., K. A. Krogfelt, P. S. Cohen, L. K. Poulsen, J. Urbance, and S. Molin. 1996. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect. Immun. 64:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licht, T. R., T. Tolker-Nielsen, K. Holmstrøm, K. A. Krogfelt, and S. Molin. 1999. Inhibition of Escherichia coli precursor 16S rRNA processing by mouse intestinal contents. Environ. Microbiol. 1:23-32. [DOI] [PubMed] [Google Scholar]
- 24.McCormick, B. A., D. P. Franklin, D. C. Laux, and P. S. Cohen. 1989. Type 1 pili are not necessary for colonization of the streptomycin-treated mouse large intestine by type 1-piliated Escherichia coli F-18 and E. coli K12. Infect. Immun. 57:3022-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick, B. A., D. C. Laux, and P. S. Cohen. 1990. Neither motility nor chemotaxis plays a role in the ability of Escherichia coli F-18 to colonize the streptomycin-treated mouse large intestine. Infect. Immun. 58:2957-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick, B. A., B. A. D. Stocker, D. C. Laux, and P. S. Cohen. 1988. The role of motility, chemotaxis, penetration through, and growth in intestinal mucus in the ability of an avirulent strain of Salmonella typhimurium to colonize the large intestines of streptomycin-treated mice. Infect. Immun. 56:2209-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrell, D. S., D. L. Hava, and A. Camilli. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol 43:1471-1491. [DOI] [PubMed] [Google Scholar]
- 28.Metcalf, W. W., W. Jiang, L. L. Daniels, S.-K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 29.Miller, C. P., and M. Bohnhoff. 1963. Changes in the mouse's enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin-treatment. J. Infect. Dis. 113:59-66. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Moore, W. E. C., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neutra, M. R. 1984. The mechanism of intestinal mucous secretion, p. 33-41. In E. C. Boedecker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, Fla.
- 33.Newman, J. V., R. Kolter, D. C. Laux, and P. S. Cohen. 1994. The role of leuX in Escherichia coli colonization of the streptomycin-treated mouse large intestine. Microb. Pathog. 17:301-311. [DOI] [PubMed] [Google Scholar]
- 34.Orskov, F., and I. Orskov. 1961. The fertility of Escherichia coli antigen test strains in crosses with K12. Acta Pathol. Mikrobiol. Scand. 51:280-290. [PubMed] [Google Scholar]
- 35.Pfennig, P. L., and A. M. Flower. 2001. BipA is required for growth of Escherichia coli K12 at low temperature. Mol. Genet. Genomics 266:313-317. [DOI] [PubMed] [Google Scholar]
- 36.Potten, C. S., and T. D. Allen. 1977. Ultrastructure of cell loss in intestinal mucosa. J. Ultrastruct. Res. 60:272-277. [DOI] [PubMed] [Google Scholar]
- 37.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine from rRNA in situ hybridization. Infect. Immun. 62:5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pradel, E., and C. A. Schnaitman. 1991. Effect of the rfaH (sfrB) and temperature on the expression of the rfa genes of Escherichia coli K-12. J. Bacteriol. 173:6428-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quastler, H., and F. G. Sherman. 1959. Cell population in the intestinal epithelium of the mouse. Exp. Cell. Res. 17:420-438. [DOI] [PubMed] [Google Scholar]
- 40.Qi, S.-Y., Y. Li, A. Szyroki, I. G. Giles, A. Moir, and C. D. O'Connor. 1995. Salmonella typhimurium responses to a bactericidal protein from human neutrophils. Mol. Microbiol. 7:523-531. [DOI] [PubMed] [Google Scholar]
- 41.Revel, H. R. 1966. Restriction and nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology 31:688-701. [DOI] [PubMed] [Google Scholar]
- 42.Rowe, S., N. Hodson, G. Griffiths, and I. S. Roberts. 2000. Regulation of the Escherichia coli K5 capsule gene cluster: evidence for the roles of H-NS, BipA, and integration host factor in regulation of group 2 capsule gene clusters in pathogenic E. coli. J. Bacteriol. 182:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slomiany, A., S. Yano, B. I. Slomiany, and G. B. J. Glass. 1978. Lipid composition of the gastric mucus barrier in the rat. J. Biol. Chem. 253:3785-3791. [PubMed] [Google Scholar]
- 44.Stahl, D. A., and R. I. Ahman. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 45.Sweeney, N. J., P. Klemm, B. A. McCormick, E. Moller-Nielsen, M. Utley, M. A. Schembri, D. C. Laux, and P. S. Cohen. 1996. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect. Immun. 64:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sweeney, N. J., D. C. Laux, and P. S. Cohen. 1996. Escherichia coli F-18 and K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 64:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szekely, E., and M. Simon. 1983. DNA sequence adjacent to flagellar genes and evolution of flagellar-phase variation. J. Bacteriol. 155:74-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharide in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 49.Wadolkowski, E. A., D. C. Laux, and P. S. Cohen. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect. Immun. 56:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, S. M., and J. J. Mekalanos. 2000. Genetic footprinting with mariner-based transposition in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:10191-10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yethon, J. A., D. E. Heinrichs, M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273:26310-26316. [DOI] [PubMed] [Google Scholar]
- 52.Yethon, J. A., E. Vinogradov, M. B. Perry, and C. Whitfield. 2000. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J. Bacteriol. 182:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yethon, J. A., and C. Whitfield. 2001. Purification and characterization of WaaP from Escherichia coli, a lipopolysaccharide kinase essential for outer membrane stability. J. Biol. Chem. 276:5498-5504. [DOI] [PubMed] [Google Scholar]