Abstract
F18 fimbrial Esherichia coli strains are associated with porcine postweaning diarrhea and pig edema disease. Recently, the FedF subunit was identified as the adhesin of the F18 fimbriae. In this study, adhesion domains of FedF were further studied by constructing deletions within the fedF gene and expressing FedF proteins with deletions either together with the other F18 fimbrial subunits or as fusion proteins tagged with maltose binding protein. The region essential for adhesion to porcine intestinal epithelial cells was mapped between amino acid residues 60 and 109 of FedF. To map the binding domain even more closely, all eight charged amino acid residues within this region were independently replaced by alanine. Three of these single point mutants expressing F18 fimbriae exhibited significantly diminished capabilities to adhere to porcine epithelial cells in vitro. In addition, a triple point mutation and a double point mutation completely abolished receptor adhesiveness. The result further confirmed that the region between amino acid residues 60 and 109 is essential for the binding of F18 fimbriae to their receptor. In addition, the adhesion capability of the binding domain was eliminated after treatment with iodoacetamide, suggesting the formation of a disulfide bridge between Cys-63 and Cys-83, whereas Cys-111 and Cys-116 could be deleted without affecting the binding ability of FedF.
Colonization by pathogenic Escherichia coli strains and other pathogens is the first event in the infection process; it is initiated by the adherence of bacteria to host cell surfaces, which is mediated by adhesins (11, 22, 30). Many of the bacterial adhesins are associated with fimbriae and are responsible for recognizing and binding to specific receptor moieties of the host cells (23, 41, 43). F18 fimbrial Escherichia coli strains adhere to the microvilli of small intestine epithelial cells in piglets and are associated with porcine postweaning diarrhea and pig edema disease, occurring after weaning or transfer to fattening premises (3, 9, 14). There are two antigenic variants of F18 fimbriae. The F18ab variant is often expressed by E. coli O139 strains producing Shiga-like toxin and causing edema disease (3, 17). The F18ac fimbrial E. coli strains often belong to serogroup O141 or O157 and cause diarrhea by expressing enterotoxins (STa or STb) either together with or without Shiga-like toxin (34).
The protective function of antibodies raised against F18 fimbriae has been demonstrated (18, 50). Both passive and active oral immunizations based on F18 fimbrial antigens have been carried out to study the prevention of edema disease (4, 18, 50). Passive immunization of piglets with egg yolk antibodies raised against F18ac (50) or F18ab (18) reduced excretion of challenge bacteria and protected experimentally challenged pigs from diarrhea and death. Chicken anti-F18ac antibodies were shown to inhibit attachment of F18ab-positive E. coli bacteria to the pig intestinal mucosa (18).
The genetic organization of the fed gene cluster, involved in the biosynthesis of the F18 fimbriae, has been characterized (20, 40). The fed gene cluster is composed of five genes. The gene encoding the major protein, FedA, and two genes encoding the minor proteins, FedE and FedF, were first described by Imberechts et al. (19, 20). The rest of the fed genes, i.e., fedB and fedC, were recently characterized and found to encode the putative usher protein (FedB) and chaperone (FedC) of F18 fimbriae (40). According to Imberechts et al. (20), either FedE or FedF could mediate adhesion of F18 fimbriae to epithelial cells. Smeds et al. (40) recently demonstrated that anti-FedF antibodies, unlike anti-FedE serum, were able to inhibit E. coli adhesion to porcine enterocytes. Specific adhesion to enterocytes was also shown with purified FedF-maltose binding protein (MBP). So far the role of FedE is unknown.
In this work, we have further analyzed the FedF protein in order to characterize more closely the domains of FedF responsible for binding of F18-fimbriated E. coli to porcine intestinal epithelial cells. Deletions were constructed within the fedF gene, and FedF derivatives possessing those deletions were expressed either together with the other fimbrial subunits or tagged with MBP. In addition, eight separate point mutations within the putative binding domain of 50 amino acids were constructed by PCR mutagenesis. Three of the amino acid residues changed were shown to be essential for efficient adhesion of F18 fimbriae to their receptor. The triple substitution and one of the double substitutions completely eliminated F18 receptor adhesiveness.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli strains and plasmids used in this study are listed in Table 1. Plasmid pMAL-p2 (New England Biolabs [NEB]) was used for periplasmic expression of truncated FedF proteins fused to MBP. Luria broth (Difco) supplemented with ampicillin (100 μg/ml) was used as the growth medium for recombinant bacteria containing the pMAL-p2 constructs. For adhesion tests, to ensure expression of F18 fimbriae by plasmid pIH120 or its derivatives, E. coli strains were grown overnight in tryptic soy broth medium (Difco) supplemented with ampicillin (100 μg/ml).
TABLE 1.
Plasmids and bacterial strains
| Plasmid or strain | Relevant featurea | Source or reference |
|---|---|---|
| Plasmids | ||
| pIH120 | pUC18 carrying the entire F18 fimbria-encoding region | 20 |
| pMAL-p2 | Apr; E. coli expression vector | 12; NEB |
| pKTH5068 | FedFΔ1-10-encoding region in pMAL-p2 | This study |
| pKTH5069 | FedFΔ1-37-encoding region in pMAL-p2 | This study |
| pKTH5070 | FedFΔ1-52-encoding region in pMAL-p2 | This study |
| pKTH5071 | FedFΔ70-77-encoding region in pMAL-p2 | This study |
| pKTH5072 | FedFΔ88-97-encoding region in pMAL-p2 | This study |
| pKTH5073 | FedFΔ110-117-encoding region in pMAL-p2 | This study |
| pKTH5075 | FedFΔ124-280-encoding region in pMAL-p2 | This study |
| pKTH5076 | FedFΔ192-280-encoding region in pMAL-p2 | This study |
| pKTH5077 | FedFΔ247-280-encoding region in pMAL-p2 | This study |
| pKTH5094 | FedF60-123-encoding region in pMAL-p2 | This study |
| pKTH5104 | FedF (Lys72→Ala) mutation in pIH120 | This study |
| pKTH5105 | FedF (Arg75→Ala) mutation in pIH120 | This study |
| pKTH5106 | FedF (Glu76→Ala) mutation in pIH120 | This study |
| pKTH5107 | FedF (Glu82→Ala) mutation in pIH120 | This study |
| pKTH5108 | FedF (Arg84→Ala) mutation in pIH120 | This study |
| pKTH5109 | FedF (His88→Ala) mutation in pIH120 | This study |
| pKTH5110 | FedF (His89→Ala) mutation in pIH120 | This study |
| pKTH5111 | FedF (Glu96→Ala) mutation in pIH120 | This study |
| pKTH5124 | pIH120/FedFΔ1-5 | This study |
| pKTH5125 | pIH120/FedFΔ1-34 | This study |
| pKTH5126 | pIH120/FedFΔ1-52 | This study |
| pKTH5127 | pIH120/FedFΔ70-77 | This study |
| pKTH5128 | pIH120/FedFΔ88-97 | This study |
| pKTH5129 | pIH120/FedFΔ110-117 | This study |
| pKTH5130 | pIH120/FedFΔ124-280 | This study |
| pKTH5131 | pIH120/FedFΔ192-280 | This study |
| pKTH5132 | pIH120/FedFΔ247-280 | This study |
| pKTH5149 | FedF (Lys72, His88, His89→Ala) triple mutation in pIH120 | This study |
| pKTH5150 | FedF (Lys72, His88→Ala) double mutation in pIH120 | This study |
| pKTH5151 | FedF (Lys72, His89→Ala) double mutation in pIH120 | This study |
| pKTH5152 | FedF (His88, His89→Ala) double mutation in pIH120 | This study |
| E. coli strains | ||
| F107/86 | Clinical isolate with F18 fimbriae | 19 |
| DH5αF′ | F′ endA1 hsd17 (rk− mk+) supE44 thi-1 recA1 gyrA (Nalr) relA1Δ(laclZYA-argF) U169 deoR [φ80d lacΔ(lacZ) M15] | 49 |
| HB101 | Hsr− Hsm− RecA− Gal− Pro−str::r F− | 5 |
| ERF2055 | HB101 with pIH120 | 40 |
| ERF2079 | HB101 with pKTH5124 | This study |
| ERF2080 | HB101 with pKTH5125 | This study |
| ERF2081 | HB101 with pKTH5126 | This study |
| ERF2082 | HB101 with pKTH5127 | This study |
| ERF2083 | HB101 with pKTH5128 | This study |
| ERF2084 | HB101 with pKTH5129 | This study |
| ERF2085 | HB101 with pKTH5130 | This study |
| ERF2086 | HB101 with pKTH5131 | This study |
| ERF2087 | HB101 with pKTH5132 | This study |
| ERF2088 | DH5αF′ with pKTH5068 | This study |
| ERF2089 | DH5αF′ with pKTH5069 | This study |
| ERF2090 | DH5αF′ with pKTH5070 | This study |
| ERF2091 | DH5αF′ with pKTH5071 | This study |
| ERF2092 | DH5αF′ with pKTH5072 | This study |
| ERF2093 | DH5αF′ with pKTH5073 | This study |
| ERF2094 | DH5αF′ with pKTH5075 | This study |
| ERF2095 | DH5αF′ with pKTH5076 | This study |
| ERF2096 | DH5αF′ with pKTH5077 | This study |
| ERF2107 | HB101 with pUC18 | This study |
| ERF2114 | DH5αF′ with pKTH5094 | This study |
| ERF2121 | HB101 with pKTH5104 | This study |
| ERF2122 | HB101 with pKTH5105 | This study |
| ERF2123 | HB101 with pKTH5106 | This study |
| ERF2124 | HB101 with pKTH5107 | This study |
| ERF2125 | HB101 with pKTH5108 | This study |
| ERF2126 | HB101 with pKTH5109 | This study |
| ERF2127 | HB101 with pKTH5110 | This study |
| ERF2128 | HB101 with pKTH5111 | This study |
| ERF2143 | HB101 with pKTH5149 | This study |
| ERF2144 | HB101 with pKTH5150 | This study |
| ERF2145 | HB101 with pKTH5151 | This study |
| ERF2146 | HB101 with pKTH5152 | This study |
Numbers following “FedFΔ” represent positions of amino acids deleted from the mature FedF protein.
DNA isolation and cloning methods.
Plasmid DNA of E. coli clones was isolated by alkaline lysis by use of the Wizard Minipreps kit (Promega). PCR products were purified with the QIAquick PCR Purification kit (Qiagen). Restriction enzymes (Promega) and T4 DNA ligase (Roche) were used according to the manufacturers' instructions. PCR was performed under the reaction conditions recommended by the manufacturer of Dynazyme DNA polymerase (Finnzymes) or ThermalAce DNA polymerase (Invitrogen). Other standard DNA procedures were performed according to the work of Sambrook et al. (37). Oligonucleotides were ordered from MedProbe (http://www.medprobe.com or http://www.mwg-biotech.com) and are listed in Table 2.
TABLE 2.
Oligonucleotides used
| Primer | Nucleotide sequence (5′→3′) | Usea |
|---|---|---|
| 508 | CACACAGGAAACAGCTATGAC | R-PCR for internal in-frame deletions of fedF |
| 614 | GTGGAATTCACAAACTCTACTACTGCAGTG | R-PCR for internal in-frame deletions of fedF |
| 615 | GCAAGCTTATGTGGTCTACTTATTACGCG | PCR |
| 815 | ATCTTGATCATTCCGTTACTCTTG | PCR |
| 816 | TCTATGATCACACAAGTCTGTTTCATACGACTGG | PCR |
| 817 | GGAATGATCACACTACCCAAATGCCAGC | PCR |
| 818 | CAATTGATCATTCATTCCTAGTTCATCAGG | PCR |
| 819 | TACGAAGCTTGATCTCAAAACCAGTAAATCG | PCR |
| 820 | CTGAAAGCTTATAATAAACAGGCGTAGCTTGG | PCR |
| 821 | TCATAAGCTTGCTCTTAGACAAATCATACC | PCR |
| 861 | GGAACTATTTTGCAATATGCGCTCGAGTGTCGTGTG | R-PCR for internal in-frame deletions of fedF |
| 862 | GAGCGCATATTGCAAAATAGTTCCAGCCTGGCATG | R-PCR for internal in-frame deletions of fedF |
| 863 | CGTGTGAGCATTCAGTGGGGGCAACAGTCACAAGTAGG | R-PCR for internal in-frame deletions of fedF |
| 864 | TTGCCCCCACTGAATGCTCACACGACACTCGAGCG | R-PCR for internal in-frame deletions of fedF |
| 865 | GGATTCGGTACATTTACTGGTTTTGAGATCTCTTTACG | R-PCR for internal in-frame deletions of fedF |
| 866 | AAAACCAGTAAATGTACCGAATCCTACTTGTGACTG | R-PCR for internal in-frame deletions of fedF |
| 908 | ATCTGGATCCTTCCGTTACTC | PCR |
| 909 | TGTCAAGCTTTATGATCAACGCAAACACAGACAAAGAAATC | PCR |
| 910 | ACTCTGATCATAGACAAGTCTGTTTCATACGACTGG | PCR |
| 911 | GGGATGATCAACACTACCCAAATGCCAGC | PCR |
| 912 | TACATGATCAATTTCATTCCTAGTTCATCAGG | PCR |
| 974 | TGTGGAATTCTCTACTCTACAAGTAGACAAG | PCR |
| 975 | CCTGTCTAGATATGTGGTCTACTTATTACGCG | PCR |
| 976 | GTCTGAATTCTACGACTGGATCAATTCTAGTGCG | PCR |
| 977 | CACTGAATTCATGCCAGCTCTGTATACGTGG | PCR |
| 978 | CAATGAATTCTTCATTCCTAGTTCATCAGG | PCR |
| 985 | GAATTCTAGATTAGATCTCAAAACCAGTAAATCG | PCR |
| 986 | GTGCTCTAGATTAATAATAAACAGGCGTAGCTTGG | PCR |
| 987 | CTGTTCTAGATTAGCTCTTAGACAAATCATACC | PCR |
| 1108 | TTCAGAATTCACTTTGACATGCCAGGCTGG | PCR |
| 1109 | GAATTCTAGATTAGATCTCAAAACCAGTAAATCG | PCR |
| 1110 | TTTGGTATGGGCAAATGGGC | R-PCR, mutation K72A |
| 1111 | GCCCATTTGCCCATACCAAA | R-PCR, mutation K72A |
| 1112 | AAATGGGGCCGAAACCCAATATG | R-PCR, mutation R75A |
| 1113 | CATATTGGGTTTCGGCCCCATTT | R-PCR, mutation R75A |
| 1114 | ATGGGCGCGCAACCCAATATG | R-PCR, mutation E76A |
| 1115 | CATATTGGGTTGCGCGCCCAT | R-PCR, mutation E76A |
| 1116 | ATATGCGCTCGCGTGTCGTGTGAG | R-PCR, mutation E82A |
| 1117 | CTCACACGACACGCGAGCGCATAT | R-PCR, mutation E82A |
| 1118 | GCTCGAGTGTGCTGTGAGCATTCA | R-PCR, mutation R84A |
| 1119 | TGAATGCTCACAGCACACTCGAGC | R-PCR, mutation R84A |
| 1120 | GTGTGAGCATTGCCCATAGTTCTGG | R-PCR, mutation H88A |
| 1121 | CCAGAACTATGGCCAATGCTCACAC | R-PCR, mutation H88A |
| 1122 | GAGCATTCACGCTAGTTCTGGCTC | R-PCR, mutation H89A |
| 1123 | GAGCCAGAACTAGCGTGAATGCTC | R-PCR, mutation H89A |
| 1124 | GCTCCATTAATGCATCTCAGTGGG | R-PCR, mutation E96A |
| 1125 | CCCACTGAGATGCATTAATGGAGC | R-PCR, mutation E96A |
| 1135 | GTGGAAGCTTCATAGACAACCCTTCGATGC | R-PCR |
| 1325 | GTGTGAGCATTGCCGCTAGTTCTGG | R-PCR, double mutation H(88, 89)A |
| 1326 | CCAGAACTAGCGGCAATGCTCACAC | R-PCR, double mutation H(88, 89)A |
R-PCR, recombinant PCR.
DNA sequencing and sequence analysis.
DNA sequencing was performed with an ABI 310 Sequencer by using the ABI PRISM BigDye Terminator Cycle Sequencing kit according to the manufacturer's instructions (Perkin-Elmer Applied Biosystems). Sequence analyses were carried out with the Sequencher 3.0 program (Gene Codes Corporation) and ClustalW.
Deletions in pIH120.
Targeted deletions at the 5′ end of fedF were made by cleaving pIH120 with BclI and HindIII. Because the BclI site is located within the signal sequence of fedF, the missing part of the signal sequence was inserted as a BamHI-HindIII fragment, amplified with primer pair 908-909 from chromosomal E. coli 107/86 DNA, thereby mutating the signal sequence at position 8 (Ile-8→Leu). PCR products amplified with primer pair 816-615, 817-615, or 818-615 by using chromosomal E. coli 107/86 DNA as a template were cloned to the adjacent BclI and HindIII sites designed within primer 909, producing pKTH5124, pKTH5125, and pKTH5126, respectively. Deletions at the 3′ end of fedF were constructed by cloning the PCR products, after amplification by primer pair 815-819, 815-820, or 815-821, into the BclI and HindIII sites of pIH120, producing pKTH5130, pKTH5131, and pKTH5132, respectively. In-frame deletions within fedF in pIH120 were constructed by using the recombinant PCR technique as described previously (16, 25). Briefly, by using E. coli F107/86 chromosomal DNA as a template, a 1.0-kb fragment was amplified with primers 614 and 862 and hybridized to a 0.7-kb fragment amplified with primer pair 861-615. The hybrid was extended by DNA polymerase in a PCR without primers under conditions described by Kylä-Nikkilä et al. (25). The double-stranded recombinant molecules formed were purified, amplified by PCR with primer pair 614-615, and finally cloned into the BclI-HindIII-cut pIH120, producing pKTH5127. Accordingly, plasmid pKTH5128 was constructed by cloning the fragment amplified with primer pair 614-615 into the BclI-HindIII-cut pIH120, by using the hybrid of two annealed PCR products, amplified with primer pairs 614-864 and 863-615, as a template. When pKTH2129 was constructed, the hybrid of the annealed PCR fragments, amplified with primer pairs 614-866 and 865-615, was used as the template in a PCR with primer pair 614-615. Recombinant E. coli strains harboring the vector constructs are listed in Table 1. To confirm the correctness of the constructs, the plasmid DNA of each recombinant strain was isolated and sequenced.
Cloning and expression of truncated fedF.
Six different PCR products generated with primer pairs 975-976, 975-977, 975-978, 974-985, 974-986, and 974-987, by using chromosomal DNA of E. coli 107/86 as a template, were separately cloned as EcoRI-XbaI fragments into pMAL-p2, producing vector constructs pKTH5068, pKTH5069, pKTH5070, pKTH5075, pKTH5076, and pKTH5077, respectively. Plasmids pKTH5127, pKTH5128, and pKTH5129 were used as templates in PCRs with primers 974 and 986. The PCR products synthesized were cloned as EcoRI-XbaI fragments into pMAL-p2 and designated pKTH5071, pKTH5072, and pKTH5073, respectively. The resulting recombinant strains were designated ERF2088 to -2096 (Table 1). A DNA fragment encoding an internal peptide of 64 amino acid residues within FedF (amino acids 60 to 123 of the mature FedF) was also cloned into EcoRI-XbaI-digested pMAL-p2, producing pKTH5094 and E. coli strain ERF2114. All inserts cloned into pMAL-p2 were in frame with malE and expressed truncated forms of FedF as fusion proteins with MBP. For overexpression of all 10 MBP-FedFΔ constructs, E. coli strains ERF2088, ERF2089, ERF2090, ERF2091, ERF2092, ERF2093, ERF2094, ERF2095, ERF2096, and ERF2114 were grown to the exponential phase (optical density at 600 nm [OD600], 0.5), followed by induction in the presence of 0.3 mM isopropylthiogalactoside (IPTG) and propagation for 3 additional hours. Induced fusion proteins were purified from sonicated crude extracts by affinity chromatography as recommended by the manufacturer of the Protein Fusion and Purification system (NEB). A protease inhibitor mix (Complete; Boehringer Mannheim) was used throughout the purification procedure. The amount and purity of the fusion proteins were estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), by using Coomassie R-250 as a dye and increasing amounts of MBP (NEB) as a standard.
Immunoblotting.
Western blotting was carried out by the technique described by Towbin et al. (46). Briefly, truncated FedF proteins fused to MBP were separated by SDS-PAGE and blotted onto a nitrocellulose membrane. After being blocked with 3% blocking reagent (Roche) for 1 h at room temperature (RT), the membrane was incubated with rabbit anti-MBP antibodies (NEB) (diluted 1:50,000 in 3% blocking reagent) at 4°C for 16 h. The membrane was washed with 0.05% Tween 20 in phosphate-buffered saline (PBS) and then incubated with a horseradish peroxidase-conjugated secondary antibody (diluted 1:100,000 in 3% blocking reagent) for 1 h at RT. After a wash with 0.05% Tween 20 in PBS, the membrane was incubated for 5 min with SuperSignal West Dura Extended Duration substrate (Pierce), and the chemiluminescence formed was detected with a molecular imager (GS-525; Bio-Rad).
Directed point mutations of charged amino acid residues in pIH120.
Point mutations directed at the eight charged amino acid residues within the region covering amino acid residues 60 to 109 of FedF were performed by the recombinant PCR technique as described previously (16, 25), by using oligonucleotides possessing the specific mutation(s). The primers used in the primer-directed mutagenesis (1110 to 1125, 1325, and 1326) are specified in Table 2. Recombinant molecules arising from the recombinant PCR step were then amplified with primers 614 and 1135 and cloned into pIH120 digested with NheI and HindIII, producing pKTH5104, pKTH5105, pKTH5106, pKTH5107, pKTH5108, pKTH5109, pKTH5110, pKTH5111, pKTH5149, pKTH5150, pKTH5151, and pKTH5152 (Table 1). The corresponding E. coli strains were designated ERF2121 to -2128 and ERF2143 to -2146, respectively (Table 1). The correctness of the point mutation constructs was verified by DNA sequencing.
In vitro adhesion to porcine epithelial cells.
Porcine small intestine epithelial cells were isolated and prepared as described previously (2, 40). Microscopic examination of the adherence of F18 fimbrial E. coli to jejunal epithelial cells was performed essentially as described by Alwan et al. (2). In order to obtain a semiquantitative estimation of the adhesiveness of E. coli cells, the number of bacteria adhering to 50 randomly chosen epithelial cells was counted after different incubation times (10, 30, and 60 min). Typically, at least two adhesion assays were independently performed with each bacterial strain.
Alkylation of cysteines.
Two hundred microliters (0.5 mg/ml) of MBP-FedFt60-123 protein (amino acids residues 60 to 123 of mature FedF) expressed from pKTH5094 was incubated with an equal volume of a solution containing 0.2 M ammonium carbonate and 20 mM dithiotreitol at 56°C for 30 min. Iodoacetamide was added to a final concentration of 55 mM, followed by incubation for 15 min (at RT) in the dark. The solution was desalted with Ultrafree-15 columns (Millipore) before the adhesion tests.
Indirect immunofluorescence microscopy.
Thawed and washed porcine small intestine epithelial cells (106/ml) were pelleted, resuspended in 250 μl of PBST (PBS containing 1% bovine serum albumin and 0.01% Tween 20), and incubated with an equal volume of proteins (0.1 mg/ml) for 1 h at 37°C. Unattached proteins were removed from the epithelial cells by three washes with PBST, and the cells were resuspended in 250 μl of PBST. The fusion proteins were then incubated with 250 μl of rabbit anti-MBP (NEB), diluted 1:50 in PBST for 1 h at RT, washed twice (with PBST), and resuspended in 250 μl of PBST. A 250-μl volume of fluorescein isothiocyanate-labeled anti-rabbit antibodies (Sigma) was added to the resuspension and incubated for 1 h at RT. After two washing steps, the epithelial cells were resuspended in 60 μl of PBS and air dried on microscopy slides. Adhesion was evaluated by immunofluorescence microscopy using Fluoprep (BioMérieux) as the mounting medium. For inhibition studies, the epithelial cells were preincubated with E. coli ERF2055 cells in PBS (OD600, 0.5) prior to incubation with proteins.
RESULTS
Adhesion capabilities of E. coli strains with mutated FedF.
The FedF protein has recently been identified as the adhesin of F18 fimbriae of E. coli (40). To characterize more closely the region in FedF involved in adhesion to the microvilli of porcine intestinal epithelial cells, a total of nine FedF deletion constructs were generated in plasmid pIH120, expressing the fed gene cluster for the F18 fimbriae in E. coli. Deletions were made at three different sites in the 5′ end of fedF and at three different sites in the 3′ end, and three deletions were targeted within fedF on the sites of four cysteine codons (Table 1). Adhesion tests performed with isolated porcine jejunal epithelial cells showed that all nine recombinant E. coli strains carrying deletions in pIH120 (ERF2079, ERF2080, ERF2081, ERF2082, ERF2083, ERF2084, ERF2085, ERF2086, and ERF2087) had lost their adhesion capabilities (data not shown). A whole-cell enzyme-linked immunosorbent assay (ELISA) with antisera raised against F18 fimbriae (19) revealed that all the recombinant strains expressed amounts of F18 fimbriae on the cell surface similar to that expressed by the E. coli control strain ERF2055. However, in whole-cell ELISA experiments with an anti-MBP-FedF antiserum (40), only one-fifth of the amount of FedF detected for ERF2055 could be detected for the recombinant strains (ERF2079 to ERF2087) (data not shown), suggesting either inefficient assembly or decreased stability of the truncated FedF subunits.
Adhesion of purified truncated FedF proteins expressed as MBP fusions.
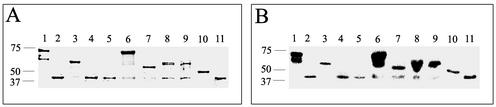
Since the deletions made in pIH120 seemed to affect the interaction of FedF with the F18 fimbrial structure rather than receptor recognition, the fedF deletion derivatives were cloned into vector pMAL-p2 and expressed as fusion proteins with MBP in E. coli (Fig. 1). The fusion proteins were purified directly from the crude extracts by affinity chromatography. E. coli strains ERF2088 (FedFΔ1-10), ERF2090 (FedFΔ1-52), ERF2093 (FedFΔ110-117), ERF2094 (FedFΔ124-280), ERF2095 (FedFΔ192-280), ERF2096 (FedFΔ247-280), and ERF2114 (FedFΔ1-59, 124-280) produced MBP-FedF chimerasof predicted sizes, although degradation products were also detected in addition to the full-length proteins (Fig. 2A). Immunoblotting with anti-MBP antibodies confirmed that all bands detected by Coomassie blue staining were MBP derivatives (Fig. 2B). The fusion proteins produced by E. coli strains ERF2089 (FedFΔ1-37), ERF2091 (FedFΔ70-77), and ERF2092 (FedFΔ88-97) were all very unstable and appeared to be almost completely degraded at the end of the purification (Fig. 2A, lanes 2, 4, and 5).
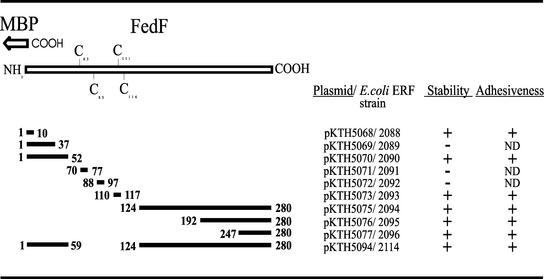
FIG. 1.
MBP-FedF deletion constructs and their adhesion capabilities. Deleted regions of FedF are shown as solid bars with flanking numbers, which refer to amino acid residues of the mature FedF. The pKTH plasmids expressing truncated MBP-FedF fusions are listed, and the stability of each fusion protein is indicated. Adhesiveness refers to the ability of a fusion protein to adhere to porcine small intestine epithelial cells.
FIG. 2.
SDS-PAGE and Western blot analyses of purified truncated MBP-FedF proteins. Fusion proteins were isolated from E. coli strains carrying pKTH5068, pKTH5069, pKTH5070, pKTH5071, pKTH5072, pKTH5073, pKTH5075, pKTH5076, pKTH5077, or pKTH5094 (lanes 1 to 10, respectively). MBP was used as a control (lane 11). (A) Proteins were run on an SDS-polyacrylamide gel and stained with Coomassie dye. (B) Immunoblots with the corresponding samples using an MBP antiserum.
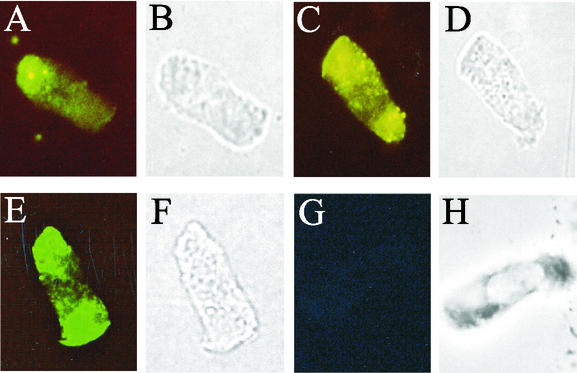
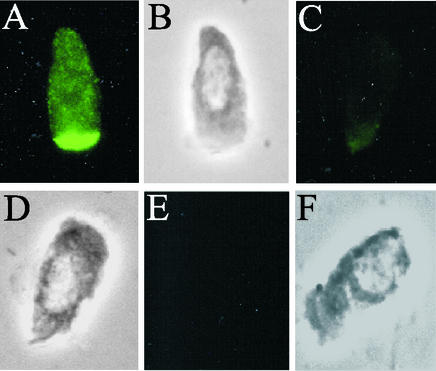
The adhesion capabilities of the fusion proteins purified from ERF2088, ERF2090, ERF2093, ERF2094, ERF2095, ERF2096, and ERF2114 were tested with isolated porcine jejunal epithelial cells. The adhesion of each fusion protein was viewed by fluorescence microscopy, and under the conditions used, all the fusion proteins exhibited bright fluorescence, distinct from that with MBP. Representative images are shown from adhesion analyses with the fusion proteins expressed from pKTH5070 (the longest 5′ deletion of fedF), pKTH5073 (a deletion within fedF), and pKTH5075 (the longest 3′ deletion of fedF) (Fig. 3A, C, and E). MBP was used as a negative control in the adhesion experiments (Fig. 3G). The longest 5′ and 3′ deletions suggested that the N and C termini are not significantly involved in the binding of FedF to its receptor. To confirm this, an MBP fusion construct (pKTH5094) that carried only the FedF internal peptide with amino acid residues 60 to 123 was created. Indeed, immunofluorescence microscopy revealed that MBP-FedFt60-123 bound efficiently to porcine jejunal cells (Fig. 4A). To examine the specificity of the FedFt60-123 adhesion, the ability of MBP-FedFt60-123 to adhere to porcine jejunal epithelial cells, preincubated with ERF2055 bacteria, was evaluated. ERF2055 cells (OD600 in PBS, 0.5) efficiently hindered the attachment of MBP-FedFt60-123 (Fig. 4C).
FIG. 3.
Indirect immunofluorescence of porcine jejunal epithelial cells after incubation with purified fusion proteins expressed from pKTH5070 (A and B), pKTH5073 (C and D), or pKTH5075 (E and F) or with MBP alone (G and H). (A, C, E, and G) Adhesion viewed by fluorescence microscopy; (B, D, F, and H) corresponding fields viewed by phase-contrast microscopy.
FIG. 4.
Indirect immunofluorescence of porcine jejunal epithelial cells after incubation with purified fusion proteins expressed from pKTH5094. (A and B) No pretreatment. (C and D) The porcine jejunal epithelial cells were preincubated with E. coli ERF2055 cells. (E and F) Fusion proteins were alkylated. (A, C, and E) Adhesion viewed by fluorescence microscopy; (B, D, and F) corresponding fields viewed by phase-contrast microscopy.
Since the short internal deletion construct consisting of amino acid residues 110 to 117, including two of the four cysteine residues of FedF, also did not abolish FedF adhesion, the role of the putative disulfide bridges was investigated more closely. When MBP-FedFt60-123 was alkylated with iodoacetamide to prevent it from forming disulfide bridges, the adhesion capability of FedFt60-123 was eliminated (Fig. 4E). This suggests that formation of a disulfide bridge between residues Cys-63 and Cys-83 is essential for the adhesiveness of FedF.
Point mutations at the putative binding region of FedF.
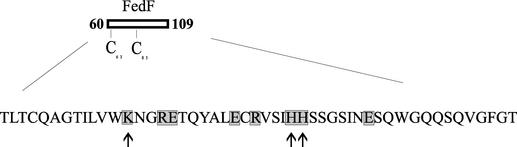
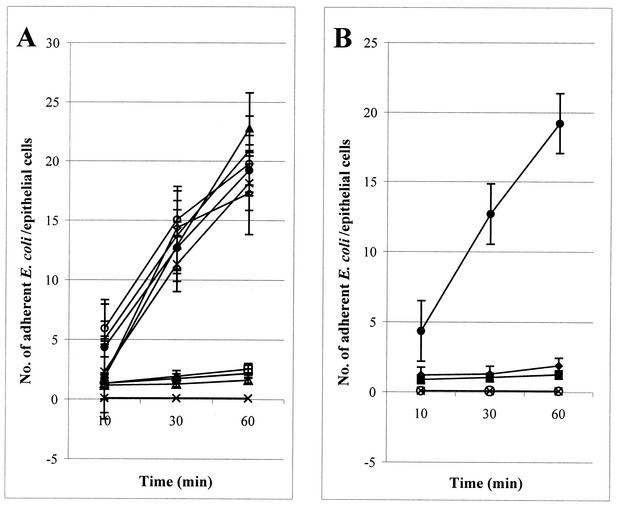
Since the MBP-FedFt60-123 and MBP-FedFΔ110-117 proteins expressed from pKTH5094 and pKTH5073, respectively, mediated adhesion to porcine jejunal epithelial cells in vitro, the region spanning amino acid residues 60 to 109 of the mature FedF was further studied. All the charged amino acids within this region were separately mutated by alanine substitution (Fig. 5). The adhesion capabilities of the resulting E. coli recombinant strains, with F18 fimbriae possessing a single, double, or triple amino acid residue substitution in FedF, are presented (Fig. 6). The abilities of recombinant strains ERF2121 (Lys-72→Ala), ERF2126 (His-88→Ala), ERF2127 (His-89→Ala) (Fig. 6A), ERF2145 (Lys-72 and His-89→Ala), and ERF2146 (His-88 and His-89→Ala) (Fig. 6B) to adhere to isolated porcine jejunal epithelial cells decreased substantially, whereas the triple mutation of ERF2143 (Lys-72, His-88, and His-89→Ala) and the double mutation of ERF2144 (Lys-72 and His-88→Ala) completely abolished adhesion to porcine enterocytes (Fig. 6B). With the other point mutations, the adhesion capability remained unaffected (Fig. 6A). The expression of F18 fimbriae and the amount of FedF in the F18 fimbrial filament for all the recombinant strains possessing the single, double, or triple point mutation in fedF were determined by whole-cell ELISA with anti-F18 fimbriae and anti-MBP-FedF sera. No significant differences in the FedF and F18 fimbrial signals were observed for any of the strains (data not shown), indicating that the point mutation constructs did not interfere with the synthesis of FedF or its attachment to the other F18 fimbrial molecules. By sequencing all the inserts harboring the point mutation, it was further confirmed that the recombinant strains with altered adhesion capability carried only the intended single, double, or triple mutation.
FIG. 5.
Point mutations of charged amino acid residues within the region spanning positions 60 to 109 of the mature FedF. Amino acid residues replaced with alanine are shaded, and amino acid residues affecting the ability of F18 fimbriae to adhere to porcine jejunal epithelial cells are indicated by arrows.
FIG. 6.
Numbers of E. coli bacteria adhering to porcine jejunal epithelial cells after 10, 30, and 60 min of incubation. (A) E. coli strains ERF2121 (open triangles), ERF2122 (solid triangles), ERF2123 (open diamonds), ERF2124 (plus signs), ERF2125 (stars), ERF2126 (horizontal lines), ERF2127 (open squares), and ERF2128 (open circles) each carry a single point mutation in FedF. (B) ERF2143 (multiplication signs) harbors a triple point mutation, and ERF2144 (multiplication signs), -2145 (solid squares), and -2146 (solid diamonds) each harbor a double mutation in FedF. E. coli ERF2055 (solid circles) and the nonfimbriated strain ERF2107 (multiplication signs) were used as positive and negative adhesion controls, respectively. For clarity, the same symbol (multiplication sign) was used for the mutant strains ERF2143 and ERF2144 and the negative-control strain ERF2107, because all these strains were nonadhesive. Results are shown as group means with 95% confidence intervals.
Sequencing of the fedF gene from clinical isolates.
To study the genetic variation in fedF, we also sequenced the fedF gene from 15 independent Finnish clinical E. coli isolates carrying either F18ab or F18ac fimbriae. Alignment of the deduced FedF amino acid sequences from the isolates revealed the highly conserved nature of FedF. All 15 FedF sequences were at least 97% identical with the sequence of the E. coli strain used in this work, F107/86 (data not shown). Table 3 shows the differences found in the predicted amino acid sequences of these FedF proteins. None of these amino acid differences in the FedF proteins affected the ability of the corresponding E. coli isolate to adhere to porcine jejunal epithelial cells (data not shown). Eight of the E. coli isolates carried a mutation at position Asn-73, which is located within the proposed binding domain of FedF (Table 3).
TABLE 3.
Amino acid differences between the F18 FedF proteins of Finnish clinical E. coli isolates and that of E. coli 107/86
| E. coli isolate (serotype) | Amino acid difference(s) |
|---|---|
| ERF2056 (O139) | None |
| ERF2057 (O139) | E151→Q |
| ERF2058 (O139) | None |
| ERF2059 (O139) | E122→K, T219→A |
| ERF2060 (O139) | E122→K |
| ERF2061 (O139) | N73→D, M211→T |
| ERF2062 (O139) | S20→N, N73→D, A110→T |
| ERF2063 (O139) | N73→D |
| ERF2064 (O139) | N73→D |
| ERF2065 (O141) | N73→D, E122→G, S144→R |
| ERF2066 (O141) | Q47→K, N73→G, N153→K |
| ERF2067 (O141) | Q47→K, N73→G, N153→K, T237→R |
| ERF2068 (O141) | T14→V, Q47→K, N73→G, N153→K |
| ERF2069 (O138) | Q47→K, T69→M, A110→G, D170→H, D235→N |
| ERF2070 (O157) | D6→G, Q47→K, V105→A, A110→G, T133→I, K142→I, D170→H, T201→S |
DISCUSSION
The genetic organization of the subunits involved in the biosynthesis of F18 fimbriae and the role of FedF as the adhesin of F18 fimbriae have been reported recently (40). In this study, the region (amino acid residues 60 to 109 of FedF) responsible for in vitro binding of F18 fimbriae to porcine small intestine epithelial cells was localized. In addition, three amino acid residues (Lys-72, His-88, and His-89) important for receptor binding were found within this region.
None of the recombinant strains harboring in-frame deletions of fedF within the F18 fimbria-expressing plasmid pIH120 were adhesive under the test conditions used, although the amount of F18-cross-reacting materials remained unchanged on the cell surface. Two C-terminal deletions of FedF have been constructed previously by Imberechts et al. (20). Truncation of FedF from amino acid residue 125 eliminated the adhesion of E. coli mutant strains to isolated porcine villi, even though F18 fimbriae were visible with an electron microscope. On the other hand, FedF with a C-terminal deletion from amino acid residue 204 was still able to mediate adhesion to porcine villi in vitro, suggesting a nonessential function of the C terminus for receptor recognition and binding (20). This result was not supported by the present work, as FedF (expressed from pKTH5132) truncated from amino acid residue 247 had lost its adhesiveness for isolated porcine small intestine cells. Whole-cell ELISA experiments with an MBP-FedF antiserum suggest that the lack of adhesiveness of the F18 fimbriae expressed by these recombinant strains is associated with the assembly or stability of fimbrial subunits rather than with changes directly affecting receptor recognition. The C-terminal motif of the E. coli type 1 and P fimbrial subunits, essential for subunit-subunit interactions, typically includes a series of alternating hydrophobic amino acid residues, which are flanked by a glycine, located 14 residues upstream of the C terminus, and a penultimate tyrosine (42). The F18 fimbrial subunits also have a highly hydrophobic C-terminal region including the features described for a putative binding region for fimbrial chaperones. The only noticeable exception is that FedF has an isoleucine residue instead of the penultimate tyrosine. The penultimate tyrosine is also absent in the P-type adhesin (PapG), which possesses another amino acid residue with an aromatic side chain (phenylalanine) at that position (42).
Since the FedF-MBP fusion protein unexpectedly adhered to isolated porcine epithelial cells without the involvement of the F18 fimbrial chaperone, FedC (40), adhesion tests were carried out with truncated MBP-FedF proteins in order to assay the adherence more directly. A fragment encoding a 64-amino-acid peptide of FedF tagged with MBP (MBP-FedFt60-123) still adhered to isolated jejunal epithelial cells. In fact, all the stable truncated fusion proteins exhibited fluorescence intensities close to that of full-length FedF tagged with MBP in adhesion tests. Although we strove for a semiquantitative estimate of the number of fusion proteins attached to the porcine epithelial cells, we cannot exclude the possibility that all (or some) of the deletions in fedF affected the adhesion capability to some extent, since minor changes in the intensity of the fluorescence were undetectable. Characterization of the binding constants would be more informative, but that is not feasible until the receptor structure is determined. The receptor for F18 fimbriae is thus far unknown, but the activity of α(1,2)-fucosyltransferase (FUT1) in the epithelia of piglets has been reported to be essential for adhesion of F18 fimbrial bacteria to small intestine mucosa (29).
Because the truncated fusion proteins expressed from both pKTH5073 and pKTH5094 were able to attach to porcine intestinal epithelial cells, the remaining region (amino acid residues 60 to 109 of FedF) was investigated more closely by site-directed point mutations. Three amino acid residues essential for efficient receptor binding were found (Lys-72, His-88, and His-89). It has previously been demonstrated that a single amino acid substitution associated with receptor binding can be crucial for adhesion capability (23, 35, 39). Replacement of a lysine residue with alanine inhibited attachment of 987P fimbriae to glycolipid receptors on the brush borders of piglet enterocytes (7), and the involvement of histidine residues in the binding of alpha-bungarotoxin to the Torpedo californica or the Homo sapiens receptor has been described (47).
Lysine and arginine residues are essential for the binding of K99 pili and S fimbriae to their receptors, which contain negatively charged sialic acid (21, 33). However, no stretches of positively charged amino acid residues similar to those in the K99 fibrillar adhesin or SfaS that have been found to mediate binding are present in the binding domain of FedF, suggesting a different receptor structure. Furthermore, the possibility that the decreased adhesion capability caused by three substitutions of the amino acid residues in FedF is related to changes in the tertiary structure of the binding domain, rather than to direct interaction between the receptor and these residues, cannot be excluded. The binding domain of FedF is located at the amino-terminal half of the protein, as in many other bacterial adhesins (8, 15, 24, 36, 38, 44, 45). In many cases, adhesins are folded into two domains, an N-terminal receptor binding domain and a C-terminal chaperone binding domain, involved in pilus assembly (8, 15, 44).
The binding domain of FedF contains two cysteine residues, as do those of the PapG, FimH, DraE, and GafD adhesins. Nuclear magnetic resonance studies of the binding domain of PapGII revealed that the cysteines form a disulfide bridge (44). To evaluate whether the cysteines (Cys-63 and Cys-83) found in the binding domain of FedF form a disulfide bridge essential for adhesion, they were alkylated with iodoacetamide prior to adhesion analysis. Not surprisingly, alkylation eliminated the adhesion capability of the binding domain, indicating the significance of a putative disulfide structure. Similar results have been obtained with DraE, where formation of a disulfide bond was reported to be essential for adhesion of the Dr fimbriae to their receptor (6). Formation of a second disulfide bridge between the two remaining cysteines in FedF (at positions 111 and 116) appeared not to be essential for adhesiveness, since they could be deleted without affecting the adhesion capability of FedF.
We were unable to inhibit in vitro adhesion of the MBP-FedFt64-123 fusion protein to porcine epithelial cells by use of a 1:10-diluted anti-MBP-FedF antiserum (40) (data not shown). Possibly this small region (amino acid residues 64 to 123) of FedF was not accessible for efficient antibody recognition or the titer of specific antibodies in the MBP-FedF serum was too low. ERF2055 cells, in contrast, efficiently inhibited the attachment of MBP-FedFt64-123 fusion proteins to porcine epithelial cells, indicating the same binding specificity. However, the MBP-FedFt64-123 fusion protein was unable to inhibit the attachment of ERF2055 bacteria to isolated porcine epithelial cells (data not shown), thus suggesting a stronger affinity of native FedF than of MBP-FedFt64-123 for the host receptor.
F18 E. coli infections of weaned porcines on farms are usually fatal and cause great economic losses (3, 14). Since there are no commercial vaccines available (32), FedF, or rather the binding domain of FedF, could be an attractive vaccine candidate. The ability of adhesins such as FimH (26, 27, 28) and SfbI (13) to prevent the establishment of bacterial infection has been demonstrated. Polymorphism in major antigen components is thought to reduce the efficiency of vaccines against respiratory pathogens (31); therefore, presenting a minimal binding domain to the immune system may be advantageous. Adhesins recognize invariant host receptors and are therefore highly conserved proteins (1, 10, 48). This was also supported by our study. Alignment of the deduced amino acid sequences of the fedF genes sequenced from 15 clinical isolates also revealed that FedF is highly conserved among F18 fimbrial E. coli strains.
Acknowledgments
The Academy of Finland, the Ministry of Agriculture and Forestry, and the Faculty of Veterinary Medicine, University of Helsinki, supported this work.
We are grateful to Sinikka Ahonen and Ulla Viitanen for excellent technical assistance. Ilkka Palva is acknowledged for valuable discussions and critical reading of the manuscript.
Editor: V. J. DiRita
REFERENCES
- 1.Abraham, S. N., D. Sun, J. B. Dale, and E. H. Beachey. 1988. Conservation of the d-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature 336:682-684. [DOI] [PubMed] [Google Scholar]
- 2.Alwan, A., T. Deignan, M. O'Sullivan, J. Kelly, and C. O'Farrelly. 1998. Quantitative assay of Salmonella adherence to intestinal epithelial cells: a new method for assessing novel intervention products. J. Microbiol. Methods 33:163-170. [Google Scholar]
- 3.Bertschinger, H. U., and C. L. Gyles. 1994. Oedema disease of pigs, p. 193-219. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Oxford, United Kingdom.
- 4.Bertschinger, H. U., V. Nief, and H. Tschäpe. 2000. Active oral immunization of suckling piglets to prevent colonization after weaning by enterotoxigenic Escherichia coli with fimbriae F18. Vet. Microbiol. 71:255-267. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in E. coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Carnoy, C., and S. L. Moseley. 1997. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol. Microbiol. 23:365-379. [DOI] [PubMed] [Google Scholar]
- 7.Choi, B. K., and D. M. Schifferli. 1999. Lysine residue 117 of the FasG adhesin of enterotoxigenic Escherichia coli is essential for binding of 987P fimbriae to sulfatide. Infect. Immun. 67:5755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury, D. A., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 9.Gaastra, W., and A.-M. Swennerholm. 1996. Colonization factors of human entero-toxigenic Escherichia coli. Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 10.Gerlach, G. F., S. Clegg, and B. L. Allen. 1989. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J. Bacteriol. 171:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons, R. J. 1973. Bacterial adherence in infection and immunity. Rev. Microbiol. 4:48-60. [Google Scholar]
- 12.Guan, C., P. Li, P. D. Riggs, and H. Inouye. 1987. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene 67:21-30. [DOI] [PubMed] [Google Scholar]
- 13.Guzmán, C. A., R. S. Talay, G. Molinary, E. Medina, and G. S. Chhatwal. 1999. Protective immune response against Streptococcus pyogenes in mice after intranasal vaccination with the fibronectin-binding protein SfbI. J. Infect. Dis. 179:901-906. [DOI] [PubMed] [Google Scholar]
- 14.Hampson, D. J. 1994. Postweaning Escherichia coli diarrhoea in pigs, p. 171-191. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Oxford, United Kingdom.
- 15.Haslam, D. B., T. Boren, P. Falk, D. Ilver, A. Chou, Z. Xu, and S. Normark. 1994. The amino-terminal domain of the P-pilus adhesin determines receptor specificity. Mol. Microbiol. 14:399-409. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. Academic Press, San Diego, Calif.
- 17.Imberechts, H., H. U. Bertschinger, M. Stamm, T. Sydler, P. Pohl, H. De Greve, J.-P. Hernasteens, M. Van Montagu, and P. Lintermans. 1994. Prevalence of F107 fimbriae on Escherichia coli isolated from pigs with oedema disease or postweaning diarrhoea. Vet. Microbiol. 40:219-230. [DOI] [PubMed] [Google Scholar]
- 18.Imberechts, H., P. Deprez, E. Van Driessche, and P. Pohl. 1997. Chicken egg yolk antibodies against F18ab fimbriae of Escherichia coli inhibit shedding of F18 positive E. coli by experimentally infected pigs. Vet. Microbiol. 54:329-341. [DOI] [PubMed] [Google Scholar]
- 19.Imberechts, H., H. De Greve, C. Schlicker, H. Bouchet, P. Pohl, G. Charlier, H. U. Bertschinger, P. Wild, J. Vandekerckhove, J. Van Damme, M. Van Montagu, and P. Lintermans. 1992. Characterization of F107 fimbriae of Escherichia coli 107/86, which causes edema disease in pigs, and nucleotide sequence of the F107 major fimbrial subunit gene, fedA. Infect. Immun. 60:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imberechts, H., P. Wild, G. Charlier, H. De Greve, P. Lintermans, and P. Pohl. 1996. Characterization of F18 fimbrial genes fedE and fedF involved in adhesion and length of enterotoxemic Escherichia coli strain 107/86. Microb. Pathog. 21:183-192. [DOI] [PubMed] [Google Scholar]
- 21.Jakobs, A. A. C., B. H. Simons, and F. K. E. J. De Graaf. 1987. The role of lysine-132 and arginine-136 in the receptor-binding domain of the K99 fibrillar subunit. EMBO J. 6:1805-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, G. W. 1997. The attachment of bacteria to the surface of animal cells, p. 139-176. In J. Reissig (ed.), Microbial interactions. Chapman and Hall, London, United Kingdom.
- 23.Klemm, P., and M. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 24.Korhonen, T. K., V. Vaisanen-Rhen, M. Rhen, A. Pere, J. Parkkinen, and J. Finne. 1984. Escherichia coli fimbriae recognizing sialyl galactosides. J. Bacteriol. 159:762-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kylä-Nikkilä, K., M. Hujanen, M. Leisola, and A. Palva. 2000. Metabolic engineering of Lactobacillus helveticus CNRZ32 for production of pure l-(+)-lactic acid. Appl. Environ. Microbiol. 66:3835-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langermann, S., R. Möllby, J. E. Burlein, S. R. Palaszynski, C. G. Auguste, A. DeFusco, R. Strouse, M. A. Schenerman, S. J. Hultgren, J. S. Pinkner, J. Winberg, L. Guldevall, M. Söderhäll, K. Ishikawa, S. Normark, and S. Koenig. 2000. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J. Infect. Dis. 181:774-778. [DOI] [PubMed] [Google Scholar]
- 27.Langermann, S., and W. R. Ballou, Jr. 2001. Vaccination utilizing the FimCH complex as a strategy to prevent Escherichia coli urinary tract infections. J. Infect. Dis. 183:84-86. [DOI] [PubMed] [Google Scholar]
- 28.Langermann, S., S. Palaszynski, M. Barnhart, G. Auguste, J. S. Pinkner, J. Burlein, P. Barren, S. Koenig, S. Leath, C. H. Jones, and S. J. Hultgren. 1997. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276:607-611. [DOI] [PubMed] [Google Scholar]
- 29.Meijerink, E., S. Neuenschwander, R. Fries, A. Dinter, H. U. Bertschinger, G. Stranzinger, and P. Vogeli. 2000. A DNA polymorphism influencing α(1,2)fucosyltransferase activity of the pig FUT1 enzyme determines susceptibility of small intestinal epithelium to Escherichia coli F18 adhesion. Immunogenetics 52:129-136. [DOI] [PubMed] [Google Scholar]
- 30.Moch, T., H. Hoschützky, J. Hacker, K. D. Kroncke, and K. Jann. 1987. Isolation and characterization of the α-sialyl-β-2,3-galactosyl-specific adhesin from fimbriated Escherichia coli. Proc. Natl. Acad. Sci. USA 84:3462-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mooi, F. R., H. van Oirschot, K. Heuvelman, H. G. J. van der Heide, W. Gaastra, and R. J. L. Willems. 1998. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxoid in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 66:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon, H. W., and T. O. Bunn. 1993. Vaccines for preventing enterotoxigenic Escherichia coli infections in farm animals. Vaccine 11:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morschhäuser, J., H. Hoschützky, K. Jann, and J. Hacker. 1990. Functional analysis of the sialic acid-binding adhesin SfaS of pathogenic Escherichia coli by site-specific mutagenesis. Infect. Immun. 58:2133-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy, B., S. C. Whipp, H. Imberechts, H. U. Bertschinger, E. A. Dean-Nystrom, T. A. Casey, and E. Salajka. 1997. Biological relationship between F18ab and F18ac fimbriae of enterotoxigenic and verotoxigenic Escherichia coli from weaned pigs with oedema disease or diarrhoea. Microb. Pathog. 22:1-11. [DOI] [PubMed] [Google Scholar]
- 35.Pouttu, R., T. Puustinen, R. Virkola, J. Hacker, P. Klemm, and T. K. Korhonen. 1999. Amino acid residue Ala-62 in the FimH fimbrial adhesin is critical for the adhesiveness of meningitis-associated Escherichia coli to collagens. Mol. Microbiol. 31:1747-1757. [DOI] [PubMed] [Google Scholar]
- 36.Riegman, N., R. Kusters, H. van Vaggel, H. Bergmans, P. van Bergenen Henegouwen, J. Hacker, and I. van Die. 1990. FIC fimbriae of a uropathogenic Escherichia coli: genetic and functional organization of the foc gene cluster and identification of minor components. J. Bacteriol. 172:1114-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schembri, M. A., H. Hasman, and P. Klemm. 2000. Expression and purification of the mannose recognition domain of the FimH adhesin. FEMS Microbiol. Lett. 188:147-151. [DOI] [PubMed] [Google Scholar]
- 39.Schembri, M. A., L. Pallesen, H. Connell, D. L. Hasty, and P. Klemm. 1996. Linker insertion analysis of the FimH adhesin of type 1 fimbriae in an Escherichia coli fimH-null background. FEMS Microbiol. Lett. 137:257-263. [DOI] [PubMed] [Google Scholar]
- 40.Smeds, A., K. Hemmann, M. Jakava-Viljanen, S. Pelkonen, H. Imberechts, and A. Palva. 2001. Characterization of the adhesin of Escherichia coli F18 fimbriae. Infect. Immun. 69:7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smyth, C. J., M. B. Marron, J. M. G. J. Twohig, and S. G. J. Smith. 1996. Fimbrial adhesins: similarities and variations in structure and biogenesis. FEMS Immun. Med. Microbiol. 16:127-139. [DOI] [PubMed] [Google Scholar]
- 42.Soto, G. E., K. W. Dodson, D. Ogg, C. Liu, J. Heuser, S. Knight, J. Kihlberg, C. H. Jones, and S. J. Hultgren. 1998. Periplasmic chaperone recognition motif of subunits mediates quaternary interactions in the pilus. EMBO J. 17:6155-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung, M., K. Fleming, H. A. Chen, and S. Matthews. 2001. The solution structure of PapGII from uropathogenic Escherichia coli and its recognition of glycolipid receptors. EMBO Rep. 2:621-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanskanen, J., S. Saarela, S. Tankka, N. Kalkkinen, M. Rhen, T. K. Korhonen, and B. Westerlund-Wikström. 2001. The gaf fimbrial gene cluster of Escherichia coli express a full-size and a truncated soluble adhesin protein. J. Bacteriol. 183:512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venera, G. D., F. D. Testai, C. Pena, H. D. Lacorazza, and M. J. Biscoglio De Jimenez Bonino. 1997. Involvement of histidine 134 in the binding of alpha-bungarotoxin to the nicotinic acetylcholine receptor. Neurochem. Int. 31:151-157. [DOI] [PubMed] [Google Scholar]
- 48.Wizemann, T. M., J. E. Adamou, and S. Langermann. 1999. Adhesins as targets for vaccine development. Emerg. Infect. Dis. 5:395-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodcock, D., P. Crowther, J. Doherty, S. Jefferson, E. De Cruz, M. Noyer-Weidner, S. Smith, M. Michael, and M. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokoyama, H., T. Hashi, K. Umeda, M. Kuroki, Y. Ikemori, and Y. Kodama. 1997. Effect of oral egg antibody in experimental F18+ Escherichia coli infection in weaned pigs. J. Vet. Med. Sci. 59:917-921. [DOI] [PubMed] [Google Scholar]