Abstract
Escherichia coli is one of the most common gram-negative bacteria that cause meningitis in neonates. Our previous studies have shown that outer membrane protein A (OmpA) of E. coli interacts with a 95-kDa human brain microvascular endothelial cell (HBMEC) glycoprotein, Ecgp, for invasion. Here, we report the identification of a gene that encodes Ecgp by screening of an HBMEC cDNA expression library as well as by 5′ rapid amplification of cDNA ends. The sequence of the Ecgp gene shows that it is highly similar to gp96, a tumor rejection antigen-1, and contains an endoplasmic reticulum retention signal, KDEL. Overexpression of either Ecgp or gp96 in both HBMECs and CHO cells increases E. coli binding and invasion. We further show that Ecgp gene-transfected HBMECs express Ecgp on the cell surface despite the presence of the KDEL motif. Northern blot analysis of total RNA from various eukaryotic cells indicates that Ecgp is significantly expressed in HBMECs. Recombinant His-tagged Ecgp blocked E. coli invasion efficiently by binding directly to the bacteria. These results suggest that OmpA of E. coli K1 interacts with a gp96-like molecule on HBMECs for invasion.
Escherichia coli K1 is the most frequent causative agent of neonatal meningitis. The pathogenic mechanisms of E. coli have been studied by utilizing brain microvascular endothelial cells (BMEC) as an in vitro blood-brain barrier (BBB) model (17, 19, 20). These studies suggest that S fimbriae mediate attachment to BMEC via NeuAc2,3-Gal epitopes of BMEC surface glycoproteins; however, they do not play a significant role in invasion (25). More-intimate attachment by the bacterial outer membrane protein A (OmpA) mediates the invasion process (15). A similar phenomenon has been identified in the pathogenesis of Neisseria gonorrhoeae, where pili promote initial adherence followed by Opa-mediated interaction for invasion (10). In addition to OmpA, other bacterial factors such as IbeA, IbeB, TraJ, and CNF also play roles in E. coli invasion of BMEC; however, OmpA appears to be the most important factor (2, 6, 7, 9). OmpA+ E. coli induces actin rearrangements at the site of bacterial entry, which are completely abolished by treatment of the bacteria with GlcNAcβ1-4GlcNAc polymers, which are receptor analogs (16, 17). Computer simulation studies of the interactions between OmpA and GlcNAcβ1-4GlcNAc epitopes indicate that these sugars have more favorable energy than any other sugar molecule tested in our experiments (4). These results are in good agreement with earlier studies in which GlcNAcβ1-4GlcNAc moieties showed significant blocking of E. coli invasion both in vitro and in vivo (16).
In support of the role of OmpA in E. coli K1 invasion, studies have also demonstrated that OmpA+ E. coli induces the phosphorylation of focal adhesion kinase (FAK) and its interaction with phosphatidylinositol 3-kinase (PI3K) (20, 21). Furthermore, GlcNAcβ1-4GlcNAc polymers blocked the activation of FAK, although at higher concentrations, indicating the role of the human BMEC (HBMEC) receptor for OmpA in transducing signals for internalization of E. coli. In addition, it was shown that E. coli also induces the activation of protein kinase C alpha (PKC-α) in an OmpA-dependent manner (27). The activated PKC-α is recruited to the plasma membrane, where it interacts with caveolin-1, a protein marker for caveolae, for internalization of E. coli (28). Several receptors, such as epidermal growth factor and fibroblast growth factor (14), have been shown to accumulate in caveolae, suggesting that the OmpA receptor could be part of caveolae during E. coli internalization.
Subsequently, it was demonstrated that OmpA binds to a 95-kDa HBMEC receptor, Ecgp, for E. coli invasion (18). OmpA-Ecgp interaction occurs via GlcNAc1,4-GlcNAc epitopes as well as the protein backbone of the receptor. The presence of Ecgp on HBMEC, but not on nonbrain endothelial cells, as shown by anti-Ecgp antibody immunocytochemistry, indicates the potential role of Ecgp in the neurotropic nature of E. coli. In addition, Ecgp clusters at the bacterial entry site, similar to actin and caveolin-1 accumulation, underscoring the importance of Ecgp in signaling for the uptake of E. coli (18). The N-terminal amino acid sequence of the 95-kDa protein was identical to that of gp96, a cell surface glycoprotein related to heat shock proteins (12). In contrast, the 65-kDa fragment, which could be an internal fragment, showed no significant homology to any known proteins in the database.
Here, we report the identification of the gene encoding Ecgp by screening of an HBMEC cDNA expression library with an anti-Ecgp antibody and by 5′ rapid amplification of cDNA ends (5′ RACE). DNA sequencing of the gene indicated that Ecgp is a gp96 homologue and that the 65-kDa cleavage product is an internal portion of Ecgp. In both HBMEC and CHO cells, overexpression of the Ecgp gene resulted in a threefold increase in invasion, suggesting a role for Ecgp in E. coli invasion of HBMEC. In support of this notion, recombinant Ecgp (rEcgp) significantly blocked E. coli invasion by directly binding to the bacteria.
MATERIALS AND METHODS
Bacteria.
Strain E44 is a spontaneous rifampin-resistant mutant of E. coli K1 strain RS 218 (serotype O18:K1:H7) isolated from the cerebrospinal fluid of a neonate with meningitis. E91 is a noninvasive strain derived from E44 in which the ompA gene has been disrupted (15). All bacterial media were purchased from Difco Laboratories (Detroit, Mich.). Bacteria were grown overnight in brain heart infusion broth with appropriate antibiotics.
E. coli invasion assays.
E. coli invasion of HBMEC was performed as described previously (15, 16). Briefly, confluent endothelial cell monolayers in 24-well plates were incubated with 107 E. coli bacteria (multiplicity of infection, 1:100) in experimental medium (M199-Ham F-12 medium [1:1] containing 5% heat-inactivated fetal bovine serum, 2 mM glutamine, and 1 mM pyruvate) for 1.5 h at 37°C. Monolayers were then washed with RPMI and further incubated with experimental medium containing gentamicin (100 μg/ml) for 1 h to kill extracellular bacteria. Monolayers were washed again and lysed in 0.5% Triton X-100. The released intracellular bacteria were enumerated by plating on sheep blood agar plates. In duplicate experiments, total cell-associated bacteria were determined as described above except that the gentamicin step was omitted. For inhibition studies, antibodies and protein fractions at different concentrations were incubated with either OmpA+ or OmpA− E. coli on ice before being added to the monolayers. The effects of the inhibitors on HBMEC were assessed by the trypan blue dye exclusion method, and their effects on bacterial viability were assessed by colony plating (15).
Construction of an HBMEC cDNA library.
The HBMEC cDNA library was generated with >99% pure HBMEC isolated as described earlier with no detectable contamination by nonendothelial cells (e.g., glial cells and pericytes). Briefly, total mRNA was isolated by the guanidine isothiocyanate method (5), and poly(A)+ mRNA was purified from total mRNA by using an oligo(dT)-cellulose column (Stratagene). The purified mRNA was used for construction of a cDNA expression library in the Lambda ZAP Express vector at the EcoRI restriction site (which was done at Stratagene). Moloney murine leukemia virus reverse transcriptase was used for first-strand synthesis, and DNA polymerase I and RNase H were used for second-strand synthesis. The cDNA synthesis reaction was primed with oligo(dT) and random primers. The primary plaques were about 3.2 × 106 PFU with an estimated background of 9% nonrecombinants. The quality of the cDNA library was assessed by examining three markers known to be present in HBMEC, for example, gamma glutamyl transpeptidase (GGT; specific to brain cells), ICAM-1, and VCAM-1 (present on endothelial cells).
The following primers were designed for PCR: (i) 5′ primer 5′-CAGAGTTCAACTGGAGAC-3′ and 3′ primer 5′-CTCATAGCCTCGGATCTC-3′ for a 0.72-kb GGT fragment (positions 195 to 913); (ii) 5′ primer 5′-TGGCAGCCAGTGGGCAAGAACC-3′ and 3′ primer 5′-GGTTCCCTTCTGAGACCTC-3′ for a 0.6-kb ICAM-1 fragment (positions 427 to 1035); and (iii) 5′ primer 5′-AGCTTTTAAAATCGAGAC-3′ and 3′ primer 5′-AAGGTGAGAGTTGCATTTC-3′ for a 0.76-kb VCAM-1 fragment (positions 189 to 953).
Screening of the HBMEC cDNA library with an anti-Ecgp antibody.
The HBMEC cDNA library was screened through three rounds with an anti-Ecgp antibody (dilution, 1:1,000) as described previously (8). An unrelated antibody was used as a negative control. The positive plaques after the third round of screening were excised from the agar and transferred to a sterile microcentrifuge tube containing 500 μl of SM buffer (1 M Tris-HCl buffer [pH 7.5] containing NaCl, MgSO4, and 2% gelatin) and 20 μl of chloroform. The plaques were left at 4°C overnight to diffuse into the buffer. The number of plaques in the buffer was calculated for each clone. Then the cDNA inserts from each clone were rescued in the Bluescript SK-phagemid by the in vivo excision protocol of Stratagene. Subsequent screens were performed by using the cDNA insert of clone c (7c) as a probe. All of the clones were sequenced by using primers T7 and T3.
5′ RACE.
Total RNAs from both HBMEC and human umbilical vein endothelial cells (HUVEC) were subjected to 5′ RACE by using a kit from Life Technologies, Inc. We used a specific primer with the sequence 5′-GACATCGACTATGGCGACCGACATAAG-3′ and an abridged anchor primer provided by the manufacturer for the 5′ RACE. The PCR product was ligated into the pAMP vector (Life Technologies, Inc.) and used to transform DH5α-competent cells. The 5′ RACE product was sequenced by using primers SP6 and T7.
Northern and Southern blot analysis.
The cDNA insert of clone 7c was labeled with biotin-dCTP by using a random 5′ primer DNA labeling kit (Amersham) and was used in Northern blot analysis. For Southern hybridization, a degenerate oligonucleotide obtained from the N-terminal amino acid sequence of the 65-kDa protein was labeled with [32P]dCTP by using a kit (Boehringer Mannheim). Prehybridization, hybridization, and washing were carried out as described by Sambrook et al. (24). Conditions for the final wash were 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 55°C.
HBMEC and CHO cell culture maintenance and transfections.
HBMEC were isolated and cultured as described previously (25). HBMEC cultures were routinely grown in RPMI 1640 containing 10% heat-inactivated fetal bovine serum, 10% Nu-serum, 2 Mm glutamine, 1 Mm sodium pyruvate, penicillin (100 U/ml), streptomycin (100 μg/ml), essential amino acids, and vitamins. CHO cells were obtained from the American Type Culture Collection (clone CCL-61) and cultured by using Ham F-12 medium with 10% heat-inactivated fetal bovine serum. Both HBMEC and CHO cells were transfected with mammalian expression vectors by using Lipofectamine as described elsewhere (27). Briefly, DNA-Lipofectamine in RPMI 1640 was added to 50% confluent HBMEC monolayers. After 6 h of incubation at 37°C, the cells were washed with RPMI-1640 and respective media were added. Three days following transfection, the cells were transferred to media containing the antibiotic G418 (400 μg/ml) for 3 weeks, and antibiotic-resistant colonies were either pooled or separated into clones for further analysis.
Purification of recombinant histidine-tagged Ecgp.
Ecgp-transfected CHO cells were grown in large quantities and resuspended in phosphate-buffered saline (PBS) containing 0.5% Triton X-100 and protease inhibitors, and cell lysates were prepared as described elsewhere (16). Cell lysates were passed through a Ni-nitrilotriacetic acid (NTA) Sepharose column (2 by 10 cm) over a period of 1 h at 4°C, followed by thorough washing with loading buffer. Bound proteins were eluted with a 0.1 M imidazole solution and dialyzed against PBS overnight at 4°C. The protein solution was concentrated by using Centricon tubes, and the protein content was estimated by the Bio-Rad method. Purified rEcgp (10 μg) was incubated with either OmpA+ or OmpA− E. coli for 1 h on ice, followed by four washes with PBS. The bacteria were treated with Laemmli buffer for 5 min and centrifuged at 10,000 rpm in a microcentrifuge. The supernatant was collected carefully, boiled for 2 min, and loaded onto a sodium dodecyl sulfate (SDS)-polyacrylamide gel. After separation, the proteins were transferred to a nitrocellulose filter and immunoblotted with an anti-His antibody (Covance Inc., Princeton, N.J.).
Flow cytometry.
Confluent HBMEC or CHO cells, either nontransfected or transfected, were lifted from the flasks with experimental medium containing 1% EGTA and washed thoroughly, and approximately 106 cells/ml were incubated with an anti-Ecgp antibody (dilution, 1:1,000) in RPMI containing 3% normal goat serum (RPMI-NGS) for 1 h on ice. Cells were then washed with RPMI containing 0.1% EGTA and further incubated with a fluorescein isothiocyanate-conjugated secondary antibody in RPMI-NGS for 30 min on ice, followed by four washes with RPMI. After a final wash, cells were resuspended in PBS and subjected to fluorescence-activated cell sorter analysis on a Becton Dickinson instrument.
RESULTS
Identification of a gp96-like cDNA clone by screening of the HBMEC cDNA expression library with an anti-Ecgp antibody.
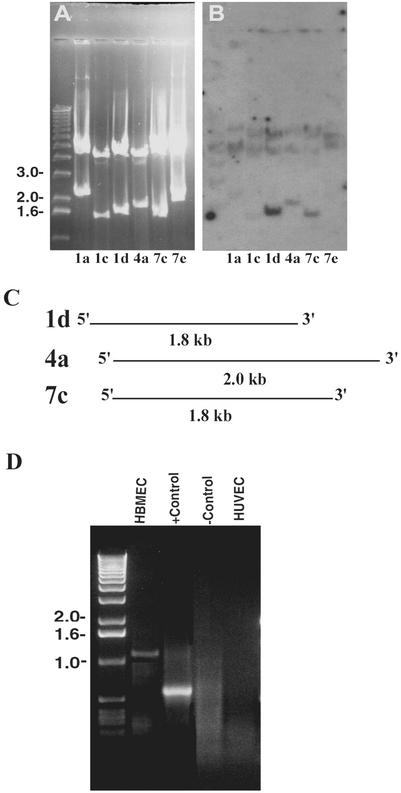
Previous studies have shown that OmpA interacts with an approximately 95 kDa HBMEC surface receptor for invasion (18). We have developed a polyclonal antibody that specifically recognizes Ecgp; thus, we set out to use an HBMEC cDNA expression library to identify the gene that encodes Ecgp. The cDNA library constructed in the Lambda Zap Express vector was screened with an anti-Ecgp antibody (Ecgp-Ab). After three rounds of screening, we identified several cDNA clones. The Ecgp-Ab showed strong reactivity to clones 4a and 7c. All the clones were verified for the cDNA inserts by EcoRI digestion and were categorized into six groups based on the size of the insert. As shown in Fig. 1A, the insert sizes range from 0.8 to 2.2 kb in these clones. The band of approximately 4 kb is the Lambda Zap vector.
FIG. 1.
Analysis of cDNA inserts from phagemid clones and 5′ RACE. (A) Phagemids obtained after screening of the HBMEC cDNA library with Ecgp-Ab were digested with EcoRI and separated on a 1% agarose gel. A 1-kb DNA marker is loaded on the left. (B) The DNA contents from the agarose gel were transferred to a nylon membrane and subjected to Southern hybridization using a degenerate oligonucleotide probe obtained from the N-terminal amino acid sequence of gp65. (C) Schematic representation of the overlapping regions of different cDNA clone sequences. (D) Agarose gel electrophoresis of the 5′ RACE product using RNAs of HBMEC and HUVEC. The primers and DNA supplied by the manufacturer were used as a positive control (+ control), and HBMEC RNA without primers was used as a negative control (− control).
Since our previous studies revealed that Ecgp cleaves into a 65- and a 30-kDa protein, and the 65-kDa protein showed a novel sequence, we next examined, by Southern hybridization using a degenerate oligonucleotide obtained from the partial N-terminal amino acid sequence of the 65-kDa protein, which of these six clones contain the sequence of the 65-kDa protein. The labeled oligonucleotide probe reacted with three clones (1d, 4a, and 7c); however, clone 1d showed the strongest reactivity (Fig. 1B). These three clones were sequenced from both T3 and T7 directions, and when blasted against the GenBank nucleotide database, they showed approximately 95% similarity to tumor rejection antigen 1 (TRA1) or gp96 (12). Clones 4a and 7c showed sequences overlapping that of 1d (Fig. 1C). We used this sequence information to generate a 2.0-kb sequence, which showed a stop codon toward the 3′ region and also contained a poly(A) tail, indicating that it encodes the C-terminal portion of Ecgp. Interestingly, none of these clones showed a putative signal peptide sequence, in contrast to our speculation that Ecgp is a transmembrane protein. Therefore, to obtain full-length cDNA, the HBMEC cDNA library was rescreened with the cDNA insert of clone 7c. We identified five more clones after the third screening. However, sequencing of these clones revealed the same sequence as clone 7c. We were unsuccessful in obtaining full-length cDNA even after another round of screening.
To identify the 5′ end of clone 7c, 5′ RACE was carried out with primers generated from the 5′ end of clone 7c, which shows a sequence different from that of gp96. As shown in Fig. 1D, a 1.1-kb fragment was generated from HBMEC total RNA. In contrast, when HUVEC RNA was used for 5′ RACE, no detectable PCR product was observed. The RNA sample and the primers supplied by the manufacturer were used as a positive control, and an HBMEC sample without primers was used as a negative control. The amplified PCR fragment was purified and cloned into plasmid CloneAmp pAmp1 (Life Technologies Inc.). The insert was sequenced with primers SP6 and T7 from both sides. The DNA database search of this sequence showed 98% sequence homology to the gp96 molecule. These results suggest that Egcp is a gp-96 like protein that acts as a receptor for OmpA.
Ecgp is a gp96 homologue that is expressed significantly in HBMEC.
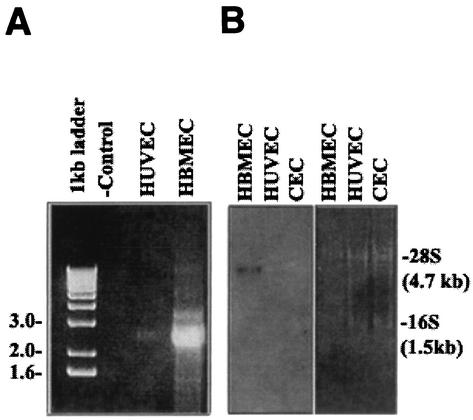
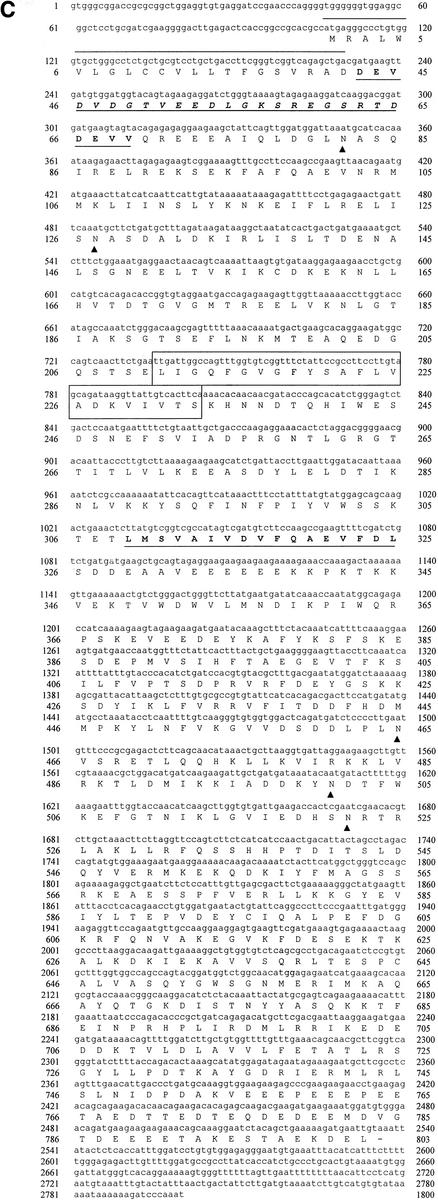
Since the sequences of clone 7c and the 5′ RACE product showed significant sequence homology to gp96, we generated a PCR fragment by reverse transcription-PCR (RT-PCR) of HBMEC mRNA using primers (Ecgp1 [5′-GAGGTGTGAGGATCCGAACC-3′] and Ecgp2 [5′-CTGTGACCCATAATCCCACA-3′]) from both the 5′ and 3′ ends with BamHI restriction site overhangs. As shown in Fig. 2A, we obtained a 2.7-kb fragment, whereas a very minute quantity of a fragment of the same size was obtained from HUVEC RNA. The 2.7-kb fragment was purified and cloned into BamHI sites of the pcDNA3 vector (Invitrogen). After the orientation was verified by restriction digestion analysis, the cloned fragment was sequenced. The DNA sequence of the PCR fragment was almost similar to the sequence we derived from both the 5′ RACE product and clone 7c. A single open reading frame was identified which translates into a protein of 803 amino acids, with a predicted molecular mass of 95.4 kDa (Fig. 2C), which is 98% identical to the gp96 protein. It is interesting that the N-terminal portion of the predicted Ecgp sequence has a heat shock protein element in addition to a signal peptide sequence (amino acids 1 to 21). It also contains a putative transmembrane domain, as determined by both the PROSITE and TopPred 2 programs (amino acids 191 to 212). The sequence between amino acids 280 and 297 showed considerable difference between Ecgp and gp96 at both the DNA level and the protein level. In addition, a KDEL site followed by a stop codon and a poly(A) tail (12 bp) was also present at the 3′ end. These results suggest that a very closely related gp96-like sequence encodes Ecgp.
FIG. 2.
Generation of the Ecgp gene by RT-PCR, and the nucleotide sequence of Ecgp. (A) Total RNAs of HBMEC and HUVEC were used to generate an RT-PCR product by using primers generated from the gp96 sequence. HBMEC RNA without primers was used as a negative control (− control). (B) Northern blot analysis of total RNA from HBMEC, HUVEC, and CEC by use of the labeled 7c clone as a probe. The gel was stained with ethidium bromide before transfer to a nylon membrane. Molecular sizes of 28S and 16S RNA are shown on the right. (C) Ecgp nucleotide and derived amino acid sequences obtained from cDNA sequencing and 5′ RACE. The initiation methionine was identified as the first ATG in the open reading frame and obeys Kozak's consensus. A 21-amino-acid putative signal peptide (overlined), potential N-glycosylation sites (triangles), and 23 hydrophobic amino acids representing a putative transmembrane domain (boxed) were found. An endoplasmic retention sequence (KDEL) before the stop codon and a poly(A) tail at the end of the sequence were found. The protein sequences obtained by partial N-terminal amino acid analyses of 95- and 65-kDa proteins in our previous study are boldfaced and underlined.
To further examine whether differences in the expression of Ecgp in various cells cause the differences in susceptibility to E. coli invasion, Northern blot analysis of total RNAs from HBMEC, HUVEC, and conjunctival epithelial cells (CEC) was carried out by using the 2.7-kb Ecgp gene as a probe. As shown in Fig. 2B, significant expression of Ecgp gene message (∼3 kb) in HBMEC was observed compared to that in HUVEC and CEC. To verify equal loading of total RNA, the gel was stained with ethidium bromide prior to transfer to a nylon membrane, which showed two major bands corresponding to 16S and 28S RNA. These results are in agreement with the results of 5′ RACE, in which very small or no detectable quantities of the product were generated from HUVEC.
Increased surface expression of either Ecgp or gp96 in transfected HBMEC and CHO cells.
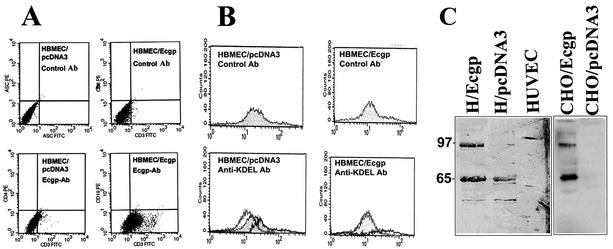
Previous studies showed that E. coli invades HBMEC at a frequency of only 0.1% of total bacteria added (15). In addition, immunocytochemical analysis with Ecgp-Ab showed that only 10 to 15% of cultured HBMEC express Ecgp on their surfaces, suggesting that a subset of the HBMEC population expresses Ecgp. Thus, to determine if increased expression of Ecgp is associated with increased E. coli invasion, HBMEC were transfected with the pcDNA3 vector containing the Ecgp gene. In addition, another set of HBMEC was transfected with the pcDNA3 vector containing gp96 cDNA. We also transfected CHO cells, which are less susceptible to E. coli invasion than HBMEC, with these plasmids. After 3 weeks of selection in G418, the resistant HBMEC were characterized for Ecgp expression by flow cytometry. One representative histogram each is shown for Ecgp and gp96 transfectants (Fig. 3A). Approximately 60% more cells were positive with Ecgp-Ab in Ecgp-transfected HBMEC than in pcDNA3-transfected HBMEC, suggesting that Ecgp is expressed on the surfaces of HBMEC (Fig. 3A). Interestingly, Ecgp-transfected CHO cells showed only a 30% increase in expression of Ecgp over that in control cells. We also analyzed these transfectants by using anti-KDEL antibodies to investigate the possibility that gp96 molecules come out of the cell completely and attach to the membrane via the glycocalyx of the plasma membrane. Interestingly, we observed no increase in the number of cells positive with the anti-KDEL antibody in Ecgp-transfected versus pcDNA3-transfected HBMEC (Fig. 3B). The membrane proteins of these HBMEC, when subjected to Western blot analysis with Ecgp-Ab, showed 95- and 65-kDa proteins, which were expressed in quantities significantly higher than those in pcDNA3-transfected HBMEC. However, three times more proteins from CHO cells were needed to obtain a similar intensity of Ecgp, which also showed 95- and 65 kDa proteins (Fig. 3C). In contrast, the HUVEC membrane fraction showed no detectable proteins in that molecular range. These results strongly suggest that Ecgp or gp96 is expressed at the cell surfaces of HBMEC in detectable quantities and that the C-terminal portion of the protein, which contains the KDEL, might still reside in the cytoplasm.
FIG. 3.
Flow cytometry and Western blot analysis of HBMEC overexpressing Ecgp. (A and B) HBMEC transfected with the Ecgp gene were released from culture flasks with a medium containing EDTA, washed, and incubated with either Ecgp-Ab (A) or an anti-KDEL antibody (B), followed by fluorescein isothiocyanate (FITC)-labeled secondary antibodies. The cells were then processed for flow cytometry analysis. HBMEC transfected with pcDNA3 were used as a control. (C) Total membrane proteins (10 μg) from HBMEC (H/Ecgp) or total membrane proteins (30 μg) from CHO cells (CHO/Ecgp) transfected with the Ecgp gene were subjected to immunoblotting with Ecgp-Ab. Molecular sizes (in kilodaltons) are indicated on the left.
Overexpression of either Ecgp or gp96 in HBMEC enhances E. coli invasion.
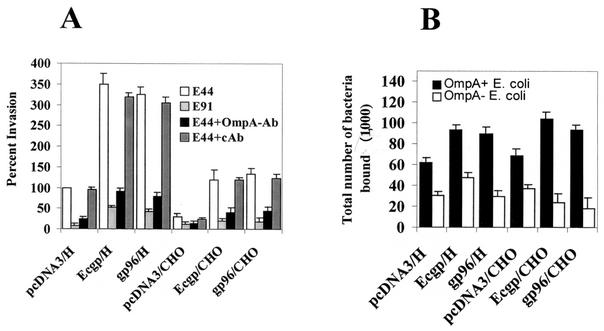
Both Ecgp- and gp96-transfected HBMEC were used in E. coli invasion assays in order to determine their functional characteristics. HBMEC transfected with either Ecgp or gp96 showed a threefold increase in the invasion (Fig. 4A) over that for pcDNA3-transfected HBMEC (26,500 ± 2,375 versus 7,500 ± 1,575 CFU/well, respectively; P < 0.001). Transfected CHO cells also showed a fourfold increase in the invasion of E. coli (2,350 ± 570 CFU/well for pcDNA3-transfected CHO cells versus 10,575 ± 1,250 CFU/well for Ecgp-transfected CHO cells). In contrast, the noninvasive OmpA− E. coli showed similar background levels (<10% of OmpA+ E. coli) in all transfectants. To verify whether the increase in E . coli invasion was due to increased binding of the bacteria to these transfected cells, we examined the total cell-associated bacteria. There was an approximately 40% increase in the binding of OmpA+ E. coli to both Ecgp- and gp96-transfected HBMEC or CHO cells over that to control cells, whereas OmpA− E. coli showed no significant increase (Fig. 4B). Despite similar levels of cell-associated OmpA+ E. coli in HBMEC and CHO cell transfectants, the invasion of CHO cells expressing either Ecgp or gp96 was three times lower than that of HBMEC expressing Ecgp. Increased association of the bacteria with HBMEC could also be a factor for this increased invasion. These results indicate that surface-expressed Ecgp functions as a receptor for OmpA+ E. coli for enhanced invasion of HBMEC. These results also point out that gp96 provides the same specificity as Ecgp for E. coli invasion; thus, minor differences in the amino acid sequence of Ecgp may be tolerable during the interaction with OmpA.
FIG. 4.
Invasion and binding by OmpA+ and OmpA− E. coli of HBMEC and CHO cells transfected with either Ecgp or gp96. (A) Confluent monolayers of HBMEC transfected with either pcDNA3, Ecgp, or gp96 were used in E. coli invasion assays with OmpA+ E. coli (E44) or OmpA− E. coli (E91) as described in Materials and Methods. In some experiments, E44 was preincubated with Fab fragments of either an anti-OmpA antibody or a control antibody (cAb) for 1 h on ice before addition to the monolayers. The experiments were carried out at least twice in triplicate, and results are expressed as percent invasion, with invasion by E. coli of pcDNA3-transfected HBMEC taken as 100%. Error bars, standard deviations. (B) Experiments were carried out like those for which results are shown in panel A except that the gentamicin step was omitted. Results are expressed as total numbers of bacteria per well (in thousands). Error bars, standard deviations.
To further examine the specificity of the interaction of OmpA+ E. coli with the transfected HBMEC, the E. coli bacteria were pretreated with Fab′ fragments of the anti-OmpA antibody prior to their addition to HBMEC monolayers. An unrelated polyclonal antibody was used as a control. The anti-OmpA antibody significantly blocked the invasion by E. coli of both Ecgp- and gp96-transfected HBMEC, whereas the control antibody had no effect (18,250 ± 1,790 CFU/well with the control antibody versus 3,590 ± 890 CFU/well with the anti-OmpA antibody; P < 0.001) (Fig. 4A). These data suggest that the increase in E. coli invasion of HBMEC is due to a specific interaction of OmpA with either Ecgp or gp96 in these transfected cells.
rEcgp blocks E. coli invasion.
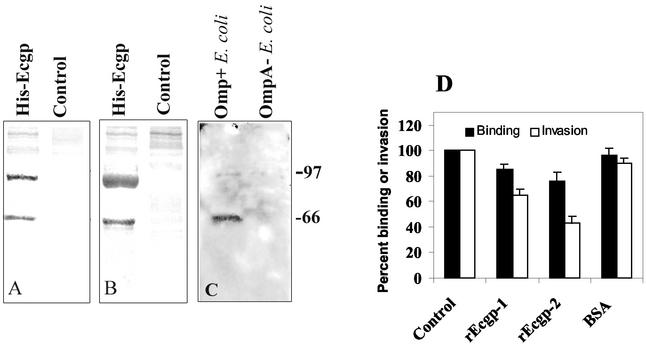
Previous studies showed that Ecgp that had been partially purified by lectin affinity chromatography significantly inhibited E. coli invasion (18). Thus, we next examined the effect of rEcgp on invasion. The Ecgp gene was cloned into the pcDNA3-His B vector, which was then transfected into CHO cells, and the His-tagged rEcgp was isolated by Ni-NTA Sepharose column chromatography. Since the histidine tag is on the N-terminal side of the Ecgp gene, the mature protein should not contain the His tag, as it is cleaved before it inserts into the membrane. However, we speculated that some portion of the gp96 escapes the endoplasmic reticulum (ER) and is available in the cytoplasm. Our results suggest that small quantities of His-rEcgp can be isolated from Ecgp-overexpressing CHO cells. Purified rEcgp showed two major bands (95- and 65-kDa proteins) by Coomassie blue staining (Fig. 5A). A duplicate blot obtained by probing with Ecgp-Ab revealed that these two proteins reacted to the antibody (Fig. 5B). In contrast, pcDNA3-transfected CHO total-cell lysates, when passed over a Ni-NTA Sepharose column, showed negligible amounts of proteins in both the Coomassie gel and the immunoblot. Then invasion assays were performed by using OmpA+ and OmpA− E. coli strains incubated with different concentrations of rEcgp. As shown in Fig. 5D, invasion of E. coli was significantly blocked in a dose-dependent manner, with 50% inhibition achieved by 10 μg of rEcgp/ml (9,230 ± 650 CFU/well for bovine serum albumin [BSA] versus 4,150 ± 790 CFU/well for rEcgp protein; P < 0.01). We achieved only 70% inhibition, even at a protein concentration of 30 μg/ml, relative to invasion by BSA-treated bacteria. OmpA− E. coli showed similar levels of background invasion even in the presence of His-Ecgp (data not shown). rEcgp showed neither a cytotoxic effect nor an anti-bactericidal effect under the conditions employed. Interestingly, the total number of cell-associated bacteria was 20 to 30% lower in the presence of these proteins than in the presence of BSA-treated bacteria. The decrease in E. coli invasion after treatment with rEcgp may be due to a decrease in the number of bacteria that bound to HBMEC, which also suggests that Ecgp is required for both binding and invasion of E. coli.
FIG. 5.
Inhibition of OmpA+ E. coli invasion by rEcgp. (A) rEcgp (5 μg) isolated from CHO cells transfected with Ecgp was separated by SDS-10% polyacrylamide gel electrophoresis and stained with Coomassie blue. (B) A duplicate gel was transferred to a nitrocellulose membrane and immunoblotted with an anti-His antibody. (C) Purified rEcgp (10 μg) was incubated with either OmpA+ or OmpA− E. coli for 1 h on ice and washed thoroughly, and bound proteins were released with Laemmli buffer. The proteins, after separation by SDS-10% polyacrylamide gel electrophoresis, were immunoblotted with an anti-His antibody. (D) Various concentrations of rEcgp were incubated with OmpA+ E. coli for 1 h on ice prior to addition to HBMEC monolayers. Levels of both total cell-associated bacteria (Binding) and intracellular bacteria (Invasion) were calculated as described in Materials and Methods. The experiments were carried out at least twice in triplicate, and results are expressed as percent binding or percent invasion, with the control invasion taken as 100%. Error bars, standard deviations from the means.
To further examine whether the inhibition of E. coli invasion by rEcgp is due to direct binding of Ecgp to the bacteria, OmpA+ and OmpA− E. coli strains were incubated with 5 μg of rEcgp for 1 h on ice. The bacteria were then washed with PBS, and the bound protein was released by Laemmli buffer. The proteins, when probed with an anti-His antibody, revealed a 95-kDa and a 65-kDa (minor) protein bound only to OmpA+ E. coli, not to OmpA− E. coli (Fig. 5C). These results clearly suggest that the inhibition of E. coli invasion of HBMEC is due to the blocking of OmpA binding sites by rEcgp.
DISCUSSION
The data provided in this study indicate that OmpA binds to a highly similar gp96 homologue, Ecgp, a process that may activate signal transduction pathways such as the tyrosine phosphorylation of FAK and paxillin, leading to actin rearrangement. Despite the role of other E. coli virulence factors such as IbeA, IbeB, and CNF, the interaction of OmpA with HBMEC via Ecgp appears to be crucial for the invasion process. The evidence for this, obtained via several independent approaches from our laboratory, is as follows: (i) the effects of Ecgp expression in either HBMEC or CHO cells on the invasion of E. coli, (ii) the ability of rEcgp to bind to OmpA+ E. coli but not to OmpA− E. coli, (iii) inhibition of E. coli invasion by rEcgp, (iv) protein tyrosine phosphorylation of FAK, paxillin, and PKC by OmpA+ E. coli, and (v) convergence of FAK and PKC at the OmpA+ E. coli entry site in HBMEC. In addition, accumulation or clustering of Ecgp at the OmpA+ E. coli attachment sites, but not at the OmpA− E. coli attachment sites, strongly suggests a role for Ecgp in E. coli invasion. However, other mechanisms of E. coli entry mediated by factors other than OmpA might exist, although they would contribute to invasion to a lesser extent.
rEcgp bound directly to OmpA+ E. coli, as shown by Western blot analysis; however, rEcgp blocked E. coli invasion by only 70%. This may be due to a change in the conformation of Ecgp in solution from that of Ecgp expressed on the HBMEC surface. It is interesting that gp96, despite slight differences from Ecgp in amino acid sequence, also mediates the invasion of HBMEC. Structural analysis of both proteins indicates no significant differences at and around the N-glycosylation sites, suggesting that minor differences in other portions of Ecgp are not important for the function. Although the detailed mechanism of OmpA binding to Ecgp to induce actin rearrangements remains to be elucidated, it is worth noting the apparent differences in the invasion process reported for other bacteria. Shigella flexneri binds to α5β1 integrins to elicit actin remodeling via secreted Ipa proteins (30). The interaction of Shigella with integrins induced the clustering of integrins, similar to Ecgp clustering. Listeria monocytogenes, another causative agent of neonatal meningitis, invades by binding to E-cadherin on epithelial cells via internalin A (InlA) (11). Another protein, InlB, is responsible for entry into hepatocytes and HBMEC, but its receptors are not known (3). The InlA interaction induces the activation of PI3K, but not FAK, for cytoskeletal rearrangements. We also recently observed that a group B streptococcus, another pathogen that causes meningitis, does not require FAK activation during its invasion of HBMEC (unpublished data). In contrast, OmpA-mediated invasion induces both FAK and PI3K activation, which likely occurs via Ecgp signaling and is different from that of other bacterial pathogens.
Ecgp contains a C-terminal KDEL motif similar to that of gp96, which is a signature marker for an ER retention signal expressed on the cell surface. However, it has been demonstrated that several eukaryotic cells express gp96 on their surfaces (1, 22); therefore, it is possible that a portion of the Ecgp generated in the cell could be directed to the cell surface as a transmembrane protein. In agreement with this, flow cytometry analysis with an anti-KDEL antibody showed that the KDEL motif is not expressed on the cell surface, indicating that the motif could be in the cytoplasmic domain of Ecgp inside the cell. In fact, an ER-resident molecule, calnexin, is expressed on the surfaces of immature thymocytes in a similar fashion (13). The surface expression of these ER retention molecules is not a property of all cells but rather appears to be selective. The presence of such receptors on HBMEC may have very important immunological consequences, since they are able to transduce differentiative signals vital for preserving BBB integrity. In addition, Ecgp or gp96 shows homology to heat shock proteins such as Hsp90, which play an important role in immune response in infectious diseases and other diseases. Thus, it is tempting to speculate that the multiplying E. coli organisms during the bacteremia stage induce (stress-induced) expression of Ecgp on the surfaces of HBMEC, which in turn are utilized as receptors by E. coli in crossing the BBB. In a different scenario, certain neonates, due to either physiological or genetic stress conditions, may express Ecgp more significantly on the BBB, thus becoming more susceptible for the incidence of meningitis if they are infected with E. coli. The immunological relevance of the peptide binding capacity of gp96 was first described for tumor systems (23, 29). Recent studies have shown that gp96-peptide complexes can elicit immune responses and a potent cytotoxic T-lymphocyte response, suggesting that gp96 can bind to a broad array of peptides (19). Therefore, the binding of OmpA peptide regions to Ecgp is of interest for developing vaccination strategies against E. coli. The key question is whether E. coli exploits Ecgp to avoid an immunologic response at the area of colonization or whether E. coli simply takes the opportunity to bind Ecgp for invasion.
Taken together, our previous and present results strongly suggest that OmpA binds to a gp96 homologue, Ecgp (a 95-kDa protein), and a 65-kDa protein, which could be a cleavage product of Ecgp. We are presently investigating whether there is another 65-kDa OmpA binding protein on HBMEC. This is the first report to show that overexpression of an E. coli K1 receptor in a eukaryotic cell line generally resistant to invasion enhances invasion by bacteria.
Acknowledgments
We thank Scott Filler for critical reading of the manuscript.
This work was supported by NIH grants AI40567 and HD 41525 (to N.V.P).
Editor: J. N. Weiser
REFERENCES
- 1.Altmeyer, A., R. G. Maki, A. M. Feldweg, M. Heike, V. P. Protopopov, S. K. Masur, and P. K. Srivastava. 1996. Tumor-specific cell surface expression of the-KDEL containing, endoplasmic reticular heat shock protein gp96. Int. J. Cancer 69:340-349. [DOI] [PubMed] [Google Scholar]
- 2.Badger, J. L., C. A. Wass, and K. S. Kim. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol 36:174-182. [DOI] [PubMed] [Google Scholar]
- 3.Braun, L., and P. Cossart. 2000. Interactions between Listeria monocytogenes and host mammalian cells. Microbes Infect. 2:803-811. [DOI] [PubMed] [Google Scholar]
- 4.Datta, D., N. Vaidehi, W. B. Florino, K. S. Kim, N. V. Prasadarao, and W. A. Goddard III. 2003. Interaction of E. coli outer membrane protein A with sugars on the receptors of the brain microvascular endothelial cells. Proteins 50:213-221. [DOI] [PubMed] [Google Scholar]
- 5.Davis, L. G., M. D. Dibner, and J. F. Battey. 1986. Guanidine isothiocyanate preparation of total RNA, p. 130-135. In L. G. Davis (ed.), Basic methods in molecular biology. Elsevier Science Publishing Co., Inc., New York, N.Y.
- 6.Huang, S. H., C. A. Wass, Q. Fu, N. V. Prasadarao, M. F. Stins, and K. S. Kim. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63:4470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, S. H., Y. H. Chen, Q. Fu, M. F. Stins, Y. Wang, C. A. Wass, and K. S. Kim. 1999. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect. Immun. 67:2103-2109., [DOI] [PMC free article] [PubMed]
- 8.Kershaw, D. B., P. E. Thomas, B. L. Wharram, M. Goyal, J. E. Wiggins, C. I. Whiteside, and R. C. Wiggins. 1995. Molecular cloning, expression, and characterization of podocalyxin-like protein 1 from rabbit as a transmembrane protein of glomerular podocytes and vascular endothelium. J. Biol. Chem. 270:29439-29446. [DOI] [PubMed] [Google Scholar]
- 9.Khan, N. A., Y. Wang, K. J. Kim, J. W. Chung, C. A. Wass, and K. S. Kim. 2002. Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J. Biol. Chem. 277:15607-15612. [DOI] [PubMed] [Google Scholar]
- 10.Kupsch, E. M., B. Knepper, T. Kuroki, I. Heuer, and T. F. Meyer. 1993. Variable opacity (Opa) outer membrane proteins account for the cell tropisms displayed by Neisseria gonorrhoeae for human leukocytes and epithelial cells. EMBO J. 12:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lecuit, M., S. Dramsi, C. Gottardi, M. Fedor-Chaiken, B. Gumbiner, and P. Cossart. 1999. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18:3956-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maki, R. G., L. J. Old, and P. K. Srivastava. 1990. Human homologue of murine tumor rejection antigen gp96: 5′-regulatory and coding regions and relationship to stress-induced proteins. Proc. Natl. Acad. Sci. USA 87:5658-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okazaki, Y., H. Ohno, K. Takase, T. Ochiai, and T. Saito. 2000. Cell surface expression of calnexin, a molecular chaperone in the endoplasmic reticulum. J. Biol. Chem. 275:35751-35758. [DOI] [PubMed] [Google Scholar]
- 14.Parton, R. G. 1996. Caveolae and caveolins. Curr. Opin. Cell Biol. 8:542-548. [DOI] [PubMed] [Google Scholar]
- 15.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. H. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasadarao, N. V., C. A. Wass, and K. S. Kim. 1996. Endothelial cell GlcNAcβ1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect. Immun. 64:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasadarao, N. V., C. A. Wass, M. F. Stins, H. Shimada, and K. S. Kim. 1999. Outer membrane protein A-promoted actin condensation of brain microvascular endothelial cells is required for Escherichia coli invasion. Infect. Immun. 67:5775-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasadarao, N. V. 2002. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect. Immun. 70:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randow, F., and B. Seed. 2001. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat. Cell Biol. 3:891-896. [DOI] [PubMed] [Google Scholar]
- 20.Reddy, M. A., C. A. Wass, K. S. Kim, D. D. Schlaepfer, and N. V. Prasadarao. 2000. Involvement of focal adhesion kinase in Escherichia coli invasion of human brain microvascular endothelial cells. Infect. Immun. 68:6423-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddy, M. A., N. V. Prasadarao, C. A. Wass, and K. S. Kim. 2000. Phosphatidylinositol 3-kinase activation and interaction with focal adhesion kinase in Escherichia coli K1 invasion of human brain microvascular endothelial cells. J. Biol. Chem. 275:36769-36774. [DOI] [PubMed] [Google Scholar]
- 22.Robert, J., A. Menoret, and N. Cohen. 1999. Cell surface expression of the endoplasmic reticular heat shock protein gp96 is phylogenetically conserved. J. Immunol. 163:4133-4139. [PubMed] [Google Scholar]
- 23.Robert, J., A. Menoret, P. K. Srivastava, and N. Cohen. 2001. Immunological properties of heat shock proteins are phylogenetically conserved. Adv. Exp. Med. Biol. 484:237-249. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Stins, M. F., N. V. Prasadarao, L. Ibric, C. A. Wass, P. Luckett, and K. S. Kim. 1994. Binding characteristics of S fimbriated Escherichia coli to isolated brain microvascular endothelial cells. Am. J. Pathol. 145:1228-1236. [PMC free article] [PubMed] [Google Scholar]
- 26.Stins, M. F., N. V. Prasadarao, C. A. Wass, and K. S. Kim. 1999. Escherichia coli binding to and invasion of brain microvascular endothelial cells derived from humans and rats of different ages. Infect. Immun. 67:5522-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sukumaran, S. K., and N. V. Prasadarao. 2002. Regulation of protein kinase C-α in Escherichia coli K1 invasion of human brain microvascular endothelial cells. J. Biol. Chem. 277:12253-12262. [DOI] [PubMed] [Google Scholar]
- 28.Sukumaran, S. K., M. J. Quon, and N. V. Prasadarao. 2002. Escherichia coli K1 internalization via caveolae requires caveolin-1 and protein kinase C-α interaction in human brain microvascular endothelial cells. J. Biol. Chem. 277:50716-50724. [DOI] [PubMed] [Google Scholar]
- 29.Udono, H., and P. K. Srivastava. 1994. Comparison of tumor-specific immunogenicities of stress-induced proteins gp96, hsp90, and hsp70. J. Immunol. 152:5398-5403. [PubMed] [Google Scholar]
- 30.Watari, M., S. Funato, and C. Sasakawa. 1996. Interaction of Ipa proteins of Shigella flexneri with α5β1 integrin promotes entry of the bacteria into mammalian cells. J. Exp. Med. 183:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]