Abstract
Borrelia burgdorferi is the etiologic agent of Lyme disease, the most prevalent arthropod-borne disease in the United States. The genome of the type strain, B31, consists of a 910,725-bp linear chromosome and 21 linear and circular plasmids comprising 610,694 bp. During its life cycle, the spirochete exists in distinctly different environments, cycling between a tick vector and a mammalian host. Temperature is one environmental factor known to affect B. burgdorferi gene expression. To identify temperature-responsive genes, genome arrays containing 1,662 putative B. burgdorferi open reading frames (ORFs) were prepared on nylon membranes and employed to assess gene expression in B. burgdorferi B31 grown at 23 and 35°C. Differences in expression of more than 3.5 orders of magnitude could be readily discerned and quantitated. At least minimal expression from 91% of the arrayed ORFs could be detected. A total of 215 ORFs were differentially expressed at the two temperatures; 133 were expressed at significantly greater levels at 35°C, and 82 were more significantly expressed at 23°C. Of these 215 ORFs, 134 are characterized as genes of unknown function. One hundred thirty-six (63%) of the differentially expressed genes are plasmid encoded. Of particular interest is plasmid lp54 which contains 76 annotated putative genes; 31 of these exhibit temperature-regulated expression. These findings underscore the important role plasmid-encoded genes may play in adjustment of B. burgdorferi to growth under diverse environmental conditions.
Lyme disease, the most common arthropod-borne infection in the United States, is a multisystemic infectious disease caused by spirochetes of the Borrelia burgdorferi sensu lato complex (39, 61). If untreated, B. burgdorferi can survive for extended periods of time in the human host, resulting in dermatitis, carditis, arthritis, and neuritis (35, 51). B. burgdorferi is maintained in nature by a complex enzootic cycle involving Ixodes ticks and mammalian hosts, typically rodents (32). To perpetuate this enzootic cycle, spirochetes must be transmitted between two dramatically different environments. Within the unfed tick, B. burgdorferi resides in the midgut. At the onset of tick feeding, however, B. burgdorferi recognizes environmental changes that initiate its migration through the midgut epithelium and hemolymph to the salivary glands before ultimately being deposited in the mammalian host. The environment of the tick midgut changes rapidly during tick feeding with regard to temperature, pH, nutrient levels, and mammal-specific factors contained in the blood meal. Together, these environmental changes combine to trigger spirochete migration. The rapid increase in temperature that occurs upon feeding appears to be an important signal for changes in spirochete gene expression and subsequent migration to the salivary glands (48). In this regard, many studies have used organisms shifted from 23 to 35°C during in vitro cultivation as a surrogate for tick feeding. This has led to the identification of several proteins thought to be important in the borrelial transmission process (25-27, 49, 55).
B. burgdorferi has a unique genome consisting of a 910-kbp linear chromosome and numerous linear and circular plasmids comprising an additional 600 kbp (7, 22). The annotated genome sequence for B. burgdorferi sensu stricto strain B31 contains 1,689 putative open reading frames (ORFs), of which nearly two-thirds have no known function. Although a number of intriguing findings resulted from elucidation of the full genome sequence (e.g., the large numbers of putative lipoprotein-encoding genes and paralogous genes and the paucity of identifiable genes encoding metabolic activities), few clues to the mechanisms underlying the host-pathogen interaction and pathogenicity are forthcoming from examination of the genome sequence alone (22). Recent results indicate that many of the genes and proteins differentially expressed by temperature are plasmid encoded, suggesting that plasmids are an important repository for genes integral to B. burgdorferi transmission to the mammalian host (2, 3, 25-27, 49, 53, 55, 66). Furthermore, B. burgdorferi also expresses some plasmid-encoded genes exclusively during mammalian infection, which are thought to be important in disease establishment and pathogenesis (1, 3, 13, 19, 20, 27, 57, 60). Consistent with these notions, it has been demonstrated that the loss of specific plasmids correlates with B. burgdorferi attenuation or loss of virulence in mice (31, 34, 43, 50). However, it has not been experimentally determined if the absence of specific plasmids or genes also can arrest the migration of B. burgdorferi at different points within the tick as it is transmitted to the mammalian host. To determine which genes facilitate borrelial migration through the tick and establishment in the mammalian host, a logical first step is to identify molecules induced by signals encountered in the tick and mammalian environments, such as temperature.
Global gene expression profiling is a powerful tool for assessment of bacterial gene expression under different physiological conditions (58, 62) and has the potential to identify unknown virulence genes (11). Here, we describe the construction of a B. burgdorferi genome array and its application for profiling global changes in B. burgdorferi gene expression resulting from the environmental cue of increased temperature.
MATERIALS AND METHODS
ORF selection and PCR primer design.
ORF selection for a B. burgdorferi whole genome array poses a unique problem, especially for the plasmids, due to the presence of a substantial number of paralogous sequences. There are 175 paralogous gene families in the genome, with some families containing genes with virtually identical sequences. In addition, the plasmids contain at least 152 pseudogenes (i.e., genes that appear to have been mutationally inactivated) and a number of small ORFs that overlap longer ORFs (7). To simplify array design, each sequence annotated as a gene in the sequence database, including all paralogous genes, was represented on the array. In addition, pseudogenes were included, as were several regions on the linear plasmids with unique sequences that appear to lack any discernible ORFs, since it is of interest whether these regions are transcribed.
Pseudogenes and overlapping genes were managed by manually curating the published strain B31-MI ORF list. First, all pseudogenes were incorporated into the ORF list; this involved identifying all regions of nucleotide similarity among the plasmids and the plasmid-like rightmost 7.2 kbp of the chromosome (7, 14). Then, all overlapping ORFs (including the pseudogenes) were compared, and among any overlapping ORF set, only the largest was retained. Finally, primer sets were included for the largest of several unique plasmid-encoded regions several kilobase pairs in length that appear to contain no ORFs. This resulted in a list of 1,697 “gene features” (see supplementary Table 1 [www.nymc.edu/iraschwartz/borreliaarrays]). Primers were designed by Sigma-Genosys (Houston, Tex.) based on the following criteria: (i) a minimum length of 18 bases, (ii) a G+C content that was within the range of 30 to 70%, (iii) a melting temperature of at least 55°C, and (iv) a G or a C present in one of the last three 3′ base positions. In this fashion, suitable primers were found that would amplify a significant portion of the ORF in the majority of cases. For longer ORFs, sequences were truncated from the 5′ end, such that most PCR products would not exceed 2,000 bp. For ORFs of less than 250 bp, primers were designed to amplify the entire coding sequence, from start to stop codons. Hence, these shorter ORFs would represent the maximal hybridizable domain for array studies.
TABLE 1.
Oligonucleotides used for real-time quantitative RT-PCR
| ORF | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| BB0147 | TCAAGCGTCTTGGACTTTAAGAGTT | CACCAGAGAAAAGATTTGCAACATTA |
| BB0240 | AAGTCCCGAAATACCAGGAGAAAT | TTCTTGCTGCTGTGTAAATACCAAA |
| BB0241 | AAGTTGGGTGGAACATGATCCT | CGTCAATTTCATTGGGCAAA |
| BB0242 | TGTTTGGCAAGCAAATAAATCAATT | TTACCTTTTCAAAAGGTTTAGACAGTTT |
| BB0647 | CCACCCTATTCAACTTGACGATATTA | TGCAATGCCCTGAGTAAATGATT |
| BB0690 | ATTTCTTTGTTATTCACAAAAAAACTCAAA | AATTCAGAATCATATCCAAGCATTCTT |
| BB0838 | AGAGCACTATTCTGACCCGTATGTT | TGCTAGTTTGCTCTTTAACCGAATCT |
| BBA15 | ATGTTAGCAGCCTTGACGAGAAA | GATCGTACTTGCCGTCTTTGTTTT |
| BBA16 | AAATGGGAAGACAGTACTAGCACTTTAA | TGTATTGTTGTACTGTAATTGTACCATCTGTT |
| BBA65 | ACTGATATTTTCAATTTAGCAGAGATTGTAA | TGTTGGATTCGTATACCACCTGATATT |
| BBB19 | CTGATGCAAAAGAAGCCATTTTAA | TGCTTTTGACAAGACCTCTACTGATT |
| BBE19 | TAAAATAGTTGCCATTTGCTCAATTAA | TTAGAAGAACTTTATATTTTTTAGACAAGAGTGTT |
| BBE22 | TCAGAATTTCCTAATGATCTAAATACGAA | TGCAATCATCATAAAATCCACTGTAAC |
| BBG05 | AATTATCATACAAATCAGAGTGGTATGGAT | TCTTAGAGTCGTATTTTTAATGTGACAACTAC |
| BBL40 | ATTACAGGGCCTGTATATGATGATTTTACT | AGCTTTCCTAATCCTTCGTCGTTA |
| BBR41 | ACAGAAAAATTACAATGGACCAATGAA | AAAGCGCACCTTCTGAAATAAGTAATA |
Strains and growth conditions.
B31 is the type strain of B. burgdorferi, and B31-MI is the designation given to the B31 derivative for which the genomic sequence was determined (7, 22). B31-MI was passed through an experimental mouse-tick infectious cycle and reisolated from a mouse that was infected by needle inoculation (18). This B31-MI reisolate was directly moved to a plate, and a clone (B31-MI P1.1) that contained all the plasmids identified in the genomic sequence was derived from an individual colony picked and cultivated in liquid Barbour-Stoenner-Kelley II (BSK-II) medium. Frozen glycerol stocks were made directly from the primary outgrowth of this clone. Plating of B31-MI and determination of plasmid content were performed as previously described (16). B. burgdorferi was grown in liquid and solid BSK-II medium prepared in the laboratory; commercial BSK-H medium obtained from Sigma did not support growth of B31-MI P1.1 at a comparable rate or to the same final density.
B31-MI P1.1 was inoculated directly to BSK-II medium from the glycerol stock and grown to a moderate density at 35°C. This culture was diluted to 105 bacteria/ml and grown at 23°C. When this culture reached 2.5 × 107 bacteria/ml, it was diluted to a density of 2.5 × 105 bacteria/ml and separate cultures were incubated at 23°C and at 35°C. The latter cultures were diluted at mid-exponential phase to a density of 5 × 105/ml, and the incubation continued at 35°C. This was done to extend the period of growth at 35°C. Cell densities were measured daily, and at the point of harvest for RNA isolation, both 23 and 35°C cultures were in mid-exponential phase and at the same density (5 × 107 bacteria/ml). The pH of the culture medium remained neutral (∼7.4) throughout growth. Although B. burgdorferi grows more slowly at 23°C than at 35°C (relative doubling times of 33 versus 8 h, respectively), spirochetes at both temperatures exhibited similar morphologies and attained comparable densities of >2 × 108 bacteria/ml in stationary phase. Aliquots were taken of all cultures for protein analysis.
RNA and DNA isolation.
For each condition (23 and 35°C), RNA was extracted from spirochetes in mid-exponential growth phase (5 × 107 bacteria per ml) from four separate 100-ml cultures, using the Ultraspec-II RNA isolation system (BIOTECX, Houston, Tex.). RNase-free chemicals and material (tubes, barrier pipette tips, gloves, diethyl pyrocarbonate-water, RNase-Zap [Ambion, Austin, Tex.]) were used throughout the process. Subsequently, RNA from each culture or condition was treated with DNase I (DNA-free kit; Ambion). Quality and concentration of the RNA, as well as the absence of contaminating DNA, were determined with the Agilent 2100 Bioanalyzer (Agilent Technologies). The 23S/16S rRNA ratio of all samples was approximately 1, which indicates high-quality RNA. No contaminating DNA was detected. Cultures at 23°C yielded approximately 30% less RNA than the same number of spirochetes grown at 35°C. The RNAs from the four cultures grown under each condition were then pooled. Equal volumes of these two pools were aliquoted, and the RNA quality and concentration were redetermined for each aliquot. Each aliquot was precipitated for storage with 0.3 M sodium acetate and 2.5 volumes of 100% ethanol. Template DNA for PCR amplification was prepared from B31-MI with a Qiagen (Valencia, Calif.) genomic DNA kit according to the manufacturer's directions.
PCR amplification.
PCR was carried out in a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, a 100 μM concentration of each of the four deoxynucleoside triphosphates, 1.25 U of Taq DNA polymerase, and 20 to 30 pmol of each primer. Typically, amplification was carried out for 35 cycles at temperatures appropriate for the primers employed. The success of the PCRs was monitored by electrophoresis of a 10-μl sample of the reaction mixture through a 1 to 2% agarose gel in 1× Tris-borate-EDTA buffer. The amount of amplified product was determined by densitometric analysis of agarose gels using known quantities of molecular size markers. An amplification reaction was scored as successful if a single product was within 5% of the expected size of the ORF predicted from the genomic sequence. Unsuccessful PCRs were of three types: (i) no product observed by ethidium bromide staining; (ii) multiple bands observed; or (iii) reactions resulting in single products of unexpected size. Reactions that failed were repeated using different amplification conditions, for example, adjustment of annealing temperature. PCR failed for 26 ORFs after several attempts, and these ORFs are not represented on the arrays. These ORFs are among the 61 listed in supplementary Table 2.
TABLE 2.
Most highly expressed genes in B. burgdorferi cultivated at 35°C
| Gene name | Description of gene product (homologous gene) | Relative transcript level | Rank at temp (°C)
|
|
|---|---|---|---|---|
| 35 | 23 | |||
| BB0440 | Ribosomal protein L34 (rpmH) | 104.42 | 1 | 1 |
| BBA59 | Lipoprotein | 100.38 | 2 | 2 |
| BB0049 | Hypothetical protein | 83.91 | 3 | 4 |
| BB0804 | Ribosomal protein S15 (rpsO) | 64.75 | 4 | 6 |
| BB0233 | Ribosomal protein S20 (rpsT) | 57.39 | 5 | 3 |
| BB0162 | Hypothetical protein | 53.80 | 6 | 16 |
| BB0500 | Ribosomal protein S13 (rpsM) | 48.66 | 7 | 11 |
| BBA62 | Lipoprotein | 38.51 | 8 | 7 |
| BB0232 | hbbU protein | 36.14 | 9 | 13 |
| BB0294 | Flagellar basal-body rod protein (flgB) | 33.76 | 10 | 10 |
| BB0559 | PTSa system, glucose-specific IIA component (crr) | 33.07 | 11 | 5 |
| BBA15 | Outer surface protein A (ospA) | 31.32 | 12 | 12 |
| BB0300 | Cell division protein (ftsA) | 29.06 | 13 | 9 |
| BB0778 | Ribosomal protein L21 (rplU) | 28.75 | 14 | 14 |
| BBB19 | Outer surface protein C (ospC) | 28.47 | 15 | 109 |
| BB0480 | Ribosomal protein L23 (rplW) | 23.07 | 16 | 21 |
| BB0486 | Ribosomal protein L29 (rpmC) | 22.28 | 17 | 17 |
| BB0704 | Acyl carrier protein | 20.90 | 18 | 23 |
| BB0365 | Lipoprotein LA7 | 20.78 | 19 | 61 |
| BBA32 | Hypothetical protein | 20.75 | 20 | 114 |
| BBF20 | Conserved hypothetical protein | 20.70 | 21 | 32 |
| BB0741 | Chaperonin (groES) | 19.39 | 22 | 20 |
| BBA16 | Outer surface protein B (ospB) | 19.27 | 23 | 19 |
| BB0256 | Ribosomal protein S21 (rpsU) | 18.45 | 24 | 25 |
| BBS37 | Conserved hypothetical protein | 17.71 | 25 | 24 |
| BBQ42 | Conserved hypothetical protein | 17.04 | 26 | 22 |
| BB0355 | Transcription factor, putative | 16.17 | 27 | 18 |
| BBE10 | Hypothetical protein | 16.13 | 28 | 47 |
| BBA74 | Outer membrane porin (oms28) | 15.55 | 29 | 127 |
| BBA28 | Hypothetical protein | 15.41 | 30 | 64 |
| BB0175 | Conserved hypothetical protein | 15.14 | 31 | 219 |
| BB0631 | Hypothetical protein | 13.78 | 32 | 26 |
| BB0229 | Ribosomal protein L31 (rpmE) | 13.47 | 33 | 34 |
| BB0562 | Hypothetical protein | 13.16 | 34 | 29 |
| BB0501 | Ribosomal protein S11 (rpsK) | 12.70 | 35 | 39 |
| BB0703 | Ribosomal protein L32 (rpmF) | 12.51 | 36 | 54 |
| BB0538 | Conserved hypothetical protein | 12.33 | 37 | 40 |
| BB0803 | tRNA pseudouridine 55 synthase (truB) | 11.90 | 38 | 30 |
| BBB28 | Hypothetical protein | 11.60 | 39 | 48 |
| BB0541 | Hypothetical protein | 11.42 | 40 | 31 |
| BB0190 | Translation initiation factor 3 (infC) | 10.74 | 41 | 41 |
| BBL35 | Conserved hypothetical protein | 10.73 | 42 | 37 |
| BB0785 | Stage V sporulation protein G | 10.72 | 43 | 51 |
| BBH29 | Conserved hypothetical protein | 10.30 | 44 | 43 |
| BB0048 | Hypothetical protein | 10.21 | 45 | 74 |
| BBM38 | erpK protein (erpK) | 10.20 | 46 | 44 |
| BB0441 | Ribonuclease P protein component (rnpA) | 10.04 | 47 | 35 |
| BB0423 | Hypothetical protein | 10.01 | 48 | 28 |
| BB0782 | Conserved hypothetical protein | 9.97 | 49 | 56 |
| BB0664 | Hypothetical protein | 9.90 | 50 | 60 |
PTS, phosphotransferase.
Preparation of DNA array.
The B. burgdorferi membrane arrays described here were produced by Sigma-Genosys. Ten nanograms of PCR-amplified ORFs was spotted in duplicate onto positively charged nylon membranes. Following spotting, each array was cross-linked with UV light. Other technical aspects related to array printing and layout have been described elsewhere.
Probe synthesis.
A mixture of all 3′ ORF-specific primers was used to generate the hybridization probes by standard cDNA synthesis. Five micrograms of RNA was mixed with dTTP, dGTP, and dCTP (final concentration, 0.4 mM each). Primers were annealed to the RNA by heating at 65°C for 10 min and cooling to 42°C. Two hundred units of Superscript II (Life Technologies), 10 U of RNase inhibitor, and 20 μCi of [α-33P]dATP (Amersham) were added in a final volume of 20 μl of first-strand buffer. The final cDNA synthesis reaction mixture was incubated at 42°C for 2 h. The labeling efficiency was determined by trichloroacetic acid precipitation. Under the above conditions, specific activities of input RNA of 2 × 106 to 5 × 106 cpm/μg were achieved.
Hybridization.
The membrane array was placed in a prehybridization solution (ExpressHyb; Clontech) and incubated at 55°C for 1 h. The 33P-labeled cDNA was added to 10 ml of ExpressHyb solution and was used to probe the membrane array by incubation overnight at 55°C. After hybridization, the membrane was washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.05% sodium dodecyl sulfate (SDS) for 30 min at room temperature and then with 0.1× SSC-0.1% SDS at 65°C for 30 min. After washing, the arrays were exposed to a PhosphorImager screen (Molecular Dynamics, Sunnyvale, Calif.) for 48 h. For comparison of samples from cells grown at different temperatures, the same membranes were probed reciprocally (i.e., switching the membranes) after stripping in alkaline stripping solution.
Array data analysis.
The procedures for array data analysis have been described in detail elsewhere (38), and a brief description is provided here. The exposed PhosphorImager screen was scanned with a pixel size of 50 or 100 μm on a Storm 840 PhosphorImager (Molecular Dynamics). The image files were analyzed using ArrayVision software (Imaging Research, St. Catharines, Ontario, Canada). A template which contains the spot layout of the array was overlaid on the phosphorimage, and the pixel intensity of each spot on the array was determined. The background-corrected pixel intensities for each spot were exported from ArrayVision into ArrayStat (Imaging Research) for statistical analysis. These values were log transformed, common error was determined, and outliers were removed. The values were then normalized by the mean across conditions by an iterative process, and a false-positive error correction is achieved by application of the false-discovery rate method (4). The z test for two independent conditions using the false-discovery rate method (nominal alpha, P ≤ 0.01) was used as the basis for computing confidence intervals.
Alternatively, ArrayVision data was analyzed using macro functions found within the Microsoft Excel spreadsheet program as described previously (38). Significance of differential expression was determined by a two-tailed, unpaired Student t test at a P of <0.01.
Real-time quantitative RT-PCR.
To independently confirm the differential expression data generated by microarray analysis, real-time quantitative reverse transcription (RT)-PCR was utilized to determine the relative expression of specific genes at the two different temperatures. Primers were designed using PRIMER EXPRESS software (PE Biosystems, Foster City, Calif.) (Table 1). All primer pairs produced a single amplicon of the expected size when B. burgdorferi total genomic DNA was used as the template for PCR. cDNA was generated using 2 μg of total RNA and 3′-specific primers as described above. Real-time PCR was performed in triplicate on 5 ng of cDNA with the SYBR Green master mix using a Perkin-Elmer ABI Prism 7700 sequence detection system following the manufacturer's instructions (PE Biosystems). To ensure that there were no genomic contamination of cDNA and no primer dimer artifact, reactions containing cDNA generated without reverse transcriptase and reactions containing primers alone also were included. The constitutively expressed flaB gene was used to normalize input cDNA for all experiments. Differential expression was determined by calculating the change in threshold cycle (ΔCt) of each ORF with RNA isolated from cells grown at the different temperatures.
Supplementary data.
Supplementary data can be found in supplementary Tables 1 to 4 posted at www.nymc.edu/iraschwartz/borreliaarrays.
TABLE 4.
Genes with increased expression at 35°C relative to 23°C
| Gene name | Description of gene product (homologous gene) | Paralogous family | Fold induction |
|---|---|---|---|
| BBA25 | Decorin binding protein B (dbpB) | 74 | 6.81 |
| BBA73 | Antigen, P35, putative | 54 | 6.26 |
| BBA72 | Hypothetical protein | 6.23 | |
| BBA66 | Antigen, P35, putative | 54 | 5.48 |
| BBB19 | Outer surface protein C (ospC) | 5.48 | |
| BB0175 | Conserved hypothetical protein | 4.91 | |
| BBA24 | Decorin binding protein A (dbpA) | 74 | 4.56 |
| BBJ37 | Hypothetical protein | 4.51 | |
| BBP28 | Lipoprotein (mlpA) | 113 | 4.33 |
| BBA65 | Hypothetical protein | 54 | 3.85 |
| BBA32 | Hypothetical protein | 3.61 | |
| BBS30 | Lipoprotein (mlpC) | 113 | 3.25 |
| BBM26 | Conserved hypothetical protein | 143 | 3.15 |
| BBA74 | Outer membrane porin (oms28) | 171 | 3.00 |
| BBL26 | Conserved hypothetical protein | 143 | 2.96 |
| BBA71 | Hypothetical protein | 54 | 2.93 |
| BB0536 | Zinc protease, putative | 2.83 | |
| BB0365 | Lipoprotein LA7 | 2.73 | |
| BBK07 | Hypothetical protein | 59 | 2.73 |
| BBA37 | Hypothetical protein | 2.72 | |
| BB0844 | Hypothetical protein | 12 | 2.70 |
| BBQ33 | Conserved hypothetical protein | 143 | 2.70 |
| BBA57 | Hypothetical protein | 2.66 | |
| BBS26 | Conserved hypothetical protein | 143 | 2.61 |
| BBI43 | Conserved hypothetical protein | 55 | 2.54 |
| BBJ43 | Hypothetical protein | 90 | 2.54 |
| BBP26 | Conserved hypothetical protein | 143 | 2.48 |
| BB0077 | Hypothetical protein | 2.45 | |
| BBI42 | Outer membrane protein, putative | 52 | 2.39 |
| BBM28 | Lipoprotein (mlpF) | 113 | 2.29 |
| BBR26 | Conserved hypothetical protein | 143 | 2.24 |
| BBA36 | Lipoprotein | 2.17 | |
| BBB05 | PTSa system, cellobiose-specific IIA component (celC) | 2.14 | |
| BB0740 | Conserved hypothetical protein | 2.10 | |
| BBA64 | Antigen, P35 | 54 | 2.10 |
| BBM25 | Conserved hypothetical protein | 112 | 2.05 |
| BBJ39.1 | Multidrug-efflux transporter | 54 | 2.04 |
| BBR23 | Pore-forming hemolysin (blyA) | 109 | 2.04 |
| BB0040 | Chemotaxis protein methyltransferase (cheR-1) | 33 | 2.02 |
| BBB06 | PTS system, cellobiose-specific IIB component (celA) | 2.01 | |
| BBP25 | Conserved hypothetical protein | 112 | 2.01 |
| BB0162 | Hypothetical protein | 1.99 | |
| BBO26 | Conserved hypothetical protein | 143 | 1.99 |
| BBK01 | Hypothetical protein | 12 | 1.98 |
| BBQ89 | Hypothetical protein | 166 | 1.97 |
| BBQ20 | Hypothetical protein | 154 | 1.96 |
| BB0459 | Hypothetical protein | 1.95 | |
| BBD01 | Hypothetical protein | 176 | 1.94 |
| BBQ32 | Conserved hypothetical protein | 112 | 1.94 |
| BBS25 | Conserved hypothetical protein | 112 | 1.93 |
| BBA67 | Hypothetical protein | 1.92 | |
| BBL25 | Conserved hypothetical protein | 112 | 1.92 |
| BBP23 | Pore-forming hemolysin (blyA) | 109 | 1.92 |
| BB0203 | Lambda CII stability-governing protein (hflK) | 1.89 | |
| BBN26 | Outer surface protein, putative | 143 | 1.88 |
| BBA69 | Hypothetical protein | 54 | 1.87 |
| BBM24 | Hemolysin accessory protein (blyB) | 111 | 1.83 |
| BBQ30 | Pore-forming hemolysin (blyA) | 109 | 1.83 |
| BB0279 | Flagellar protein (fliL) | 1.82 | |
| BBR25 | Conserved hypothetical protein | 112 | 1.82 |
| BB0013 | Hypothetical protein | 1.81 | |
| BBM23 | Pore-forming hemolysin (blyA) | 109 | 1.81 |
| BBS23 | Pore-forming hemolysin (blyA) | 109 | 1.80 |
| BB0211 | DNA mismatch repair protein (mutL) | 1.79 | |
| BB0467 | Conserved hypothetical protein | 1.78 | |
| BBF33 | Surface antigen lipoprotein (vlsE) | 170 | 1.78 |
| BB0075 | Hypothetical protein | 1.77 | |
| BBA61 | Conserved hypothetical protein | 1.77 | |
| BBS12 | Hypothetical protein | 153 | 1.77 |
| BB0447 | Na+/H+ antiporter (napA) | 1.76 | |
| BBN25 | Conserved hypothetical protein | 112 | 1.76 |
| BB0218 | Phosphate ABC transporter, ATP-binding protein (pstB) | 4 | 1.75 |
| BBO25 | Conserved hypothetical protein | 112 | 1.75 |
| BB0237 | Apolipoprotein N-acyltransferase | 1.73 | |
| BBH02 | Conserved hypothetical protein | 76 | 1.73 |
| BBQ03 | Outer membrane protein, putative | 52 | 1.73 |
| BB0377 | Autoiducer-2 (AI-2) synthesis, luxS | 1.72 | |
| BBA04 | Antigen, S2 | 44 | 1.72 |
| BBK32 | Immunogenic protein P35 | 1.72 | |
| BBL23 | Pore-forming hemolysin (blyA) | 109 | 1.72 |
| BB0751 | Hypothetical protein | 1.71 | |
| BB0017 | Conserved hypothetical integral membrane protein | 1.70 | |
| BBQ28 | Conserved hypothetical protein | 141 | 1.70 |
| BBS14 | Hypothetical protein | 155 | 1.70 |
| BB0457 | Excinuclease ABC, subunit C (uvrC) | 1.69 | |
| BBL17 | Hypothetical protein | 159 | 1.69 |
| BBM08 | Conserved hypothetical protein | 107 | 1.69 |
| BB0376 | S-Adenosylmethionine synthetase (metK) | 1.65 | |
| BBQ24 | Hypothetical protein | 159 | 1.65 |
| BBR12 | Hypothetical protein | 153 | 1.65 |
| BB0126 | Hypothetical protein | 1.64 | |
| BB0374 | Hypothetical protein | 1.64 | |
| BBF32 | vls recombination cassettes Vls2-Vls16 | 170 | 1.64 |
| BBO23 | Pore-forming hemolysin (blyA) | 109 | 1.64 |
| BBR24 | Hemolysin accessory protein (blyB) | 111 | 1.64 |
| BBS22 | Conserved hypothetical protein | 142 | 1.64 |
| BBM02 | Hypothetical protein | 147 | 1.63 |
| BBP17 | Hypothetical protein | 159 | 1.63 |
| BBS34 | Conserved hypothetical protein | 50 | 1.63 |
| BBH20 | Outer membrane porin, authentic frameshift (oms28) | 171 | 1.62 |
| BBP12 | Hypothetical protein | 153 | 1.62 |
| BBP24 | Hemolysin accessory protein (blyB) | 111 | 1.61 |
| BBS01 | Hypothetical protein | 146 | 1.61 |
| BB0689 | Hypothetical protein | 1.60 | |
| BBM17 | Hypothetical protein | 159 | 1.59 |
| BBR33 | Plasmid partition protein, putative | 32 | 1.59 |
| BBS24 | Hemolysin accessory protein (blyB) | 111 | 1.59 |
| BB0086 | Conserved hypothetical protein | 1.58 | |
| BB0771 | RNA polymerase sigma factor (rpoS) | 89 | 1.58 |
| BBH01 | Conserved hypothetical protein | 166 | 1.58 |
| BB0405 | Hypothetical protein | 35 | 1.57 |
| BBK50 | Immunogenic protein P37 | 75 | 1.56 |
| BBN24 | Hemolysin accessory protein (blyB) | 111 | 1.56 |
| BBS31 | Conserved hypothetical protein | 161 | 1.56 |
| BB0308 | Hypothetical protein | 1.55 | |
| BB0446 | Aspartyl-tRNA synthetase (aspS) | 1.54 | |
| BB0812 | Pantothenate metabolism flavoprotein (dfp) | 1.54 | |
| BBH37 | Hypothetical protein | 12 | 1.53 |
| BBR17 | Hypothetical protein | 159 | 1.53 |
| BB0759 | Hypothetical protein | 1.52 | |
| BB0832 | Hypothetical protein | 1.52 | |
| BB0827 | ATP-dependent helicase (hrpA) | 1.50 | |
| BBQ13 | Conserved hypothetical protein | 149 | 1.50 |
| BBQ17 | Conserved hypothetical protein | 151 | 1.50 |
| BB0216 | Phosphate ABC transporter, permease protein (pstC) | 41 | 1.49 |
| BB0415 | Protein-glutamate methylesterase (cheB-1) | 132 | 1.49 |
| BB0431 | Chromosome segregation protein, putative | 32 | 1.49 |
| BBO24 | Hemolysin accessory protein (blyB) | 111 | 1.49 |
| BB0819 | Conserved hypothetical protein | 1.48 | |
| BB0210 | Surface-located membrane protein 1 (lmp1) | 1.47 | |
| BBN21 | Hypothetical protein | 141 | 1.45 |
| BBM36 | Conserved hypothetical protein | 144 | 1.44 |
| BBR19 | Conserved hypothetical protein | 139 | 1.42 |
PTS, phosphotransferase.
RESULTS AND DISCUSSION
Array description.
A list of 1,697 gene features was generated as described in Materials and Methods, and primers were designed to amplify these regions (see supplementary Table 1). Amplified DNAs were spotted in duplicate on positively charged nylon membranes. The array also included 29 control spots derived from PCR mixtures that either lacked DNA template or contained only spotting buffer. Sheared genomic DNA was labeled by random priming and was used to probe the arrays in triplicate. The noise level was determined by averaging the pixel values in each of the control spots after subtraction of background levels (62). Sixty-one ORFs did not yield a pixel value exceeding the noise level by a factor of 2 because they either failed PCR or did not bind sufficient probe in the hybridization step. Thus, the final array can yield informative data for 1,628 (96.4%) of the putative ORFs. The 23 chromosomal and 38 plasmid ORFs which are not represented on the array are listed in supplementary Table 2.
Generation of a B. burgdorferi strain B31-MI clonal isolate and growth conditions for transcriptional profiling.
Several studies have shown that uncloned isolates of B. burgdorferi often contain subpopulations of organisms with different plasmid contents, which can result in phenotypic differences (18). Therefore, before undertaking a global analysis of the transcriptional profiles of B. burgdorferi at different temperatures, a clonal isolate of the B31-MI strain was generated. This was accomplished by infecting a mouse with the parental, uncloned B31-MI strain and cultivating organisms from an ear punch biopsy specimen for subsequent plating on solid BSK-II medium (18). A single, well-separated colony, designated B31-MI P1.1, was selected and screened by PCR using plasmid-specific primers to ensure it harbored all 21 plasmids previously identified (data not shown).
This B31-MI clone was cultivated in BSK-II medium at 23 and 35°C as described in Materials and Methods. Aliquots of the culture were analyzed for the characteristic induction of OspC protein after temperature shift by SDS-polyacrylamide gel electrophoresis and immunoblot analysis. As expected, no OspC was detected in organisms cultivated at 23°C, whereas prominent induction of OspC was observed in temperature-shifted organisms (Fig. 1).
FIG. 1.
Total protein profile and OspC expression of temperature-shifted cultures used for RNA isolation. (A) SDS-polyacrylamide gel electrophoresis of whole-cell lysates stained with Sypro Ruby. A 24-kDa band corresponding to OspC is missing in B31-MI P1.1 grown at 23°C but present in spirochetes shifted to 35°C (arrow). Each lane contains borrelial lysate equivalent to 3.6 × 107 cells. Molecular masses are indicated on the left. (B) Immunoblot of whole-cell lysates probed with rabbit polyclonal antiserum to OspC. Lysates were identical to those in panel A.
Transcript analysis.
RNA was isolated from four separate 100-ml cultures grown at each temperature. After DNase I treatment and evaluation of RNA quality (see Materials and Methods), the RNAs isolated from the four cultures at each temperature were pooled. In this manner potential variability in the transcript levels introduced during growth or RNA isolation of individual cultures was minimized. Radioactive probes were prepared by cDNA synthesis using 3′ ORF-specific primers as described in Materials and Methods and hybridized to the membrane arrays. Probe synthesis and array hybridization were performed eight separate times in two different laboratories (at the New York Medical College and at the University of Oklahoma Health Sciences Center). Pixel densities in each spot were determined by phosphorimager analysis and statistically analyzed using the ArrayStat software package. Thus, 16 data points were analyzed for each ORF.
A total of 146 ORFs (45 chromosomal ORFs, 99 plasmid ORFs, and 2 plasmid intergenic probes) did not give detectable signals at either temperature. These are listed in supplementary Table 3. Of these, 57 ORFs (31 chromosomal and 26 plasmid) do not appear to be obviously defective by sequence analysis and gene arrangement. These may well be functional genes that are not expressed at detectable levels under the growth conditions used. However, also in this unexpressed class of ORFs are 44 pseudogenes and 45 unusually short plasmid ORFs (<300 bp). Such short ORFs are mostly found in regions rich in pseudogenes where ORFs are not tightly packed and were previously suspected not to encode any protein product (see references 6 and 7 and supplementary material therein). The substantial overrepresentation of unexpressed ORFs in these pseudogene and short-gene classes strongly suggests that these are not functional genes. Of eight intergenic regions that appeared to lack obvious ORFs, only two (BBH16.1 and BBD06.1) showed moderate levels of expression at either temperature. This might be the result of transcriptional readthrough from the upstream ORFs. Thus, over 90% of the arrayed ORFs had at least minimal expression under one of the growth conditions.
TABLE 3.
Most highly expressed genes in B. burgdorferi cultivated at 23°C
| Gene name | Description of gene product (homologous gene) | Relative transcript level | Rank at temp (°C)
|
|
|---|---|---|---|---|
| 23 | 35 | |||
| BB0440 | Ribosomal protein L34 (rpmH) | 137.55 | 1 | 1 |
| BBA59 | Lipoprotein | 129.97 | 2 | 2 |
| BB0233 | Ribosomal protein S20 (rpsT) | 77.39 | 3 | 5 |
| BB0049 | Hypothetical protein | 75.82 | 4 | 3 |
| BB0559 | PTSa system, glucose-specific IIA component (crr) | 75.40 | 5 | 11 |
| BB0804 | Ribosomal protein S15 (rpsO) | 57.07 | 6 | 4 |
| BBA62 | Lipoprotein | 48.50 | 7 | 8 |
| BB0300 | Cell division protein (ftsA) | 40.53 | 8 | 13 |
| BB0294 | Flagellar basal-body rod protein (flgB) | 40.22 | 9 | 10 |
| BB0500 | Ribosomal protein S13 (rpsM) | 40.03 | 10 | 7 |
| BBA15 | Outer surface protein A (ospA) | 32.51 | 11 | 12 |
| BB0232 | hbbU protein | 30.50 | 12 | 9 |
| BB0778 | Ribosomal protein L21 (rplU) | 26.96 | 13 | 14 |
| BB0047 | Conserved hypothetical protein | 25.27 | 14 | 52 |
| BB0162 | Hypothetical protein | 22.54 | 15 | 6 |
| BB0486 | Ribosomal protein L29 (rpmC) | 21.46 | 16 | 17 |
| BB0355 | Transcription factor, putative | 20.49 | 17 | 27 |
| BBA16 | Outer surface protein B (ospB) | 20.08 | 18 | 23 |
| BB0741 | Chaperonin (groES) | 20.00 | 19 | 22 |
| BB0480 | Ribosomal protein L23 (rplW) | 19.06 | 20 | 16 |
| BBQ42 | Conserved hypothetical protein | 18.94 | 21 | 26 |
| BB0704 | Acyl carrier protein | 17.05 | 22 | 18 |
| BBS37 | Conserved hypothetical protein | 17.05 | 23 | 25 |
| BB0256 | Ribosomal protein S21 (rpsU) | 16.85 | 24 | 24 |
| BB0631 | Hypothetical protein | 16.79 | 25 | 32 |
| BB0044 | Hypothetical protein | 16.69 | 26 | 66 |
| BB0423 | Hypothetical protein | 15.35 | 27 | 48 |
| BB0562 | Hypothetical protein | 14.09 | 28 | 34 |
| BB0803 | tRNA pseudouridine 55 synthase (truB) | 13.72 | 29 | 38 |
| BB0541 | Hypothetical protein | 13.61 | 30 | 40 |
| BBF20 | Conserved hypothetical protein | 12.78 | 31 | 21 |
| BB0061 | Thioredoxin (trxA) | 12.68 | 32 | 51 |
| BB0229 | Ribosomal protein L31 (rpmE) | 12.45 | 33 | 33 |
| BB0441 | Ribonuclease P protein component (rnpA) | 12.31 | 34 | 47 |
| BBE21 | Conserved hypothetical protein | 11.84 | 35 | 58 |
| BBL35 | Conserved hypothetical protein | 11.26 | 36 | 42 |
| BB0691 | Translation elongation factor G (fus-2) | 10.85 | 37 | 56 |
| BB0501 | Ribosomal protein S11 (rpsK) | 10.52 | 38 | 35 |
| BB0538 | Conserved hypothetical protein | 10.47 | 39 | 37 |
| BB0190 | Translation initiation factor 3 (infC) | 10.26 | 40 | 41 |
| BB0143 | Conserved hypothetical protein | 10.18 | 41 | 89 |
| BBH29 | Conserved hypothetical protein | 10.14 | 42 | 44 |
| BBM38 | erpK protein (erpK) | 9.96 | 43 | 46 |
| BBH16 | Hypothetical protein | 9.91 | 44 | 63 |
| BBI29 | Hypothetical protein | 9.79 | 45 | 178 |
| BBE10 | Hypothetical protein | 9.76 | 46 | 28 |
| BBB28 | Hypothetical protein | 9.44 | 47 | 39 |
| BB0338 | Ribosomal protein S9 (rpsl) | 9.29 | 48 | 61 |
| BB0489 | Ribosomal protein L24 (rplX) | 9.25 | 49 | 55 |
| BB0785 | Stage V sporulation protein | 9.21 | 50 | 43 |
PTS, phosphotransferase.
In order to obtain values for transcript abundance for each ORF, the following normalizations were performed essentially as described elsewhere (62). The relative transcript level for each ORF was determined by dividing the average pixel density for that ORF by the sum of the total pixels of all ORFs on arrays (which exceeded noise by a factor of 2) hybridized with cDNA (prepared from total RNA) to yield a percent expression value for each ORF. A similar calculation was performed for genomic DNA hybridizations, generating a percent genomic value for each ORF. Finally, dividing the percent expression value by the percent genomic value yields a measure of relative transcript level for each expressed ORF. A list of the most highly expressed genes in B. burgdorferi cultivated at either 35 or 23°C is given in Tables 2 and 3 and a complete list is presented in supplemental Table 4.
Several points are worthy of note. As expected, a substantial number of the most highly expressed genes at either temperature encode ribosomal proteins or other proteins involved in translation. The genes encoding the abundant outer surface proteins OspA, OspB, and OspC are highly expressed, indicating that the cellular levels of these proteins are regulated largely at the transcription step. Genes encoding products of no known function (i.e., annotated as hypothetical or conserved hypothetical in the sequence database) represent 40% of the 50 most highly expressed genes. This underscores the important roles likely to be played by such genes in B. burgdorferi cellular function. The lists of the 50 most highly expressed genes at the two temperatures largely overlap. Several ORFs, most notably ospC, bba32, bba74, bbi29, and bb0175, are highly expressed at 35°C but have significantly lower expression at 23°C (see below).
Expression of paralogous genes.
As described above, the B. burgdorferi genome contains 175 paralogous gene families ranging from 2 to 41 members. This greatly complicates both the construction of a genome array and subsequent analysis of array data. One approach would involve choosing some subset of paralogous genes to represent all members of the family. This would require experimental determination of the parameters governing cross-hybridization and selection of only those family members that would not cross-hybridize. Alternatively, one could ignore this problem and simply amplify all genes regardless of sequence identity. As noted earlier, the second approach was taken for construction of the arrays used in the present studies. Thus, understanding the expression of individual members of a paralogous family will require more detailed examination.
The extent of sequence identity among members of specific paralogous families varies substantially. In some cases, sequence identity among the individual family members is quite low (e.g., family 54), whereas for others the genes are indistinguishable at the sequence level (e.g., family 112). It is instructive to consider the expression data for selected cases to gain insight into the extent of cross-hybridization encountered in these studies. Family 54 consists of 14 members, most with <60% sequence identity to any other family member. In this situation, cross-hybridization is of little concern, and, indeed, family 54 genes display vastly different levels of expression, and some members are upregulated at 35°C, whereas some have higher expression at 23°C (see below). At the other extreme is family 112; all eight members have >90% sequence identity, and some are 100% identical. In this case, it would be impossible by any hybridization-based technique (including RT-PCR) to differentiate expression from any one of these identical genes. This is discussed further below with regard to the nearly identical blyAB genes. The intermediate situation is somewhat more problematic. This is exemplified by family 62, which includes nine members. Four of these—bbj19, bbd14, bbk23, and bbh26—all have significantly higher expression at 23°C relative to 35°C. bbj19, bbk23, and bbh26 are 66 to 80% identical to each other but <30% identical to bbd14. This suggests that the expression data for the latter gene are unique, but those for the other three genes are unclear. Is there transcription of only one of these ORFs, or are the expression data a composite of transcription from all three genes? In such a case, further analysis by an alternative method, such as real-time RT-PCR with gene-specific primers, can differentiate between these possibilities.
In order to get some insight into the extent of cross-hybridization under the experimental conditions employed in the present study, several genes were amplified by PCR, labeled with [α-33P]dATP, and hybridized individually to a membrane array. When labeled bba66 was used as a representative of paralogous family 54, the only significant hybridization on the array was to this ORF. All other members of the family gave signal intensities <6.6% that of bba66 (most were <3%). In contrast, using bbs25 as a representative of paralogous family 112 resulted in virtually equal signals for this gene as well as six other close paralogs (sequence identity of 91 to 100%). bba13 and bbb03, more distantly related members of the family (sequence identity <32%), yielded signal intensities <3% that of bbs25. These results are consistent with the above theoretical discussion and provide some parameters for interpretation of expression data for paralogous genes.
Expression of cp32 plasmid genes and a putative phage operon.
The seven cp32 plasmids of strain B31 are homologous throughout most of their lengths and have several features that suggest they may be prophages (7, 8, 15). Eggers et al. (16) have found that cp32 DNA is present inside phage-like particles produced by B. burgdorferi strain CA-2-11A. Analysis of cp32 sequences shows that they carry homologs of two genes that may encode two proteins that are universally required for phage virion assembly, terminase and portal protein. In all known temperate phages (>100 have been sequenced), the large terminase subunit and portal protein genes are always found in a particular juxtaposition within the operon that carries the genes for construction of the phage particle (called the “late operon”). They are virtually always adjacent, the terminase gene is transcriptionally upstream of the portal gene, and the terminase gene is the second gene in the operon (S. Casjens, unpublished data). Genes bbp42 and bbp01 (and paralogs) have homology with the large terminase subunit and portal protein, respectively; and their juxtaposition is the same as that in the known phages. Late operon genes should not be expressed (or should be expressed at very low rates) under normal culture conditions since few phage particles are being produced. To further explore the possibility that cp32 plasmids may be temperate phages, we analyzed the relative expression of putative ORFs from several regions of the cp32 plasmids (Fig. 2).
FIG. 2.
Relative transcription of selected cp32 genes. The solid black curve represents the relative mRNA levels for all B. burgdorferi genes plotted against the mRNA level rank of all the genes (Table 1 and supplementary Table 4). The portion of the curve delineated by the most highly expressed genes, which have expression levels up to 105 on this scale, is not shown; no cp32 genes lie in that portion of the curve. The expression levels for four different classes of cp32 genes are specifically shown: ▪,13 erp and bap genes; ○, 35 genes in the putative replication-partition region (bbp30 to bbp34 and their cp32 paralogs); □, 28 genes in the blyAB region (bbp23 to bbp26 and their cp32 paralogs); •, 168 putative virion morphogenetic region genes (bbp41, bbp42, and bbp01 through bbp22 and their cp32 paralogs). The open circles, filled squares, and open squares, which should lie on the black line, were artificially raised by values of 1, 2, and 3, respectively, on the vertical axis to enable the reader to distinguish the four categories of genes.
All the genes in the cp32-1 putative late operon (bbp42, bbp43, and bbp01 through bbp22) are expressed at low levels, if at all. By contrast, most genes in the putative cp32 replication-partition gene operon and the erp genes, both of which were previously shown to be expressed (14, 23, 53), have substantially higher average mRNA levels than genes in the putative late operon (Fig. 2). bbp23, bbp24, and bbp25 (and paralogs), designated the blyAB operon, are also expressed at substantially higher levels than the late operon genes (Fig. 2). These expression data are consistent with the proposal that the cp32 plasmids are prophages and genes bbp41 through about gene bbp22 (and paralogs) may be the “late operon.” The genes in these regions are extremely tightly packed, as is the case in all other phages, and are about the number of genes required to build a tailed phage. There are no conflicting homologies to nonphage assembly genes in this region, and the extreme diversity of phage sequences makes it no surprise that nearly all of the genes in this region do not match any protein in the current database. Thus, global gene expression data add support to the idea that one region of the cp32 plasmids may encode phage morphogenetic genes.
Damman et al. (12) have presented evidence that blyA and blyB (bbp23 and bbp24 and paralogs) may encode a protein involved in lysis of the cell during phage lytic growth. Lysis genes are often found in the same operon as morphogenetic genes in other phages. However, the blyAB genes are expressed at levels substantially above the median of all B. burgdorferi expressed genes and those immediately preceding them (Fig. 2). Furthermore, a large (for this region) intergene gap precedes gene bbp23 and paralogs that could harbor a promoter. This seems inconsistent with bbp23-bbp24 being part of a prophage late morphogenesis-lysis operon and perhaps indicates that the role of blyAB should be reexamined.
Temperature-induced differential gene expression.
Many studies have established that expression of a number of B. burgdorferi genes is regulated by changes in environmental conditions (1, 3, 25-27, 41, 44, 49, 55, 64). The best studied of these cues is temperature (3, 26, 27, 45, 49, 55, 66). In order to determine the global changes in temperature-regulated gene expression, arrays were hybridized with cDNA generated from RNA isolated from B. burgdorferi cultivated at either 23 or 35°C. A total of eight independent array hybridizations were carried out for each temperature in two different laboratories. Only gene expression differences between the two temperatures that were significant at a P of <0.01 are considered in the following discussion. In total, 215 ORFs (13.2% of those included on the array) displayed significantly different expression at the two temperatures. A list of these ORFs is presented in Tables 4 and 5.
TABLE 5.
Genes with increased expression at 23°C relative to 35°C
| Gene name | Description of gene product (homologous gene) | Paralogous family | Fold induction |
|---|---|---|---|
| BBJ09 | Outer surface protein D (ospD) | 13.02 | |
| BBI02 | Conserved hypothetical protein | 84 | 6.61 |
| BBA49 | Hypothetical protein | 155 | 5.38 |
| BBJ34 | Hypothetical protein | 92 | 5.28 |
| BBA44 | Hypothetical protein | 4.53 | |
| BBJ08 | Hypothetical protein | 12 | 4.08 |
| BBA51 | Hypothetical protein | 157 | 3.43 |
| BB0241 | Glycerol kinase (glpK) | 3.12 | |
| BB0707 | Hypothetical protein | 3.02 | |
| BB0423 | Hypothetical protein | 3.00 | |
| BB0242 | Hypothetical protein | 2.94 | |
| BBJ19 | Conserved hypothetical protein | 62 | 2.90 |
| BB0243 | Glycerol-3-phosphate dehydrogenase, anaerobic (glpA) | 2.88 | |
| BBA42 | Conserved hypothetical protein | 150 | 2.73 |
| BBJ45 | Hypothetical protein | 59 | 2.64 |
| BBJ36 | Hypothetical protein | 92 | 2.62 |
| BB0599 | Cysteinyl-tRNA synthetase (cysS) | 2.60 | |
| BBI29 | Hypothetical protein | 60 | 2.57 |
| BBJ21.1 | Conserved hypothetical protein | 138 | 2.55 |
| BB0344 | DNA helicase (uvrD) | 18 | 2.46 |
| BBA41 | Conserved hypothetical protein | 149 | 2.40 |
| BBB14 | Hypothetical protein | 2.33 | |
| BB0047 | Conserved hypothetical protein | 2.32 | |
| BBA40 | Hypothetical protein | 148 | 2.23 |
| BBQ09 | Conserved hypothetical protein | 50 | 2.23 |
| BB0240 | Glycerol uptake facilitator (glpF) | 2.22 | |
| BBA46 | Hypothetical protein | 152 | 2.16 |
| BB0559 | PTSa system, glucose-specific IIA component (crr) | 2.15 | |
| BB0010 | Holo-acyl-carrier protein synthase | 2.03 | |
| BBA47 | Hypothetical protein | 153 | 2.02 |
| BBE15 | Hypothetical protein | 1.97 | |
| BB0143 | Conserved hypothetical protein | 1.93 | |
| BB0651 | Conserved hypothetical protein | 1.92 | |
| BBG30 | Hypothetical protein | 1.91 | |
| BB0044 | Hypothetical protein | 1.87 | |
| BBA54 | Hypothetical protein | 158 | 1.85 |
| BBB03 | Hypothetical protein | 1.84 | |
| BB0020 | Pyrophosphate-fructose 6-phosphate 1-phosphotransferase, beta subunit (pfpB) | 6 | 1.81 |
| BBA55 | Hypothetical protein | 159 | 1.81 |
| BBA56 | Hypothetical protein | 160 | 1.80 |
| BB0690 | Neutrophil activating protein (napA) | 1.79 | |
| BB0364 | Conserved hypothetical protein | 1.75 | |
| BB0206 | Conserved hypothetical protein | 1.74 | |
| BB0425 | Hypothetical protein | 1.71 | |
| BBA38 | Hypothetical protein | 146 | 1.71 |
| BBA39 | Hypothetical protein | 147 | 1.69 |
| BBG06 | Conserved hypothetical protein | 57 | 1.69 |
| BBI14 | Hypothetical protein | 60 | 1.69 |
| BB0002 | β-N-acetylhexosaminidase | 21 | 1.68 |
| BB0531 | Hypothetical protein | 1.67 | |
| BB0328 | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (oppA-1) | 37 | 1.63 |
| BBD14 | Conserved hypothetical protein | 62 | 1.63 |
| BBN38 | erpA | 162 | 1.61 |
| BB0225 | Conserved hypothetical protein | 124 | 1.59 |
| BB0269 | minD-related ATP-binding protein (ylxH-1) | 32 | 1.59 |
| BB0366 | Aminopeptidase I (yscI) | 131 | 1.59 |
| BB0272 | Flagellar biosynthesis protein (flhB) | 1.58 | |
| BBA52 | Outer membrane protein | 1.57 | |
| BBE30 | Hypothetical protein | 49 | 1.57 |
| BBK23 | Hypothetical protein | 62 | 1.57 |
| BB0127 | Ribosomal protein S1 (rpsA) | 1.55 | |
| BBJ41 | Antigen, P35, putative | 54 | 1.55 |
| BBG07 | Conserved hypothetical protein | 50 | 1.54 |
| BBI39 | Hypothetical protein | 54 | 1.54 |
| BB0083 | Hypothetical protein | 1.53 | |
| BB0546 | Hypothetical protein | 1.53 | |
| BB0354 | Hypothetical protein | 1.52 | |
| BB0245 | Hypothetical protein | 125 | 1.51 |
| BB0168 | dnaK suppressor | 1.47 | |
| BB0445 | Fructose-bisphosphate aldolase (fba) | 1.47 | |
| BBB04 | PTS system, cellobiose-specific IIC component (celB) PTS system, maltose and glucose-specific IIABC | 1.47 | |
| BBB29 | Component (malX) | 16 | 1.47 |
| BB0236 | Hypothetical protein | 1.45 | |
| BBA50 | Hypothetical protein | 1.43 | |
| BBH26 | Hypothetical protein | 62 | 1.43 |
| BB0112 | Ribosomal protein L9 (rpII) | 1.42 | |
| BBN18 | Hypothetical protein | 160 | 1.41 |
| BB0725 | Conserved hypothetical protein | 1.40 | |
| BB0056 | Phosphoglycerate kinase (pgk) | 1.37 | |
| BB0482 | Ribosomal protein S19 (rpsS) | 1.36 | |
| BBD12 | Hypothetical protein | 1.36 | |
| BB0091 | V-type ATPase, subunit I | 1.33 |
PTS, phosphotransferase.
One-hundred thirty-three ORFs had significantly higher expression at 35°C relative to 23°C, whereas 82 genes had significantly greater expression at 23°C in comparison to 35°C. Of these, 62% (134 of 215) are annotated as unknown function. Nearly two-thirds (136 of 215) of the temperature-regulated genes are encoded on plasmids, underscoring the important roles that such genes may play in the physiology of B. burgdorferi, especially with regard to adaptation to environmental changes.
Genomic location of temperature-regulated genes.
The chromosome was found to contain 79 temperature-regulated genes, while 136 were located on the various plasmids. Of the 21 sequenced B31-MI P1.1 plasmids, only 3 (lp5, lp21, and cp9) did not encode any ORFs differentially regulated by temperature (Fig. 3A). Among the other 18 plasmids, lp54, lp38, and the circular plasmids cp26, cp32-3, and cp32-6 had more than 20% of their ORFs regulated by temperature (42, 22, 24, 24, and 21%, respectively) (Fig. 3A). For lp54, lp38, and cp26, this number is likely to be accurate, since none of the temperature-regulated genes are expected to cross-hybridize with paralogs. As noted above, the true number of regulated genes on cp32 plasmids cannot be determined with certainty, since many of these genes are members of paralogous families with substantial sequence identity. For example, for cp32-3 and cp32-6, five of the temperature-regulated genes are members of such paralogous families.
FIG. 3.
(A) Temperature-regulated genes encoded on each genetic element. The percentage of genes on the chromosome and each plasmid differentially expressed at either 23 or 35°C. (B) Temperature-regulated genes encoded on lp54. The 76 ORFs encoded on lp54 are represented as bars linearly along the x axis. Closed bars denote ORFs expressed from the positive strand, and open bars represent those transcribed off the minus strand. Genes with greater expression at 35°C are indicated as up-regulated, and those with greater expression at 23°C are shown as down-regulated. Paralogs of genes also encoded on cp32 plasmids are indicated by shaded boxes.
Linear plasmid lp54 is of particular interest. Over 80% (26 of 32) of its temperature-regulated genes are clustered in the right half of the plasmid (Fig. 3B). Moreover, all the lp54 genes that are more significantly expressed at 23°C are clustered in a single region between bba38 and bba56, and these are all transcribed in the same direction. Casjens et al. (7) have speculated an ancient insertion of a linearized cp32 plasmid into lp54, with bbp01 to bbp18 roughly corresponding to bba38 to bba56. Interestingly, none of the genes in the equivalent regions of any cp32 plasmids are upregulated at 23°C. Thus, if the paralogous regions in lp54 are indeed the result of insertion of a cp32, subsequent changes in lp54 must have included acquisition or evolution of regulatory sequences which respond to temperature. Curiously, all the remaining temperature-responsive genes on lp54 (all of which are upregulated at 35°C) lie outside the putative cp32 insertion. It is reasonable to suggest that lp54 has evolved to play an important role in the adaptation of the spirochete to growth at different temperatures.
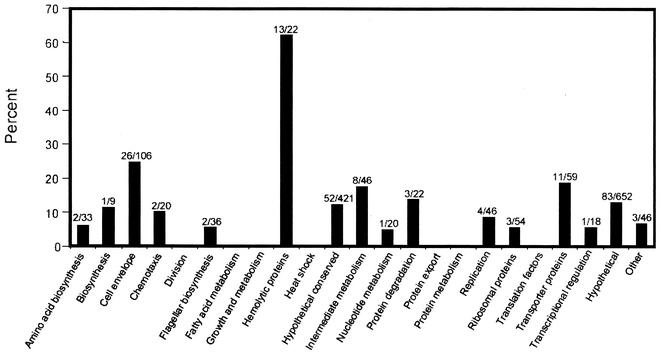
Functional categories of differentially expressed B. burgdorferi genes.
All B. burgdorferi ORFs have been grouped into 23 different functional categories based on sequence similarities to known genes and proteins from other organisms (22). Of these, 16 contain ORFs that responded differentially to a change in temperature (Fig. 4). As noted above, the most-striking observation was that of the 215 genes that are temperature regulated, 134 encode proteins of unknown function (categories HX and U). Of genes encoding proteins of known function, the greatest number was found to be putative cell envelope constituents, with 26 being differentially expressed at the two temperatures. This is consistent with the proposal that B. burgdorferi remodels its surface in response to changes in environmental conditions (3, 26, 41, 45, 49, 55, 64). Interestingly, almost all of the differentially expressed cell envelope genes (21 of 26) were found to be upregulated at 35°C. Additionally, almost 20% (11 of 59) of the genes encoding putative transport proteins were differentially expressed—five more highly at 35°C and six more highly at 23°C. Eight genes annotated as being involved in intermediary metabolism (IM) were differentially expressed; interestingly, seven were more highly expressed at 23°C. The possible role of these genes in adaptation to the lower temperature is discussed below. The only gene in this category upregulated at 35°C is bb0376, a metK (S-adenosylmethionine synthetase) homolog, which was recently shown to be essential for growth in Escherichia coli (63). Interestingly, the adjacent gene, bb0377 (annotated as a conserved hypothetical), was recently shown to be a B. burgdorferi luxS ortholog (52), and the expression of this gene is also significantly higher at 35°C. Schauder et al. have proposed that these two genes comprise an operon with bb0375 (a putative 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase) whose products would be responsible for synthesis of the quorum-sensing autoinducer, AI-2 (47). In the present study, bb0375 transcript levels were very low and differential temperature-dependent expression was not observed.
FIG. 4.
Temperature-regulated genes by functional category. ORFs were placed into 23 functional categories according to the recommendations of Fraser et al. (22). The percentage of the genes in each functional category that are differentially expressed at either 23 or 35°C is presented. The numbers above each bar indicate the fraction of genes in each category that are temperature regulated. The functional categories are as follows: ARS, amino acid biosynthesis; B, biosynthesis; CE, cell envelope; CH, chemotaxis proteins; D, cell division; F, flagellar biosynthesis; FM, fatty acid metabolism; GM, general metabolism; HE, hemolysins; HS, heat shock proteins; HX, conserved hypothetical proteins; NM, nucleotide metabolism; PD, protein degradation; PE, protein export; PM, protein metabolism; R, replication; RP, ribosomal proteins; TF, translation factors; TP, transporter proteins; TR, transcription; U, hypothetical proteins; X, other.
The other functional category for which a substantial number of genes are temperature regulated is the category of putative hemolysins; 13 are significantly upregulated (1.5- to 2-fold) at 35°C. Sixteen of the 21 putative hemolysin genes are located on cp32 plasmids. The eight blyA sequences are 100% identical, and the eight blyB sequences are 93 to 100% identical. Thus, as noted above, it is not possible to determine whether one, some, or all of these genes are expressed. The three cp32-encoded putative hemolysin genes (bbn23, bbl24, and bbq31) that did not meet the stringent statistical criteria for differential regulation (i.e., P < 0.01) were all upregulated at 35°C, with P values of 0.02, 0.015, and 0.013, respectively. It therefore appears that these genes are more significantly expressed at 35°C, and it is tempting to speculate that this may have an in vivo correlation to the temperature increase in the tick gut on acquisition of the blood meal in that expression of hemolysin may be required for its digestion.
Differential expression of cell envelope genes.
Twenty-six genes encoding putative cell envelope constituents were differentially expressed. Among these were a number of genes known to be expressed preferentially in vivo or at 35°C, including ospC and dbpAB, the levels of expression of which were elevated at 35°C by 5.5-fold and 4.5- to 7-fold, respectively. Several members of paralogous family 54—bba64, bba65, bba66, bba69, bba71, and bba73—are more highly expressed at 35°C (between 1.8- and 6.26-fold). Of these, only the bba64 protein is known to be on the cell surface (17), but bba66, bba73, bba65, and bba69 each have potential signal peptidase II sites and are predicted to be cell envelope constituents. Sequence identity between any of these genes is less than 33%, and therefore, the signals obtained for each of these genes should be specific. bba66 to bba64 and bba73 to bba71 likely constitute operons. bba64 encodes a protein designated P35, which is reported to be regulated by cell density, growth phase, or pH and is undetectable in the unfed tick (24, 29, 44, 45). This would be consistent with elevated expression at 35°C. bbk32, which encodes a fibronectin binding protein (42) and whose expression is induced in feeding ticks and mice (21), is significantly upregulated at 35°C (1.7-fold).
Three lipoprotein genes—bbm28, bbp28, and bbs30, which are all members of paralogous gene family 113 and have recently been designated Mlps (66)—are significantly upregulated at 35°C (2.3- to 4.3-fold). The mlp family is comprised of eight genes with substantial sequence identity (72 to 88%, except for bbq35). In strains 297 and B31, some members of this family were not expressed at 23°C, were expressed somewhat at 35°C, and were expressed more substantially at 37°C (41, 66). Consistent with the findings here, prior Northern blot analyses of Mlp expression revealed substantial temperature-dependent expression of mlpA (bbp28), mlpC (bbs30), mlpF (bbm28), and mlpH (bbl28); the remaining mlp genes were poorly expressed, as was the case in the present study (41). We cannot rule out the possibility that expression of some mlp genes is regulated only by mammalian host factors, as previously shown for mlp orthologs in B. burgdorferi strain 297 (1). These putative factors would not have been present under our experimental conditions. The combined data, however, strongly suggest that only a subset of the mlp genes is temperature regulated.
Another paralogous family whose members are upregulated at 35°C is family 143. These genes have no known function, but all contain a signal peptidase II modification and processing site, strongly suggesting that they encode lipoproteins. In contrast to the Mlp family, all cp32-encoded family 143 genes are temperature regulated (up to two- to threefold at 35°C). However, since all the genes share >94% sequence identity, it is not clear whether the observed expression is the result of mRNA transcript from one or more of these genes. Thus, although by database annotation the levels of expression of 21 cell envelope constituents are significantly increased at 35°C, the actual number may be somewhat higher than that (31, if one considers bba65, bba69, and the family 143 genes). Regardless of the precise number, our results underscore the notion that substantial remodeling of the spirochete's cell surface occurs as a consequence of growth at the elevated temperature (e.g., during tick feeding and transmission).
Several cell envelope genes are expressed at significantly higher levels in organisms grown at 23°C. Most noteworthy is the differential expression of plasmid lp38-encoded ospD (bbj09), which was 13-fold elevated at 23°C relative to 35°C. OspD was first characterized as a protein that was unique to low-passage-number isolates of B. burgdorferi (37). No information regarding its regulation by temperature has been reported, and its function is not known. Plasmid lp38 is missing from a subtype of B. burgdorferi human patient and animal isolates (30, 40), suggesting that it is not required for mammalian infection. By analogy to several other proteins that are known to be upregulated at 23°C and expressed in the tick midgut (e.g., OspA), this suggests that OspD may play a role during tick infection.
It previously has been shown that the various cp32s harbor genes encoding a large family of immunogenic lipoproteins, termed Erps (for OspEF-related proteins) (17, 53, 56). However, a closer examination of the Erps has revealed that they can be divided into three different and evolutionarily distinct paralogous gene families (families 162, 163, and 164). This separation is supported by the combined annotation of the complete strain B31 genome and the detailed analyses of these gene families in strains 297 and B31 (2, 5, 7, 54). It has been proposed that paralogous families 162, 163, and 164 be termed OspE-related proteins, Elps, and OspF-related proteins, respectively (2, 5). Interestingly, several studies in multiple borrelial strains have shown that some members of the 162, 163, and 164 protein families are regulated by temperature as well as other mammal-specific factors (1, 3, 26, 53, 55, 57, 60). In the present study, only expression of bbn39 (family 163) was found to be significantly altered by temperature, and surprisingly, its expression was found to be significantly greater (1.6-fold) at 23°C than at 35°C. It is worth noting in this regard that expression of bbo39 (erpL) and bbr42 (ospF) was undetectable at 23°C but was observable at 35°C (these are not listed in Table 4 because the differences in expression did not meet the required stringent statistical criteria). Prior studies have shown that the genes comprising families 162, 163, and 164 are typically upregulated at 35°C (53, 55), but the reasons for the discrepancy between the array results presented here and in earlier studies are not clear. They may be related to differences in the growth medium employed here compared to that used in prior studies (BSK-II versus BSK-H medium), which has been shown to be important in the overall regulation of these genes (65). Alternatively, it may be that expression of these various gene families is more complex than previously recognized and that factors other than temperature also influence their expression. In this regard it should be noted that prior studies of several borrelial strains, including strains N40 and 297, have shown that mammalian host factors are important in regulating transcription and translation for several family: 162, 163, and 164 orthologs (1, 3, 13, 27, 57, 60). Future studies using real-time PCR and specific primers will likely be needed to fully elucidate the complex regulation of the genes which comprise these three lipoprotein gene-encoding families.
Differential expression of putative transporter genes.
A number of genes with possible transporter functions are differentially expressed by temperature. Two operons are of particular interest. bb0216 to bb0218 encode components of a putative phosphate ABC transporter, and bb0216 and bb0218 are significantly upregulated 1.5- to 1.75-fold at 35°C. Expression of bb0217 was also greater at 35°C (1.4-fold) but with a P value of 0.019, which did not reach significance. Therefore, it is tempting to speculate that phosphate transport may be enhanced at the higher temperature.
A cellobiose/chitobiose transporter operon is encoded on circular plasmid cp26. This operon has an unusual organization; celC and celA (bbb05 and bbb06) are transcribed off the plus strand, whereas celB (bbb04) is transcribed divergently. (The cel genes were renamed chb in keeping with their role in chitobiose utilization and nomenclature in E. coli [59], but the original cel designation is used here.) Interestingly, celC and celA were found to have increased expression at 35°C (2.14- and 2.0-fold, respectively), whereas celB had significantly higher expression at 23°C. A previous study demonstrated that celB expression was higher in organisms grown at 23°C, but that of celCA was not affected by temperature (59). The celB gene product is required for utilization of chitobiose, a dimer of N-acetylglucosamine, the subunit of chitin and a component of the tick cuticle. This led to the hypothesis that chitobiose may serve as a nutrient source for the spirochete within ticks (59). This would be consistent with elevated expression of chitobiose-related genes at the lower temperature. Interestingly, bb0002, which encodes a β-N-acetylhexoseaminidase, is also significantly elevated in spirochetes cultivated at 23°C (Table 5). This enzyme could presumably be involved in the metabolism of chitobiose that is taken up via the transporter.
Glycolysis and glycerol uptake and utilization.
Among the genes annotated as being involved in intermediary metabolism, several are significantly upregulated by growth at 23°C. In particular, bb0240 to bb0243 appear to constitute an operon encoding products involved in glycerol uptake (glpF; bb0240) and utilization (glpK and glpA; bb0241 and bb0243, respectively). These gene products would mediate glycerol uptake (glpF) and its conversion to dihydroxyacetone phosphate via a glycerol 3-phosphate intermediate (glpK and glpA). All four putative genes are significantly upregulated between 2.2- and 3.1-fold at 23°C. RT-PCR analysis indicates that all four genes are cotranscribed (data not shown). bb0242 has no database match, and its role in this process is unknown. Interestingly, genes that provide a second route to dihydroxyacetone phosphate are also elevated significantly at 23°C. This would be mediated by a pyrophosphate-dependent phosphofructokinase (bb0020) and aldolase (bb0445), which are upregulated 1.5- to 1.8-fold in organisms grown at 23°C. Genes encoding the enzymes that catalyze the reactions downstream of dihydroxyacetone phosphate in glycolysis exist in B. burgdorferi. Of these, only pgk (encoding phosphoglycerate kinase) transcripts were moderately elevated at the lower temperature. Taken together, these data suggest that lower temperature induces B. burgdorferi to increase its cellular level of dihydroxyacetone phosphate, presumably to enhance the rate of energy production via glycolysis. Alternatively, these genes may be repressed at the higher temperature.
Temperature shift and global transcriptional regulation.
It was not clear previously if B. burgdorferi undergoes a heat shock response due to the rapid rise in temperature it experiences during tick feeding. In the present study, none of the genes encoding typical heat shock proteins (e.g., dnaK or groEL) are up-regulated by temperature shift. Furthermore, there was no discernible down-regulation in components of the translational machinery, such as ribosomal proteins, after the temperature shift. This global gene expression profile would suggest that a shift in temperature from 23 to 35°C does not induce a heat shock or general stress response (10). These observations lead us to propose that the altered gene expression profiles observed on temperature shift from 23 to 35°C are directly related to an adaptive response for migration of B. burgdorferi from the tick midgut to the salivary glands and/or transmission of B. burgdorferi from the tick to mammalian host.
Hubner et al. (28) recently demonstrated that the enhanced expression of ospC and dbpA in response to environmental cues such as elevated temperature and decreased pH proceeds via a novel regulatory network involving the alternate sigma factors RpoN and RpoS. In addition, RpoS protein levels were elevated in cells grown at 35°C relative to those cultivated at 23°C. Consistent with this, a 1.6-fold increase in RpoS mRNA was observed in the current study in spirochetes grown at 35°C (Table 4).
Correlation between microarrays and real-time RT-PCR.
Real-time quantitative RT-PCR was employed to validate the final microarray data. Sixteen ORFs with differential expression values ranging from approximately 3-fold-higher expression at 23°C to 5.5-fold-higher greater expression at 35°C were chosen for the real-time PCR analysis (Table 1). As shown in Fig. 5, there was a high correlation between the expression values obtained by real-time PCR analysis and those measured by array analysis (r = 0.93).
FIG. 5.
Concordance of differential expression of selected genes as measured by gene array or real-time RT-PCR. The relative expression levels for 16 genes listed in Table 5 were determined by gene array or real-time RT-PCR. The correlation coefficient for comparison of the two data sets (r) is 0.93.
Concordance with other microarray analyses.
Several recent reports have described the use of gene arrays to facilitate B. burgdorferi transcriptional analysis (33, 36, 46). Narasimhan et al. spotted clones from a strain N40 genomic library and probed the arrayed clones with a DECAL cDNA library generated from either fed or unfed tick midguts (36). In this manner they identified a number of B. burgdorferi genes expressed in either the fed- or unfed-tick state. Liang et al. limited their analysis to 137 putative lipoprotein genes and studied their expression during murine infection (33). All of these genes were expressed during in vitro growth, but a substantial number were downregulated during murine infection. The limited overlap between the genes identified in these studies (33, 36) and those in our data set is most likely the result of substantial differences in array design and transcript source. Revel et al. recently reported an analysis of global regulation of B. burgdorferi gene expression similar to that presented here (46). Of 215 genes found to be differentially regulated by temperature in the present study, only 25 (11.6%) were regulated under the fed-tick condition (that most comparable to the present study) of Revel et al. (46). The concordance was somewhat better when the comparison was limited to those genes which were upregulated at 35°C (17.3%; 23 of 133) and still better if only those genes upregulated at 35°C by twofold or more in this study were considered (41%; 17 of 41). The limited correspondence between the two data sets may be a result of several methodological differences. The temperature shift in this study was from 23 to 35°C compared to 37°C in the Revel et al. study (46). In addition, Revel et al. also altered the pH between the two conditions, so the observed changes may be a combination of both temperature and pH regulation. Technical differences such as the cDNA labeling method and membrane versus glass slide arrays may, in part, account for the observed disparity. In addition, a cloned isolate of B31-MI was used in the present study; nonclonal isolates of B31-MI have been shown to consist of subpopulations that have very different genotypes (i.e., plasmid content) with various phenotypes (18). Additional B. burgdorferi gene array studies performed under similar experimental conditions are necessary to reconcile these overlapping data sets.
Concluding remarks.
Microarray experiments are often regarded as unfocused since they typically have broad and encompassing hypotheses (9). However, much has been gleaned from microarray analyses, especially when designed to characterize differences in gene expression due to a single environmental stimulus. In the present study, we specifically chose to examine the response of a cloned isolate of B. burgdorferi to a single environmental variable (temperature) to help identify genes encoding proteins that could be important in the adaptation of B. burgdorferi for growth in the tick and the mammalian host.
It is apparent from the present study that plasmid-encoded ORFs are integral to the temperature shift response. Most prominent among the findings are the previously unrecognized roles for many lp54 genes in this response, a possible role for OspD in the tick environment, and the contribution of cp32-encoded genes to the enzootic life cycle of B. burgdorferi. The latter point is supported by the findings that (i) more than 20% of the ORFs encoded on many of the cp32s are regulated by temperature, with almost all having increased expression at 35°C; (ii) the expression of many of the putative hemolysins encoded on these plasmids was significantly up-regulated by temperature, which may be related to digestion of the blood meal; (iii) several cp32 lipoprotein genes encoding surface antigens are differentially up-regulated by temperature (e.g., mlp genes), suggesting they may be important at the host-pathogen interface; and (iv) the overall transcriptional profiles and gene homologies are consistent with the cp32s being the genetic elements that may encode a Borrelia-specific bacteriophage. This is especially noteworthy since our analysis of the cp32 transcriptomes is consistent with recent speculation that cp32-encoded bacteriophage may be involved in horizontal transfer of genetic information between spirochetes during tick feeding (i.e., at the same time the cp32 ORFs are up-regulated) (5, 14).
The combined data indicate that major alterations in the expression of borrelial outer surface antigens, cell envelope constituents, nutrient transporters, and metabolic genes take place in response to a shift in growth temperature from 23 to 35°C (as may occur during tick feeding). These changes are likely important for the parasitic strategy of B. burgdorferi as it adapts to growth in the tick environment and the mammalian host and for enabling the spirochete to traverse the tick environment during migration from the midgut to salivary glands. The results provide numerous new avenues for focused, hypothesis-based investigations to help delineate the role(s) of specific genes, operons, and regulons in the life cycle of this pathogen. In addition, several previously unrecognized temperature-regulated surface proteins were identified. These may provide new vaccine candidates or targets for novel therapeutics for the treatment or prevention of Lyme disease.
Acknowledgments
C. Ojaimi and C. Brooks contributed equally to this work.
We thank Dorothee Grimm for deriving strain B31 clone p1.1, Simon Sims for primer design, Jianmin Zhong for PCR amplification and helpful discussions, and P. Scott Hefty for critical review of the manuscript.
This work was supported by grants AI45801 (I.S.), RR15564 (D.R.A.), AI37248 (A.B.), AI27044 (J.B.), AI29735 (J.R.), and AI42345 (J.S.) from the National Institutes of Health; the American Heart Association (grant 003013N to D.R.A.); and the G. Harold and Leila Y. Mathers Charitable Foundation (I.S.). Caroline Ojaimi was supported in part by a fellowship from the Arthritis Foundation, New York Chapter, and Chad S. Brooks was supported in part by Molecular Pathogenesis Training Grant AI-07364 (NIAID).
Editor: J. T. Barbieri
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 5.Caimano, M. J., X. Yang, T. G. Popova, M. L. Clawson, D. R. Akins, M. V. Norgard, and J. D. Radolf. 2000. Molecular and evolutionary characterization of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Infect. Immun. 68:1574-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casjens, S. 2000. Borrelia genomes in the year 2000. J. Mol. Microbiol. Biotechnol. 2:401-410. [PubMed] [Google Scholar]
- 7.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 10.Cluss, R. G., A. S. Goel, H. L. Rehm, J. G. Schoenecker, and J. T. Boothby. 1996. Coordinate synthesis and turnover of heat shock proteins in Borrelia burgdorferi: degradation of DnaK during recovery from heat shock. Infect. Immun. 64:1736-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings, C. A., and D. A. Relman. 2000. Using DNA microarrays to study host-microbe interactions. Emerg. Infect. Dis. 6:513-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damman, C. J., C. H. Eggers, D. S. Samuels, and D. B. Oliver. 2000. Characterization of Borrelia burgdorferi BlyA and BlyB proteins: a prophage-encoded holin-like system. J. Bacteriol. 182:6791-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, S., S. W. Barthold, S. S. Giles, R. R. Montgomery, S. R. Telford III, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Investig. 99:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggers, C. H., M. J. Caimano, M. L. Clawson, W. G. Miller, D. S. Samuels, and J. D. Radolf. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the lyme disease spirochaete. Mol. Microbiol. 43:281-295. [DOI] [PubMed] [Google Scholar]
- 15.Eggers, C. H., S. Casjens, S. F. Hayes, C. F. Garon, C. J. Damman, D. B. Oliver, and D. S. Samuels. 2000. Bacteriophages of spirochetes. J. Mol. Microbiol. Biotechnol. 2:365-373. [PubMed] [Google Scholar]
- 16.Eggers, C. H., and D. S. Samuels. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Hage, N., K. Babb, J. A. Carroll, N. Lindstrom, E. R. Fischer, J. C. Miller, R. D. Gilmore, Jr., M. L. Mbow, and B. Stevenson. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821-830. [DOI] [PubMed] [Google Scholar]
- 18.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fikrig, E., S. W. Barthold, W. Sun, W. Feng, S. R. Telford, III, and R. A. Flavell. 1997. Borrelia burgdorferi P35 and P37 proteins, expressed in vivo, elicit protective immunity. Immunity 6:531-539. [DOI] [PubMed] [Google Scholar]
- 20.Fikrig, E., M. Chen, S. W. Barthold, J. Anguita, W. Feng, S. R. Telford III, and R. A. Flavell. 1999. Borrelia burgdorferi erpT expression in the arthropod vector and murine host. Mol. Microbiol. 31:281-290. [DOI] [PubMed] [Google Scholar]
- 21.Fikrig, E., W. Feng, S. W. Barthold, S. R. Telford, III, and R. A. Flavell. 2000. Arthropod- and host-specific Borrelia burgdorferi bbk32 expression and the inhibition of spirochete transmission. J. Immunol. 164:5344-5351. [DOI] [PubMed] [Google Scholar]
- 22.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Lara, J., M. Picardeau, B. J. Hinnebusch, W. M. Huang, and S. Casjens. 2000. The role of genomics in approaching the study of Borrelia DNA replication. J. Mol. Microbiol. Biotechnol. 2:447-454. [PubMed] [Google Scholar]
- 24.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 25.Hagman, K. E., X. Yang, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 2000. Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 68:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, and D. R. Akins. 2002. Changes in temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 70:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Indest, K. J., R. Ramamoorthy, M. Sole, R. D. Gilmore, B. J. Johnson, and M. T. Philipp. 1997. Cell-density-dependent expression of Borrelia burgdorferi lipoproteins in vitro. Infect. Immun. 65:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer, R., D. Liveris, A. Adams, J. Nowakowski, D. McKenna, S. Bittker, D. Cooper, G. P. Wormser, and I. Schwartz. 2001. Characterization of Borrelia burgdorferi isolated from erythema migrans lesions: interrelationship of three molecular typing methods. J. Clin. Microbiol. 39:2954-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane, R. S., J. Piesman, and W. Burgdorfer. 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36:587-609. [DOI] [PubMed] [Google Scholar]
- 33.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine Host. J. Exp. Med. 196:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDowell, J. V., S. Y. Sung, M. Labandeira-Rey, J. T. Skare, and R. T. Marconi. 2001. Analysis of mechanisms associated with loss of infectivity of clonal populations of Borrelia burgdorferi B31MI. Infect. Immun. 69:3670-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nadelman, R. B., and G. P. Wormser. 1998. Lyme borreliosis. Lancet 352:557-565. [DOI] [PubMed] [Google Scholar]
- 36.Narasimhan, S., F. Santiago, R. A. Koski, B. Brei, J. F. Anderson, D. Fish, and E. Fikrig. 2002. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J. Bacteriol. 184:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norris, S. J., C. J. Carter, J. K. Howell, and A. G. Barbour. 1992. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect. Immun. 60:4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ojaimi, C., C. Brooks, D. Akins, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, S. Sims, and I. Schwartz. 2002. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 358:165-177. [DOI] [PubMed] [Google Scholar]
- 39.Orloski, K. A., E. B. Hayes, G. L. Campbell, and D. T. Dennis. 2000. Surveillance for Lyme disease—United States, 1992-1998. Morb. Mortal. Wkly. Rep. Surveill. Summ. 49:1-11. [PubMed] [Google Scholar]
- 40.Palmer, N., C. Fraser, and S. Casjens. 2000. Distribution of twelve linear extrachromosomal DNAs in natural isolates of Lyme disease spirochetes. J. Bacteriol. 182:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porcella, S. F., C. A. Fitzpatrick, and J. L. Bono. 2000. Expression and immunological analysis of the plasmid-borne mlp genes of Borrelia burgdorferi strain B31. Infect. Immun. 68:4992-5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Probert, W. S., and B. J. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 30:1003-1015. [DOI] [PubMed] [Google Scholar]
- 43.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramamoorthy, R., and M. T. Philipp. 1998. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect. Immun. 66:5119-5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramamoorthy, R., and D. Scholl-Meeker. 2001. Borrelia burgdorferi proteins whose expression is similarly affected by culture temperature and pH. Infect. Immun. 69:2739-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 48.Schwan, T. G., and J. Piesman. 2002. Vector interactions and molecular adaptations of Lyme disease and relapsing fever spirochetes associated with transmission by ticks. Emerg. Infect. Dis. 8:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed]
- 50.Simpson, W. J., C. F. Garon, and T. G. Schwan. 1990. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb. Pathog. 8:109-118. [DOI] [PubMed] [Google Scholar]
- 51.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345:115-125. [DOI] [PubMed] [Google Scholar]
- 52.Stevenson, B., and K. Babb. 2002. LuxS-mediated quorum sensing in Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 70:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869-1879. [DOI] [PubMed]
- 55.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tilly, K., A. F. Elias, J. Errett, E. Fischer, R. Iyer, I. Schwartz, J. L. Bono, and P. Rosa. 2001. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183:5544-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallich, R., C. Brenner, M. D. Kramer, and M. M. Simon. 1995. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect. Immun. 63:3327-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, G., A. P. van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei, Y., J. M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei, Y., and E. B. Newman. 2002. Studies on the role of the metK gene product of Escherichia coli K-12. Mol. Microbiol. 43:1651-1656. [DOI] [PubMed] [Google Scholar]
- 64.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 65.Yang, X., T. G. Popova, M. S. Goldberg, and M. V. Norgard. 2001. Influence of cultivation media on genetic regulatory patterns in Borrelia burgdorferi. Infect. Immun. 69:4159-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, X., T. G. Popova, K. E. Hagman, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 1999. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect. Immun. 67:6008-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]