Abstract
Ion channels activated by the binding of cyclic nucleotides first were discovered in retinal rods where they generate the cell’s response to light. In other systems, however, it has been difficult to unambiguously determine whether cyclic nucleotide-dependent processes are mediated by protein kinases, their classical effector enzymes, or cyclic nucleotide-gated (CNG) ion channels. Part of this difficulty has been caused by the lack of specific pharmacological tools. Here we report the purification from the venom of the Australian King Brown snake of a peptide toxin that inhibits current through CNG channels. This toxin, which we have named Pseudechetoxin (PsTx), was purified by cation exchange and RP-HPLC and has a molecular mass of about 24 kDa. When applied to the extracellular face of membrane patches containing the α-subunit of the rat olfactory CNG channel, PsTx blocked the cGMP-dependent current with a Ki of 5 nM. Block was independent of voltage and required only a single molecule of toxin. PsTx also blocked CNG channels containing the bovine rod α-subunit with high affinity (100 nM), but it was less effective on the heteromeric version of the rod channel (Ki ≈ 3 μM). We have obtained N-terminal and partial internal sequence data and the amino acid composition of PsTx. These data indicate that PsTx is a basic protein that exhibits some homology with helothermine, a toxin isolated from the venom of the Mexican beaded lizard. PsTx promises to be a valuable pharmacological tool for studies on the structure and physiology of CNG channels.
Keywords: Pseudechis australis, Australian king brown snake; peptide toxins; snake venom; patch-clamp technique; peptide sequencing
For many years cyclic nucleotides were presumed to exert their numerous cellular effects primarily through the activation of the cyclic nucleotide-dependent protein kinases, PKA and PKG (1, 2). Recently, however, this assumption has been challenged by the advent of ion channels directly activated by the binding of cyclic nucleotides. These channels are emerging as important components of signaling systems throughout the body (3). Cyclic nucleotide-gated (CNG) channels first were identified in the sensory epithelium of the visual and olfactory systems where they generate the receptor’s electrical signal in response to light and odorants (for a review see ref. 4). In the past few years, CNG channels have been discovered in other excitable tissues such as the brain, heart, and skeletal muscle, as well as nonexcitable tissues such as the liver, kidneys, and testes. Their roles in nonsensory cells, however, remain obscure. In the retina, CNG channels have been shown to stimulate synaptic transmission in response to nitric oxide cues (5, 6), and recent results suggest that they may play a role in long-term potentiation (7). In nonexcitable tissues they may provide a regulated pathway for the entry of extracellular calcium (8).
CNG channel proteins are tetramers containing α- and β-subunits (9–15). Each subunit contains an amino-terminal transmembrane domain and pore region (16) followed by a single cyclic nucleotide-binding domain near the carboxyl terminus (17). The cooperative binding of four cyclic nucleotide molecules activates a nonspecific cation conductance (18). Although expression of the α-subunits alone generates cyclic nucleotide-dependent currents, coexpression of the β-subunits creates channels that more faithfully reproduce the properties of the native channel (13, 14).
The lack of specific pharmacological tools has made it difficult to unambiguously determine whether cyclic nucleotide-dependent processes are mediated by PKs, their classical effector enzymes, or CNG channels. Although there is a formidable array of specific blockers for sodium, calcium, and potassium channels, specific blockers for CNG channels are scarce. A number of pharmaceuticals, including l-cis-diltiazem (19), tetracaine (20), pimozide (21), and LY-58358 (22), have been reported to block current through CNG channels, but these agents have significant drawbacks. First, they exert their effects from the cytoplasmic face of the channel, making their utility questionable in more intact preparations. Second, all of these blockers have multiple targets, making results somewhat difficult to interpret. An alternative route to determine the identity of the effector enzyme is through the development of cyclic nucleotide analogs with differential effects on CNG channels and PKG. Some progress has been made in this direction in the past few years. Wei and colleagues (23) have produced a series of cGMP derivatives that combine phosphorothioate modification of the cyclic phosphate with phenyletheno-(PET) derivatization of the 2-amino group. Specifically, they have shown that the SP-8-Br-PET-cGMPS isomer is a potent activator of PKG, but a competitive inhibitor of the rod CNG channel (23). Conversely, the RP-8-Br-cGMPS activates the CNG channel, but antagonizes activation of PKG (24). Although these derivatives show some promise as selective pharmacological agents, they bind to the channel with rather low affinity and their ability to permeate the cell membrane is unclear.
In this manuscript, we report the purification of a peptide toxin that blocks current through CNG channels when applied to the extracellular face of excised outside-out membrane patches. This toxin was purified from the venom of Pseudechis australis, the Australian king brown snake, and promises to be a useful pharmacological tool to study the structure and function of CNG channels.
MATERIALS AND METHODS
Snake venoms were obtained from Venom Supplies (Tanunda, South Australia). Spider venoms were from Spider Pharm (Feasterville, PA). Other venoms were from Sigma. Calcium channel blockers were from Alamone Labs (Jerusalem, Israel). Cyclic nucleotides and tosylamidophenylethyl chloromethyl ketone-treated trypsin were from Sigma. cDNAs encoding the α-subunits of the olfactory and retinal CNG channels were the kind gifts of R. R. Reed (Johns Hopkins University), Emily Liman (Harvard University), and William Zagotta (University of Washington). The cDNA encoding the rod β-subunit was the gift of Robert Molday (University of British Columbia). l-cis-Diltiazem was the gift of E. Boehme at Hoechst-Marion-Roussel (Cincinnati, OH). All other reagents were of the highest purity available.
Purification of Pseudechetoxin (PsTx).
We have purified PsTx by using two methods. In the first method, 100 mg of dried venom from Pseudechis australis was dissolved in 1 ml of HPLC starting buffer (water containing 0.1% trifluoroacetic acid, TFA); insoluble material was removed by a 5-min centrifugation at maximum speed in an Eppendorf microcentrifuge. The clarified venom was loaded onto a Jupiter C5 column (250 × 21.2 mm; Phenomenex, Torrance, CA) by using a Hewlett-Packard 1100 HPLC system. PsTx was eluted with a 120-min gradient from 0.1% TFA to 80% acetonitrile (containing 0.08% TFA) with a 5 ml/min flow rate. Fractions containing PsTx inhibitory activity were pooled, and PsTx was further purified by isocratic elution from an analytical (250 × 4.6 mm) C4 Jupiter column (Phenomenex) by using 40% acetonitrile/water (containing 0.1% TFA).
A second purification method used an initial step of cation-exchange chromatography. Fifty milligrams of dried P. australis venom was dissolved in 50 mM Hepes, pH 7.5, and applied to a Bioscale S-5 column (Bio-Rad). The venom components were eluted from this strong cation-exchange matrix by using a 60-min linear gradient from 0–2 M KCl with 50 mM Hepes, pH 7.5 throughout. Each fraction was screened for the presence of PsTx by analytical RP-HPLC (as described below) and SDS/PAGE. PsTx subsequently was purified to apparent homogeneity by RP-HPLC on the analytical Jupiter C4 HPLC column. Proteins were eluted by using a 1 ml/min linear gradient (2% per min) from 0.1% TFA to acetonitrile (containing 0.08% TFA). PsTx eluted from this chromatography system at approximately 32.5 min, well resolved from all other proteins.
Expression of CNG Channels.
cRNAs encoding the olfactory and rod CNG channels were transcribed by using the mMessage mMachine kit (Ambion, Austin, TX) as described (25). Xenopus oocytes were injected with ≈25 ng of cRNA. For experiments using both the α- and β-subunits of the rod channel, the respective cRNAs were injected in a 1:2 molar ratio. Patch-clamp experiments were performed 3–5 days after injection.
Electrophysiological Methods.
Currents were recorded from excised patches in the outside-out configuration by using an Axopatch 200A amplifier and Digidata 1200 interface (Axon Instruments, Foster City, CA), controlled by Pclamp6.0 software. Electrodes were fabricated from borosilicate glass (Sutter Instruments, Novato, CA) and had an initial resistance from 1–2 MΩ. Gigaohm seals were obtained in control solution containing 130 mM NaCl, 3 mM Hepes, and 0.2 mM EDTA, pH 7.4. The internal pipette solution also contained 2 mM cGMP. After obtaining the original seal, the membrane was ruptured with a 5-msec pulse to 1.3 VDC. After allowing the seal resistance to stabilize, outside-out patches were obtained by slowly withdrawing the pipette from the cell. Fifty-millisecond voltage pulses ranging from −100 to 100 mV in 20-mV intervals were given from a holding potential of 0 mV. Solutions were applied to the extracellular face of the patch via the “sewer pipe” method using a RSC-100 rapid perfusion system (Molecular Kinetics, Pullman, WA). The seal resistance and “leak” current was estimated by application of 10 mM MgCl2 in control solution. This concentration of Mg2+ should block >98% of the current at −50 mV in all channels studied (26, 27). To insure that the majority of channels expressed by coinjection of cRNA encoding the rod α- and β-subunits were heteromeric, we tested their sensitivity to l-cis-ditiazem in excised inside-out patches. The heteromeric version of the channel is >100-fold more sensitive to block by this drug than the homomeric α-subunit version (10). Application of 10 μM l-cis-diltiazem blocked >85% of the current recorded from patches expressing both the α- and β-subunits. Currents from patches expressing α alone were blocked only 5–10%.
Amino Acid Analysis and Edman Degradation.
Phenylthiocarbamyl amino acid analysis was performed by using an Applied Biosystems 420H/130/920 automated sequence system (28). Edman degradation was performed on an Applied Biosystems gas phase sequencer (model 470/120/900) (29). PsTx (5 μg) was carboxyamidomethylated and then digested for 6 hr at 37°C with endoproteinase Asp-N (Boehringer Mannheim) in 1 M urea, 50 mM ammonium bicarbonate by using 2% protease by weight (29), and peptides were purified by RP-HPLC. Peptide sequences were analyzed for possible homologies by using the National Center for Biotechnology blast network (30).
Miscellaneous Methods.
SDS/PAGE was performed by the method of Laemmli (31) using a mini-Protean II system (Bio-Rad), and gels were stained with Coomassie brilliant blue G-250. Data analysis and curve fitting were done with sigmaplot software (Jandel Scientific, San Rafael, CA).
RESULTS
In our search for a toxin that blocks CNG channels, we initially screened a variety of toxins known to block calcium channels. When applied at concentrations between 1 and 10 μM, the ω-conotoxins (32, 33), agatoxin-TK (34), calcicludine (35), waglerine, and taicatoxin (36) had no effect on CNG channels. We also tested the sodium channel antagonist, μ-conotoxin-GIIIB, which has been reported to block CNG channels purified from the kidney (37). This toxin also was ineffective on the olfactory and rod CNG channels. Only the arginine polyamine, FTX (38), had a significant effect on CNG channels. When applied at a concentration of 10 μM, FTX blocked approximately 80% of the cyclic nucleotide-dependent current at −80 mV, but was relatively ineffective at positive voltages.
We next screened a variety of crude venoms for novel toxins that blocked CNG channels. Most were applied at 1–10 mg/ml. We used three criteria during initial screening to select for venoms that might contain high-affinity peptide toxins. Block should be relatively independent of voltage, develop and reverse slowly, and occur when the venom was applied to the extracellular, but not intracellular, face of the patch. We tested venoms from 30 snakes from Australia and Indonesia as well as venoms from several amphibians, lizards, and spiders. Many of the venoms tested contained compounds that produced a fast-acting and voltage-dependent block, similar to FTX. Although only a few venoms met the criteria listed above, the venom from P. australis looked promising. Application of this venom at 5 mg/ml blocked approximately 80% of the current through the olfactory CNG channel and appeared to have at least two active components. One component produced a rapid, voltage-dependent block of ≈50% of the current. With continued application, a second component blocked an additional 20–30% of the current and seemed less voltage dependent. Upon removal of toxin, ≈50% of the current recovered within seconds, whereas the remaining 20–30% recovered fully only after several minutes. The slow component of block was not seen when P. australis venom was applied to inside-out patches.
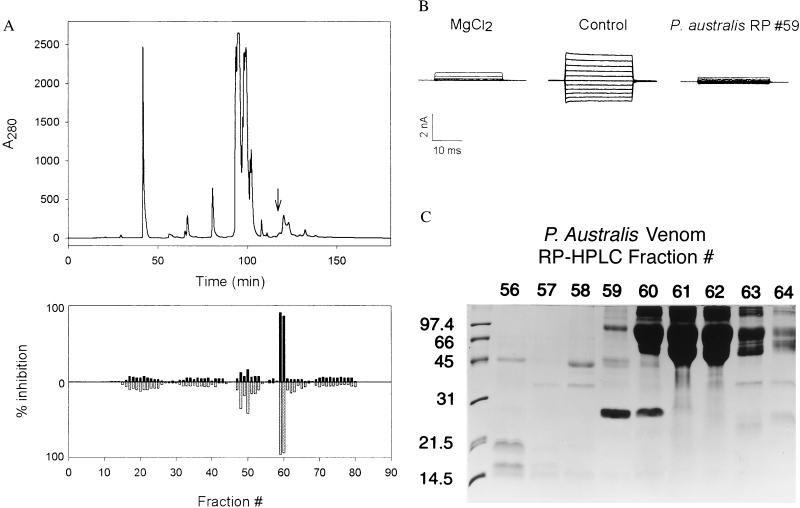
We next set out to purify the active component from P. austalis venom. Noting that many peptide toxins have been successfully purified by RP-HPLC, we applied 100 mg of venom to a C5 semipreparative column and eluted with a water-acetonitrile gradient as described in Fig. 1A. The majority of inhibitory activity was contained in fractions #59 and #60 (Fig. 1 A and B). When analyzed by SDS/PAGE (Fig. 1C), these fractions contained a protein with a Mr of approximately 24 kDa, which was absent in noninhibitory fractions. This protein, which we have named Pseudechetoxin (PsTx), was purified to apparent homogeneity by a subsequent isocratic elution from an analytical C4 reversed-phase column.
Figure 1.
(A) RP-HPLC separation of PsTx from P. australis venom. The components in 100 mg of venom were separated on a C5 column by using a water/acetonitrile gradient as described. (Upper) The column profile at 280 nm. Ten-milliliter fractions were dried and redissolved in 1 ml of water. After a 1:30 dilution into control buffer, the fractions were applied to excised outside-out patches expressing the olfactory CNG channel α-subunit. (Lower) Histogram depicts the fractional inhibition of current at +80 mV (upward, black fill) and at −80 mV (downward, gray fill). The PsTx eluted at 118 min. (indicated by an arrow, Upper); the corresponding inhibitory activity was found in fractions 59 and 60. These data suggest that PsTx is a minor component of P. australis venom, perhaps <0.1%. (B) Block of CNG channel currents by 10 mM Mg2+ and crude PsTx (reversed-phase fraction 59). CNG channel currents in excised outside-out patches were evoked by 20-ms steps from a holding potential of mV to a voltage ranging from −100 to +100 mV in 20-mV steps. Shown are current families in control solution and in the presence of 10 mM MgCl2 and PsTx applied to the extracellular face of the membrane. (C) SDS/PAGE analysis of reversed-phase fractions 56–64. A 10-μl sample of each reconstituted fraction was applied to a SDS-15% polyacrylamide gel. After separation, the gel was stained with Coomassie brilliant blue R250. Inhibitory activity was strongly correlated with the presence of a 24-kDa protein in fractions 59 and 60.
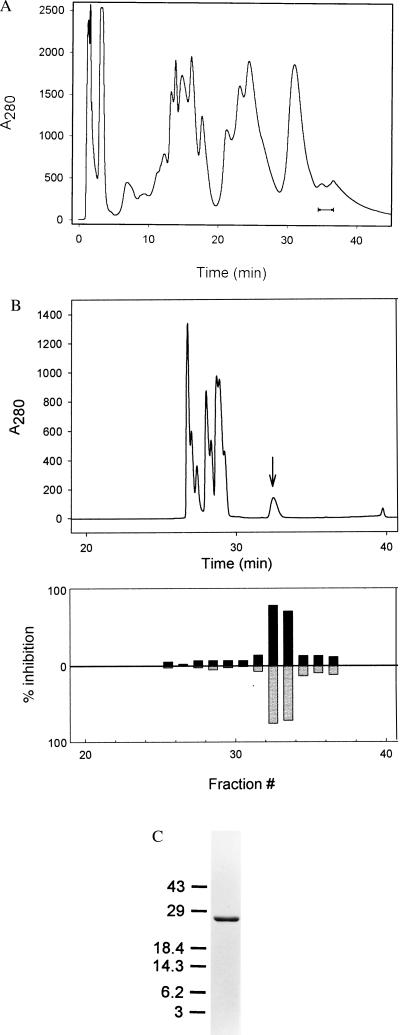
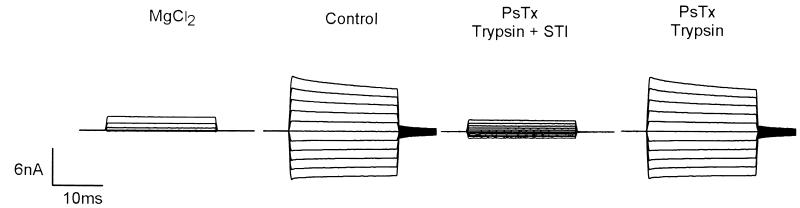
To confirm the identity of the toxin, PsTx was purified by a second method, namely cation exchange chromatography (Fig. 2A). The 24-kDa candidate protein eluted late in the gradient, and its presence was once again correlated with inhibitory activity. As shown in Fig. 2B, PsTx subsequently was purified to apparent homogeneity by RP-HPLC. Fig. 2C shows a Coomassie-stained SDS-15% polyacrylamide gel containing the final RP-HPLC-purified preparation. To ensure that the inhibitory activity present in the purified fractions was, in fact, caused by a protein, PsTx was subjected to digestion with trypsin. As shown in Fig. 3, this treatment abolished inhibitory activity.
Figure 2.
(A) Cation exchange chromatography of crude P. australis venom. Fifty milligrams of venom was applied to a Bio-Rad S-50 cation exchange matrix and eluted with a KCl gradient. Shown is the column profile at 280 nm. Inhibitory activity eluted near the end of the gradient. The indicated fractions were pooled for RP-HPLC. (B) Reversed-phase chromatography of pooled cation exchange fractions. Cation exchange fractions were pooled from 100 mg of raw venom and applied to a C4 reversed-phase column. Components were differen tially eluted with a water/acetonitrile gradient (0.1% TFA throughout). (Upper) The column profile at 280 nm. The 24-kDa component, PsTx, eluted at 32–34 min as a broad peak as indicated by the arrow. One-minute fractions were collected and dried. After being redissolved in 1 ml of water, a portion of each fraction was diluted 1:30 into control buffer containing BSA and applied to the extracellular face of a membrane patch expressing the rat olfactory channel. (Lower) the fraction current inhibition at +80 mV (upward, black) and at −80 mV (downward, gray). (C) SDS/PAGE analysis of a final RP-HPLC purified PsTx preparation. Shown is a SDS–15% polyacrylamide gel containing 1 μg of PsTx after staining with Coomassie brilliant blue.
Figure 3.
Inhibitory activity of PsTx is sensitive to trypsin proteolysis. Shown are currents recorded from outside-out patches containing the α-subunit of the rat olfactory CNG channel. Pipette contains 2 mM cGMP; the extracellular face of the patch was exposed to control solution containing the substances indicated. Voltage pulses were given as described in Fig. 1B. For the panel labeled “Trypsin,” 20 nM PsTx was incubated in control solution containing 100 μg/ml of trypsin for 36 hr. at 37°C. At that time, a 2-fold molar excess of soybean trypsin inhibitor (STI) was added to halt digestion. In the panel labeled “Trypsin + STI,” trypsin and STI were premixed before incubation.
Edman sequence analysis of the intact protein revealed a frayed N-terminal structure with the most abundant component exhibiting the sequence SNKKYQK. A second species began at the first asparagine residue, and a low abundance component began at the first lysine. Amino acid analysis of purified PsTx demonstrated that it is a highly basic protein, with Lys, Arg, and His residues totaling almost 20 mol% (Table 1). After proteolysis of purified PsTx with endoproteinase Asp-N, an internal peptide was purified by RP-HPLC and found to exhibit the sequence DKHNALRRSVKPTARNMLQ. Underlined residues are identical in a stretch of helothermine (residues 22–40), a toxin from the saliva of the Mexican beaded lizard (39). This fragment also shows high homology with the cysteine-rich secretory proteins found in testes and saliva.
Table 1.
Amino acid analysis of purified PsTx
| Amino acid | Mol% |
|---|---|
| Asx (D + N) | 10.59 ± 0.20 |
| Glx (E + Q) | 6.43 ± 0.12 |
| Ser (S) | 9.20 ± 0.10 |
| Gly (G) | 6.91 ± 0.06 |
| His (H) | 2.32 ± 0.14 |
| Arg (R) | 5.40 ± 0.12 |
| Thr (T) | 4.19 ± 0.10 |
| Ala (A) | 9.78 ± 0.05 |
| Pro (P) | 8.15 ± 0.09 |
| Tyr (Y) | 5.13 ± 0.09 |
| Val (V) | 4.85 ± 0.13 |
| Met (M) | 2.08 ± 0.23 |
| Ile (I) | 5.27 ± 0.10 |
| Leu (L) | 4.11 ± 0.09 |
| Phe (F) | 3.36 ± 0.08 |
| Lys (K) | 12.19 ± 0.06 |
Composition based on phenylthiocarbamyl amino acid analysis of HPLC purified PsTx (method 2). Values represent mol% ± the relative SD from replicate analyses (n = 5–8) and are uncorrected for partial destruction and incomplete hydrolysis. Cys and Trp were not determined.
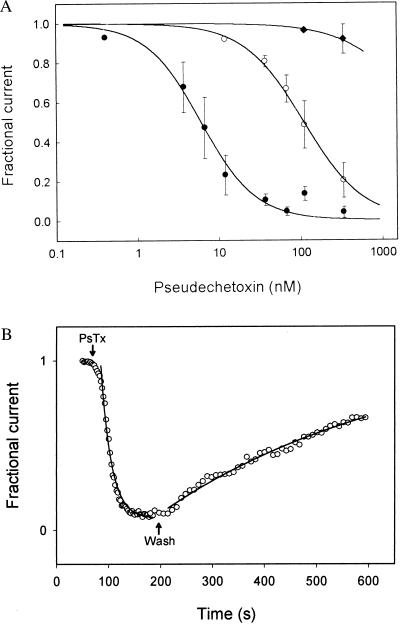
We next tested the affinity of PsTx for several types of CNG channels. Purified PsTx was quantified by amino acid analysis and then applied to excised outside-out patches containing olfactory and rod α-subunits, and the rod αβ heteromeric channel. The PsTx was diluted to concentrations ranging from 1 to 500 nM in control solution containing 50 μg/ml BSA. The dose–response relations were fit by using the Hill equation to extract the parameters of Ki, the concentration required to block half of the current, and n, a rough indication of the number of PsTx molecules required. PsTx was most effective at blocking the olfactory α-subunit channels with a Ki of 5 nM; the rod α-subunit was close behind with a Ki of 100 nM (Fig. 4A). Both dose–response relations were fit by using a Hill coefficient, n, of 1.2, suggesting that only a single molecule of PsTx is required for block. Only the highest concentration of PsTx tested (500 nM) had an effect on the heteromeric version of the rod channel, suggesting that the affinity of PsTx for this version of the rod channel (≈3 μM) is at least 30-fold lower than that for the α-subunit alone.
Figure 4.
(A) PsTx blocks CNG channels with nM affinities. Outside-out patches containing the α-subunit of the rat olfactory CNG channel (rOLF; •), the α-subunit of the bovine rod CNG channel (bRET; ○), or both the α- and β-subunits of the bovine rod CNG channel (bRETαβ; ⧫) were tested for their sensitivity to block by PsTx at −60 mV. Dose–response relations were fit by the Hill equation: I+PsTx/I−PsTx = Kin/([PsTx]n + Kin) to determine apparent affinities. The fit parameters for the curves shown were: rOLF −Ki = 5 nM, n = 1.2; bRET − Ki = 100 nM; n = 1.2; bRETαβ − Ki = 3.5 μM; n = 1.2. (B) Block by PsTx was slow to develop and reverse. Voltage pulses to +50 mV were given every 2.5 or 5 sec. to outside-out patches expressing the rat olfactory CNG channel α-subunit. Shown is the fractional current as a function of time after the application and removal of 125 nM PsTx (arrows). Thin lines show single exponential fits with rate constants kon = 4 × 105 M−1 sec−1 and koff = 2.1 × 10−3 sec−1.
As shown in Fig. 4B, block of channels composed of the rat olfactory CNG channel α-subunit had a relatively rapid onset, reaching equilibrium in 10–20 s when applied at 125 nM, and reversed slowly over 5–10 min after the removal of PsTx. These time courses indicate a bimolecular rate constant of 4 × 105 M−1 sec−1, and an off-rate of 2.1 × 10−3 sec−1. These kinetic constants predict a binding affinity of 5 nM, which agrees closely with the affinity determined by the dose–response titration.
DISCUSSION
Snake venoms traditionally have been a plentiful source of neurotoxins. The venoms of the Australian elapid snakes, such as P. australis, however, are not well characterized. Previous work has identified only long- and short-chain nicotinic neurotoxins (40, 41) as well as several phospholipases (42).
Here we report the purification of PsTx, a peptide blocker of CNG ion channels. When applied to the extracellular face of Xenopus oocyte patches expressing the olfactory and rod channel α-subunits, it blocked current with a Ki of 5–100 nM, with a 20-fold preference for the olfactory channel. Block was almost insensitive to voltage, suggesting that PsTx occludes the external vestibule, but does not significantly penetrate the transmembrane field. Its affinity for the rod αβ heteromultimer was at least an order of magnitude lower (≈3 μM). It will be interesting to test PsTx on native channels from photoreceptors and olfactory neurons, as well as on other tissues known to express CNG channels, such as the kidney and hippocampus.
At the molecular level, PsTx is a highly basic, 24-kDa protein. Five of six peptides that we have obtained from PsTx show no homology to other proteins in the database. One stretch of 19 residues, however, shows a high degree of homology to helothermine, a toxin isolated from the saliva of Heloderma horridum horridum, the Mexican beaded lizard (43). Helothermine has been reported to block a variety of ion channels, including the L-, N-, and P-type calcium channels from cerebellar granule cells (44), potassium channels in cerebellar granule cells (45), and the skeletal muscle ryanodine receptor (39). The same stretch of amino acids also showed a high homology with a family of cysteine-rich secretory proteins (46). The function of these proteins is virtually unknown at the current time. There is some data to suggest that a testes-specific member of this family is involved in sperm maturation (47). Given the presence of CNG channels in sperm, it is interesting to speculate that these proteins may serve as endogenous regulators of CNG channel function. Finally, the genes of these secreted proteins encode a signal peptide that subsequently is removed, suggesting that the N-terminal heterogeneity might arise from imprecise cleavage.
PsTx promises to be a useful pharmacological tool for the study of the structure and function of CNG channels. One obvious structural question concerns the graded affinity of PsTx for the three channel types tested here. By testing the affinity of chimeric channels, it should be possible to further define the toxin’s binding site on the extracellular face of the channel. A long-range goal is to map the structure of the channel’s outer vestibule by using complementary mutagenesis of the channel and PsTx. This type of molecular cartography has been enormously successful on potassium channels by using mutants of charybdotoxin (48, 49).
Experiments are underway to investigate the ability of PsTx to block other types of channels. Helothermine has been shown to inhibit current through a broad spectrum of channels. Although this possibility is a concern with PsTx, the lack of similarity in amino acid composition makes it unlikely that PsTx inhibits a similar spectrum of channels. If PsTx turns out to be relatively specific, it can be used as a pharmacological scalpel to dissect out the physiological roles of CNG channels.
Acknowledgments
We thank Jeff Hulmes for his help during the initial sequencing, and Drs. Jeffrey W. Karpen, MariaLuisa Ruiz, and Thomas Rich for comments on the manuscript. This work was supported by Grants EY11397 (R.L.B.) and EY06603 (J.W.C.) from the National Eye Institute and a grant from the Oregon Lions Sight and Hearing Foundation (R.L.B.).
ABBREVIATIONS
- CNG
cyclic nucleotide-gated
- PsTx
Pseudechetoxin
- PK
protein kinase
- TFA
trifluoroacetic acid
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Meinkoth J L, Alberts A S, Went W, Fantozzi D, Taylor S S, Hagiwara M, Montminy M, Feramisco J R. Mol Cell Biochem. 1993;127-128:179–186. doi: 10.1007/BF01076769. [DOI] [PubMed] [Google Scholar]
- 2.Francis S H, Corbin J D. Annu Rev Physiol. 1994;56:237–272. doi: 10.1146/annurev.ph.56.030194.001321. [DOI] [PubMed] [Google Scholar]
- 3.Finn J T, Grunwald M E, Yau K-W. Annu Rev Physiol. 1996;58:395–426. doi: 10.1146/annurev.ph.58.030196.002143. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman A L. Curr Opin Neurobiol. 1995;5:296–303. doi: 10.1016/0959-4388(95)80041-7. [DOI] [PubMed] [Google Scholar]
- 5.Rieke F, Schwartz E A. Neuron. 1994;13:863–873. doi: 10.1016/0896-6273(94)90252-6. [DOI] [PubMed] [Google Scholar]
- 6.Savchenko A, Barnes S, Kramer R H. Nature (London) 1997;390:694–698. doi: 10.1038/37803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parent A, Schrader K, Munger S D, Reed R R, Linden D J, Ronnett G V. J Neurophysiol. 1998;79:3295–3301. doi: 10.1152/jn.1998.79.6.3295. [DOI] [PubMed] [Google Scholar]
- 8.Kaupp U B. Curr Opin Neurobiol. 1995;5:434–442. doi: 10.1016/0959-4388(95)80002-6. [DOI] [PubMed] [Google Scholar]
- 9.Kaupp U B, Niidome T, Tanabe T, Terada S, Bonigk W, Stuhmer W, Cook N J, Kangawa K, Matsuo H, Hirose T. Nature (London) 1989;342:762–766. doi: 10.1038/342762a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen T Y, Peng Y W, Dhallan R S, Ahamed B, Reed R R, Yau K-W. Nature (London) 1993;362:764–767. doi: 10.1038/362764a0. [DOI] [PubMed] [Google Scholar]
- 11.Körschen H G, Illing M, Seifert R, Sesti F, Williams A, Gotzes S, Colville C, Muller F, Dose A, Godde M. Neuron. 1995;15:627–636. doi: 10.1016/0896-6273(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 12.Dhallan R S, Yau K-W, Schrader K A, Reed R R. Nature (London) 1990;347:184–187. doi: 10.1038/347184a0. [DOI] [PubMed] [Google Scholar]
- 13.Liman E R, Buck L B. Neuron. 1994;13:611–621. doi: 10.1016/0896-6273(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 14.Bradley J, Li J, Davidson N, Lester H A, Zinn K. Proc Natl Acad Sci USA. 1994;91:8890–8894. doi: 10.1073/pnas.91.19.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D T, Tibbs G R, Siegelbaum S A. Neuron. 1996;16:983–990. doi: 10.1016/s0896-6273(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 16.Zagotta W N, Siegelbaum S A. Annu Rev Neurosci. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]
- 17.Brown R L, Gramling R, Bert R J, Karpen J W. Biochemistry. 1995;34:8365–8370. doi: 10.1021/bi00026a018. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz M, Karpen J W. Nature (London) 1997;389:389–392. doi: 10.1038/38744. [DOI] [PubMed] [Google Scholar]
- 19.Haynes L W. J Gen Physiol. 1992;100:783–801. doi: 10.1085/jgp.100.5.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fodor A A, Gordon S E, Zagotta W N. J Gen Physiol. 1997;109:3–14. doi: 10.1085/jgp.109.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicol G D. J Pharmacol Exp Ther. 1993;265:626–632. [PubMed] [Google Scholar]
- 22.Leinders-Zufall T, Zufall F. J Neurophysiol. 1995;74:2759–2762. doi: 10.1152/jn.1995.74.6.2759. [DOI] [PubMed] [Google Scholar]
- 23.Wei J Y, Cohen E D, Yan Y Y, Genieser H G, Barnstable C J. Biochemistry. 1996;35:16815–16823. doi: 10.1021/bi961763v. [DOI] [PubMed] [Google Scholar]
- 24.Wei J Y, Cohen E D, Genieser H G, Barnstable C J. J Mol Neurosci. 1998;10:53–64. doi: 10.1007/BF02737085. [DOI] [PubMed] [Google Scholar]
- 25.Brown R L, Snow S D, Haley T L. Biophys J. 1998;75:825–833. doi: 10.1016/s0006-3495(98)77571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Root M J, MacKinnon R. Neuron. 1993;11:459–466. doi: 10.1016/0896-6273(93)90150-p. [DOI] [PubMed] [Google Scholar]
- 27.Frings S, Seifert R, Godde M, Kaupp U B. Neuron. 1995;15:169–179. doi: 10.1016/0896-6273(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 28.Crabb J W, West K A, Dodson W S, Hulmes J D. In: Current Protocols in Protein Science. Coligan J E, Ploegh H L, Smith J A, Speicher D W, editors. New York: Wiley; 1997. pp. 11.9.1–11.9.42. [Google Scholar]
- 29.Crabb J W, Nie Z, Chen Y, Hulmes J D, West K A, Kapron J T, Ruuska S E, Noy N, Saari J C. J Biol Chem. 1998;273:20712–20720. doi: 10.1074/jbc.273.33.20712. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Olivera B M, Rivier J, Scott J K, Hillyard D R, Cruz L J. J Biol Chem. 1991;266:22067–22070. [PubMed] [Google Scholar]
- 33.Olivera B M, Miljanich G P, Ramachandran J, Adams M E. Annu Rev Biochem. 1994;63:823–867. doi: 10.1146/annurev.bi.63.070194.004135. [DOI] [PubMed] [Google Scholar]
- 34.Teramoto T, Niidome T, Miyagawa T, Nishizawa Y, Katayama K, Sawada K. NeuroReport. 1995;6:1684–1688. doi: 10.1097/00001756-199508000-00022. [DOI] [PubMed] [Google Scholar]
- 35.Schweitz H, Heurteaux C, Bois P, Moinier D, Romey G, Lazdunski M. Proc Natl Acad Sci USA. 1994;91:878–882. doi: 10.1073/pnas.91.3.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Possani L D, Martin B M, Yatani A, Mochca-Morales J, Zamudio F Z, Gurrola G B, Brown A M. Toxicon. 1992;30:1343–1364. doi: 10.1016/0041-0101(92)90511-3. [DOI] [PubMed] [Google Scholar]
- 37.McGeoch J E, Guidotti G. J Biol Chem. 1992;267:832–841. [PubMed] [Google Scholar]
- 38.Dupere J R, Moya E, Blagbrough I S, Usowicz M M. Neuropharmacology. 1996;35:1–11. doi: 10.1016/0028-3908(95)00156-5. [DOI] [PubMed] [Google Scholar]
- 39.Morrissette J, Kratzschmar J, Haendler B, el-Hayek R, Mochca-Morales J, Martin B M, Patel J R, Moss R L, Schleuning W D, Coronado R. Biophys J. 1995;68:2280–2288. doi: 10.1016/S0006-3495(95)80410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takasaki C. J Biochem. 1989;106:11–16. doi: 10.1093/oxfordjournals.jbchem.a122797. [DOI] [PubMed] [Google Scholar]
- 41.Takasaki C, Tamiya N. Biochem J. 1985;232:367–371. doi: 10.1042/bj2320367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takasaki C, Yutani F, Kajiyashiki T. Toxicon. 1990;28:329–339. doi: 10.1016/0041-0101(90)90068-i. [DOI] [PubMed] [Google Scholar]
- 43.Mochca-Morales J, Martin B M, Possani L D. Toxicon. 1990;28:299–309. doi: 10.1016/0041-0101(90)90065-f. [DOI] [PubMed] [Google Scholar]
- 44.Nobile M, Noceti F, Prestipino G, Possani L D. Exp Brain Res. 1996;110:15–20. doi: 10.1007/BF00241369. [DOI] [PubMed] [Google Scholar]
- 45.Nobile M, Magnelli V, Lagostena L, Mochca-Morales J, Possani L D, Prestipino G. J Membr Biol. 1994;139:49–55. doi: 10.1007/BF00232674. [DOI] [PubMed] [Google Scholar]
- 46.Kratzschmar J, Haendler B, Eberspaecher U, Roosterman D, Donner P, Schleuning W D. Eur J Biochem. 1996;236:827–836. doi: 10.1111/j.1432-1033.1996.t01-1-00827.x. [DOI] [PubMed] [Google Scholar]
- 47.Mizuki N, Sarapata D E, Garcia-Sanz J A, Kasahara M. Mamm Genome. 1992;3:274–280. doi: 10.1007/BF00292155. [DOI] [PubMed] [Google Scholar]
- 48.Hidalgo P, MacKinnon R. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 49.Stampe P, Kolmakova-Partensky L, Miller C. Biochemistry. 1994;33:443–450. doi: 10.1021/bi00168a008. [DOI] [PubMed] [Google Scholar]