Abstract
Cryptosporidium meleagridis, a protozoon first observed in turkeys, has been linked by several investigators to cryptosporidiosis in humans. C. meleagridis is the only known Cryptosporidium species that infects both avian and mammalian species. We describe the successful propagation of C. meleagridis (isolate TU1867), originally purified from a patient with diarrhea, in laboratory animals including chickens, mice, piglets, and calves. TU1867 was readily transmitted from one animal host to another, maintaining genetic homogeneity and stability. The rate of infectivity and virulence of TU1867 for the mammalian species were similar to those of Cryptosporidium parvum. Laboratory propagation of genetically and phenotypically stable and well-characterized reference isolates, representing various Cryptosporidium species, particularly those infectious to humans, will improve considerably the spectrum and quality of laboratory and field investigations on this medically important protozoa.
Several species are now recognized within the genus Cryptosporidium, a protozoon associated largely with enteric infections in all classes of vertebrates (5, 7, 13, 14, 25, 26). Mammals are commonly infected by either Cryptosporidium parvum (intestinal) or Cryptosporidium muris (gastric). But only the former appears to cause natural disease in humans and in newborn ruminants (26). However, unlike the infection in ruminants, humans remain susceptible to infection throughout their life, and in immunodeficient individuals, cryptosporidiosis can become persistent, leading to life-threatening diarrhea and wasting (9). Moreover, humans appear to be susceptible to other Cryptosporidium species, including Cryptosporidium canis, Cryptosporidium felis, and Cryptosporidium meleagridis (4, 10, 11, 12, 15-18, 30; J. K. Tumwine, A. Kekitiinwa, N. Nabukeera, D. E. Akiyoshi, S. M. Rich, G. Widmer, X. Feng, and S. Tzipori, submitted for publication), which are normally associated with specific mammalian hosts (14, 23). Among these, C. meleagridis is the most curious since it is the only Cryptosporidium species known to naturally infect mammalian (humans) and avian (turkeys) species. It was first observed in turkeys in 1955 by Slavin (19), where it was reported to cause respiratory tract infections (20). C. meleagridis in humans, however, has been associated with gastrointestinal symptoms, and whether simultaneous infections of the respiratory tract occur as well, is not clear. In a recent study involving the molecular characterization of over 2,000 human stool isolates of Cryptosporidium, 19 (∼1%) were identified as C. meleagridis (17). Of these 19 individuals, 6 had a history of foreign travel, 4 were coinfected with Giardia, and 2 were human immunodeficiency virus-positive, reflecting the normal range of risk factors associated with human cryptosporidiosis. This and other studies (10, 11, 12, 30; Tumwine et al., submitted) confirms that C. meleagridis, in addition to C. parvum, is associated with human disease.
One of 4 C. meleagridis isolates, obtained from human patients with diarrhea (Tumwine et al., submitted), was recently adapted in this laboratory to continuously propagate in laboratory animals. In this study, we have investigated the phenotypic characterization of this isolate and have expanded the range of hosts susceptible to C. meleagridis. Propagation of well-characterized isolates representing various Cryptosporidium species, which are free of possible contamination, particularly those infectious to humans, in our view, will enhance the quality of laboratory and field investigations.
MATERIALS AND METHODS
Source of C. meleagridis isolates.
Approximately 1,800 stool samples from children with diarrhea were analyzed by Cryptosporidium oocyst wall protein (COWP) PCR-restriction fragment length polymorphism (PCR-RFLP). Of 444 (∼25%) stool samples which were positive for Cryptosporidium (Tumwine et al., submitted), 4 (∼1%) of them had restriction digest profiles consistent with that previously reported for C. meleagridis (16), and all 4 of these were associated with acute diarrheal illness. One of these 4 isolates, designated TU1867, was selected for further studies.
Animal passages.
Oocysts identified as C. meleagridis were purified from the stool of a child from Uganda with diarrhea. The oocysts were fed to two gnotobiotic piglets (28) and to five gamma interferon-knockout (γGKO) mice (8, 22). Gnotobiotic piglets (germfree and colostrum deprived) are routinely used in our division for infectivity studies and for propagation of specific laboratory isolates of Cryptosporidium, including the human genotype 1 isolate TU502 (1). Propagation under germfree conditions prevents cross-contamination with isolates from environmental and laboratory sources. Oocysts purified from feces and gut contents of the infected gnotobiotic piglets were confirmed as C. meleagridis, and this isolate was designated TU1867. TU1867 was subsequently fed to other piglets (106 oocysts per animal), γGKO and immunosuppressed C57BL/6 mice (103 oocysts per animal) (K. Huang, D. E. Akiyoshi, X. Feng, and S. Tzipori, submitted for publication), 2- to 7-day-old chicken and turkey poults (104 to 106 oocysts per animal), and one calf (108 oocysts). Oocyst shedding was monitored either daily (piglets and calf) or three times a week (mice, chickens, and turkeys) by microscopic examination of fecal smears stained by modified acid fast (28). The intensity of shedding was scored as previously described (22).
Purification of oocysts.
Oocysts were purified from feces of piglets or mice by using a protocol which included an ether extraction and nycodenz step gradient. Briefly, following concentration of the fecal material by centrifugation (4,000 × g, 10 to 15 min), the oocyst-containing pellet was resuspended in 0.5% Tween 80 (in water) and filtered through a single layer of sterile gauze to remove particulate fecal material. An equal volume of diethyl ether was added, and the sample was vortexed and then centrifuged (4,000 × g, 10 to 15 min). The ether and aqueous layers were aspirated off, with particular care taken to remove the fat plug present in the ether layer. The oocyst pellet was resuspended in water and centrifuged (4,000 × g, 10 to 15 min) to wash the oocyst pellet. After two water washes, the oocyst pellet was resuspended in 4 ml of water and was carefully layered on top of a nycodenz gradient composed of 2.5 ml of 10% (wt/vol in water) nycodenz (Sigma-Aldrich Corp., St. Louis, Mo.) and 2.5 ml of 25% (wt/vol in water) nycodenz. Following centrifugation (4,000 × g, 30 min), the oocyst layer, located at the interface of the two nycodenz layers, was collected and washed once with water. If the oocyst preparation had a significant amount of contaminants, it was further purified on a Percoll gradient as previously described (29). Oocysts from the feces of chicken and turkey poults were purified by using standard methods of salt flotation bleach, Percoll, and nycodenz steps (29).
In vitro infectivity studies.
The infectivity of TU1867 on Madin-Darby bovine kidney cell monolayers was compared with the standard C. parvum genotype 2 isolate GCH1 (28). Briefly, monolayers grown in 96 microtiter plates were infected with various oocyst doses per well, and after 48 h of incubation at 37°C, the cells were fixed and reacted with specific anti-C. parvum rabbit serum, and the number of immunofluorescent-labeled parasites was quantitated by using a computer-based video image analysis system (22).
Histology and EM studies.
At necropsy, representative gut sections were removed from each infected species (piglets, mice, and chicken poults) and fixed in 10% buffered formalin prior to processing of paraffin sections and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.3) for electron microscopy (EM). The animals used for these studies were (i) a 1-day-old gnotobiotic piglet orally challenged with 106 oocysts and sacrificed 5 days later; (ii) 10- to 12-week-old C57BL/6 male mice immunosuppressed with dexamethasone phosphate (Huang et al., submitted), orally challenged with 105 oocysts, and sacrificed 17 or 25 days later; and (iii) 2-day-old chicken poults orally challenged with 5 × 105 oocysts per animal and sacrificed 7 days later.
For EM, the tissues were postfixed in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.3) containing 0.8% potassium ferricyanide. After en bloc staining with 1% aqueous uranyl acetate, the tissues were dehydrated in an alcohol series and embedded in Spurr's epoxy resin. Following thin sectioning, the grids were stained with methanolic uranyl acetate, followed by Sato's lead citrate. Transmission electron photomicrographs were obtained with a JEOL 1010 electron microscope.
DNA extraction and PCR-RFLP analysis.
DNA was extracted from fecal samples as previously described by Buckholt et al. (3). Genomic DNA from purified oocysts was purified as previously described (29). COWP PCR-RFLP analysis was performed as described by Akiyoshi et al. (1). Species- and strain-specific analysis of Cryptosporidium was performed by using the nested PCR protocol of the small subunit (SSU) rRNA gene as described by Xiao et al. (31, 32), with the reverse primer substituted in the primary PCR as described by Xiao et al. (30). Genotype identification was determined by restriction digestion of the nested SSU rRNA PCR product with AseI. Standards included in each set of PCRs were C. parvum genotype 1 and 2 controls, a mock DNA extraction control, and a negative PCR control.
PCR and DNA sequence analysis.
Nested PCR products, amplified from the SSU rRNA gene from the different animal passages, including humans, pigs, chicken, mice, and calves, were cloned into the pCR4-TOPO vector (Invitrogen Inc., Carlsbad, Calif.), and the resulting clones were sequenced.
Nucleotide sequence accession numbers.
Nucleotide sequences of the COWP and SSU rRNA fragments from the TU1867 isolate were submitted to GenBank and have accession numbers AY166840 and AY166839, respectively.
RESULTS
In vitro infectivity.
Table 1 presents a comparative dose-response assay in vitro between GCH1, a genotype 2 C. parvum isolate, and TU1867, with immunofluorescence as the method of parasite detection. Parasite quantitation was measured by using a semiautomated computerized imaging system (22, 24). The data demonstrated that C. meleagridis is capable of infecting mammalian cells. While the infection rate was lower for TU1867 than GCH1 at the lower doses, at 106 oocysts per well, the rate of infection between the two isolates was comparable. Similar results were observed with the human C. parvum genotype 1 isolate TU502 (data not shown).
TABLE 1.
Comparative infectivity studies of C. parvum genotype 2 (GCH1 isolate) and C. meleagridis (TU1867 isolate) on Madin-Darby bovine kidney cells
| Isolate | No. of oocysts/mla | Parasite countb | SD |
|---|---|---|---|
| GCH1 | 125,000 | 674.61 | 94.67 |
| 250,000 | 966.43 | 180.13 | |
| 500,000 | 1,007.19 | 208.97 | |
| 1,000,000 | 1,094.78 | 197.69 | |
| TU1867 | 125,000 | 68.03 | 26.09 |
| 250,000 | 291.09 | 87.89 | |
| 500,000 | 681.73 | 202.07 | |
| 1,000,000 | 987.56 | 197.56 |
For each oocyst dose, 0.2 ml of oocysts was added per well and each dose was tested in quadruplicate.
Mean parasite counts of each quadruplicate reading for each dose are given. Only intracellular stages of the parasite are included in the count.
Animal infectivity of TU1867.
At the time of writing, TU1867 has been passaged 10 times in piglets, 8 times in mice, 4 times in chickens, 2 times in turkey poults, and 1 time in calves. Infection occurred regardless of the origin of the oocysts, indicating that C. meleagridis was readily transmissible between and among mammalian and avian animal species. Clinical signs were consistently observed in infected piglets only, in which diarrhea and oocyst excretion normally began on day 4 or 5 postinoculation and continued for 10 to 12 days. Clinical signs included diarrhea, but no appreciable signs of dehydration or mortality. The infected colostrum-fed calf developed no clinical signs, and excretion of oocysts was evident 12 days after challenge. Oocyst excretion in chickens lasted from days 4 to 12 postinoculation, and the yield was approximately 5 × 106 oocysts per animal. Similar results were obtained with turkey poults, and the onset and duration of excretion tended to depend on age, with longer incubation and shorter duration in animals aged 8 days compared with 2-day-old poults. Infection in immunosuppressed and γGKO mice was indistinguishable from that normally observed in animals infected with C. parvum genotype 2 isolates. Direct inoculation of γGKO mice with oocysts purified from patients resulted in infection, and there was no change in the genetic profile from those subsequently propagated in piglets.
Histological and EM studies.
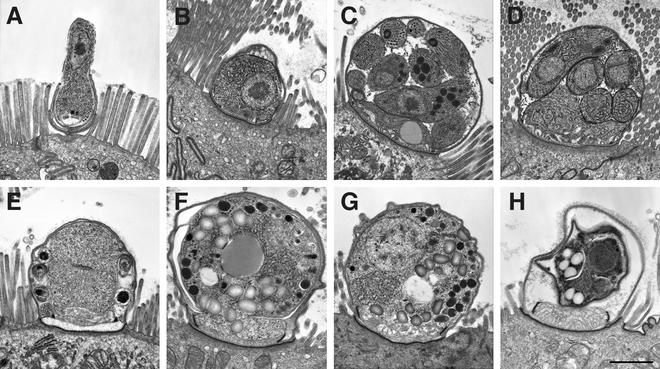
The various endogenous developmental stages of C. meleagridis are morphologically indistinguishable from those of other Cryptosporidium species in terms of internal structure, shape, size, localization, and interaction with the host cell membrane, be it of mammalian or avian origin (Fig. 1A). The distribution of the C. meleagridis infection in the chicken poults was patchy and tended to be restricted to the lower part of the small intestine and in the ceca (Fig. 1A to F and H). Mucosal changes were restricted to the infected areas and included a slight shortening of villi and an irregular epithelial surface layer. The mouse gut was more-intensely infected and was distributed throughout the small intestine and less distributed in the large intestine (Fig. 1G). Curiously, the pyloric region of the stomach was free of infection which is normally seen in mice infected with C. parvum genotype 2 (8). The intensity and distribution of infection and the mucosal alterations in piglets infected with TU1867 were milder than those seen with C. parvum genotype 2 (28) and were similar to those of C. parvum genotype 1.
FIG. 1.
Electron micrographs of sections from a mouse (A to F and H) and a chicken (G) experimentally infected with C. meleagridis. (A) Early development of a trophozoite as it is being engulfed by the parasitophorous membrane. (B) Fully developed trophozoite. (C) First-generation schizogony with 8 merozoites ready to be released into the intestinal lumen. (D) Second-generation schizogony with 4 merozoites which will develop into male or female sexual stages. (E) Dividing microgamy, showing 6 of 16 developing microgametes, the male sexual stage. (F) Fertilized macrogamete or zygote. (G) Fertilized macrogamete or zygote from an infected chicken, showing similar morphology and developmental stage to that of a zygote grown in a mouse (F). (H) Fully developed oocyst still within its parasitophorous vesicle. Bar, 1 μm.
Genotype analysis.
The C. meleagridis isolate TU1867 from the human patient was originally identified by COWP PCR-RFLP analysis. Oocysts from subsequent passages in mice, pigs, chickens, and turkeys were also genotyped by this method. The COWP PCR-RFLP profiles of DNA extracted from the oocysts isolated from these different passages (Fig. 2A) were identical to those previously reported by others for C. meleagridis, and there were no observable changes in this profile as the oocysts were passaged through different hosts. In addition, there was no indication of the presence of a subpopulation or contamination with our C. parvum laboratory isolates. The 553-bp COWP PCR fragment amplified from the human patient was cloned and sequenced. Using the BLAST algorithm (2), this sequence was compared to published sequences in the GenBank database and found to have 100% sequence identity to two other C. meleagridis COWP sequences (accession numbers AF266266 and AF248742).
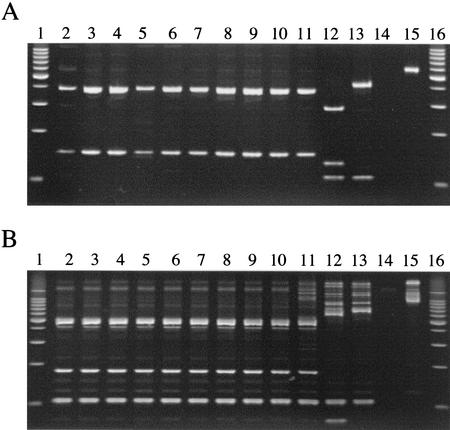
FIG. 2.
COWP (A) and SSU rRNA (B) PCR-RFLP analyses of oocysts from C. meleagridis TU1867 passaged through different host species. For COWP and SSU rRNA RFLP analysis, PCR products were digested with RsaI and AseI, respectively. Samples are from the following animal passages: human (lane 2), first mouse (lane 3), last mouse (lane 4), first pig (lane 5), last pig (lane 6), first chicken (lane 7), last chicken (lane 8), first turkey (lane 9), last turkey (lane 10), and calf (lane 11). C. parvum genotype 1 (TU502) and genotype 2 (GCH1) digested PCR products are shown in lanes 12 and 13, respectively. A negative PCR control (lane 14), uncut PCR product (lane 15), and 100-bp DNA ladder (lanes 1 and 16; Promega Corp., Madison, Wis.) are also included. The mock DNA extraction sample was negative for both PCRs (data not shown). The expected sizes of the 553-bp RsaI-digested C. meleagridis COWP fragment are 372, 147, and 34 bp, and the sizes of the 833-bp AseI-digested SSU rRNA fragment are 456, 171, 104, and 102 bp. The expected sizes of this COWP fragment are 284, 129, 106, and 34 bp for C. parvum genotype 1 and 413, 106, and 34 bp for C. parvum genotype 2. The expected sizes of the SSU rRNA fragment are 561, 104, 102, and 70 bp for C. parvum genotype 1 (837 bp, AF481962) and 628, 104, and 102 bp for genotype 2 (834 bp, AF164102).
A second confirmational PCR-RFLP analysis was performed with the SSU rRNA gene (Fig. 2B). Nested PCR was performed as described by Xiao et al. (30-32). The resulting 833-bp fragment was digested with AseI, which can distinguish at least nine Cryptosporidium species and C. parvum genotypes (32). The AseI RFLP profile of the TU1867 isolate was identical to that reported for C. meleagridis (accession number AF112574) (32) and did not change as the isolate was passaged through a different host, nor was a second Cryptosporidium population detected. The nested PCR products from the original human sample and from the four different animal host passages (mice, pigs, chickens, and calf) were cloned and sequenced. These sequences were invariant and shared 100% sequence identity to one previously published sequence (GenBank accession number AF112574) of C. meleagridis when analyzed with the BLAST algorithm.
DISCUSSION
TU1867, originally purified from a child from Uganda with diarrhea (Tumwine et al., submitted), is the first reported isolate of C. meleagridis to be continuously propagated in the laboratory and well characterized. It was readily cross-transmitted from one species to the next and appears to infect a broad range of hosts that include both avian (chicken and turkey) and mammalian (human, piglet, calf, and mouse) species. So far, we have been able to infect every animal species we have attempted, which is quite different from results with other Cryptosporidium species tried, including C. parvum, Cryptosporidium baileyi, and C. canis (unpublished data). Our data supports and extends previous reports describing host-range studies. Darabus reported the passage of one C. meleagridis isolate originally isolated from chickens into mice, rabbits, rats, and cattle (6), and later, Sreter et al. (21) reported the passage of a second isolate from turkeys into mice then into chickens.
The TU1867 isolate retained a consistent rate of infection and maintained the same genetic profile, reflecting the stability and homogeneity of this isolate during the different host passages. Our study also reports the first published series of EM photographs showing the endogenous developmental stages of C. meleagridis propagated in mammals. The life cycle and the morphology of the endogenous stages of TU1867 were indistinguishable from those of C. parvum (Fig. 1), and the sites and distribution of infection in the gut were largely similar to those observed in C. parvum-infected animals (26). Our observation that the main site of infection is the small intestine is consistent with previous studies by others (14, 20). The ability to continuously propagate TU1867 under laboratory conditions made it possible to conduct for the first time some basic comparative infectivity studies as well as to study the host range of C. meleagridis. Additional isolates will need to be characterized in a similar manner to confirm that the observations made for TU1867 are shared by some or all C. meleagridis isolates.
The ability of an avian Cryptosporidium isolate to infect mammals has significant epidemiologic implications with regard to human public health in terms of transmission and the risk associated with exposure to birds and possibly to other mammals. The diarrheal disease seen in the 4 infected children was indistinguishable from that in children with acute illness due to C. parvum (Tumwine et al., submitted). Nor were there apparent differences in animals experimentally infected with TU1867 compared to those infected with C. parvum genotype 1 (piglets and calves) (1), or genotype 2 (pigs, calves, and mice) (27). We do not believe that propagation of TU1867 in piglets had influenced the spectrum of infectivity to other animal species, as other investigators have been able to infect a range of animals with C. meleagridis (21). While natural infections with C. meleagridis have not been reported so far in chicken, rodents, cattle, or swine, the infectivity (number of oocysts required to induce infections) and virulence (intensity of infection or disease and distribution in the gut) suggest that they are more than likely to occur, much as seen in humans. The difference is that animal isolates are less-rigorously analyzed and genotyped compared to isolates from humans. The ability of C. meleagridis to infect a range of hosts, in contrast to C. parvum type 1 in particular, is curious and not understood at all.
Laboratory propagation of well-characterized isolates representing various Cryptosporidium species, particularly those infectious to humans, has been a major undertaking by this laboratory. In our view, there is a need for representative reference isolates affecting humans to be maintained in germfree animals to prevent contamination with other environmental or laboratory isolates. Access and use of genetically well-defined and stable reference isolates will improve considerably the quality of future laboratory and field investigations of these medically important pathogens.
Acknowledgments
This study was supported by NIH contract N01-AI-75321.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Akiyoshi, D. E., X. Feng, M. A. Buckholt, G. Widmer, and S. Tzipori. 2002. Genetic analysis of a Cryptosporidium parvum human genotype 1 isolate passaged through different host species. Infect. Immun. 70:5670-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Buckholt, M. A., J. H. Lee, and S. Tzipori. 2002. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 68:2595-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caccio, S., E. Pinter, R. Fantini, I. Mezzaroma, and E. Pozio. 2002. Human infection with Cryptosporidium felis: case report and literature review. Emerg. Infect. Dis. 8:85-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carreno, R. A., N. J. Polomy, H. Lee, J. T. Trevors, and S. A. De Grandis. 2001. Phenotypic and genotypic characterization of Cryptosporidium species and isolates. J. Ind. Microbiol. Biotechnol. 26:95-106. [DOI] [PubMed] [Google Scholar]
- 6.Darabus, G. 1997. Experimental studies of inter- and intraspecific transmission of Cryptosporidium parvum and C. meleagridis. Rev. Rom. Med. Vet. 7:155-160. [Google Scholar]
- 7.Fayer, R., C. A. Speer, and J. P. Dubey. 1997. The general biology of Cryptosporidium, p. 1-42. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Inc., Boca Raton, Fla.
- 8.Griffiths, J., C. Theodos, M. Paris, and S. Tzipori. 1998. The interferon-γ gene-disrupted mouse: a highly susceptible model for evaluation of therapeutic agents against Cryptosporidium parvum. J. Clin. Microbiol. 36:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths, J. K. 1998. Human cryptosporidiosis: epidemiology, transmission, clinical disease, treatment and diagnosis. Adv. Parasitol. 40:37-81. [DOI] [PubMed] [Google Scholar]
- 10.Guyot, K., A. Follet-Dumoulin, E. Lelièvre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. A. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan, U. M., L. Xiao, R. Fayer, A. A. Lal, and R. C. A. Thompson. 1999. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int. J. Parasitol. 29:1733-1751. [DOI] [PubMed] [Google Scholar]
- 14.O'Donoghue, P. J. 1995. Cryptosporidium and cryptosporidiosis in man and animals. Int. J. Parasitol. 25:139-195. [DOI] [PubMed] [Google Scholar]
- 15.Pedraza-Diaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium ‘dog type’ from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 16.Pedraza-Díaz, S., C. Amar, and J. McLauchlin. 2000. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189-194. [DOI] [PubMed] [Google Scholar]
- 17.Pedraza-Diaz, S., C. F. L. Amar, J. McLauchlin, G. L. Nichols, K. M. Cotton, P. Godwin, A. M. Iversen, L. Milne, J. R. Mulla, K. Nye, H. Panigrahl, S. R. Venn, R. Wiggins, M. Williams, and E. R. Youngs. 2001. Cryptosporidium meleagridis from humans: molecular analysis and description of affected patients. J. Infect. 42:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Pieniazek, N. J., F. J. Bornay-Llinares, S. B. Slemenda, A. J. da Silva, I. N. S. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slavin, D. 1955. Cryptosporidium meleagridis (sp. nov.). J. Comp. Pathol. 65:262-266. [DOI] [PubMed] [Google Scholar]
- 20.Sréter, T., and I. Varga. 2000. Cryptosporidiosis in birds-a review. Vet. Parasitol. 87:261-279. [DOI] [PubMed] [Google Scholar]
- 21.Sréter, T., G. Kovács, A. J. da Silva, N. J. Pieniazek, Z. Széll, M. Dobos-Kovács, K. Márialigeti, and I. Varga. 2000. Morphologic, host specificity, and molecular characterization of a Hungarian Cryptosporidium meleagridis isolate. Appl. Environ. Microbiol. 66:735-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theodos, C. M., K. L. Sullivan, J. K. Griffiths, and S. Tzipori. 1997. Profiles of healing and nonhealing Cryptosporidium parvum infection in C57BL/6 mice with functional B and T lymphocytes: the extent of gamma interferon modulation determines the outcome of infection. Infect. Immun. 65:4761-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzipori, S. 1983. Cryptosporidiosis in animals and humans. Microbiol. Rev. 47:84-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzipori, S. 1998. Cryptosporidiosis: laboratory investigations and chemotherapy. Adv. Parasitol. 40:5-36. [DOI] [PubMed] [Google Scholar]
- 25.Tzipori, S., and G. Widmer. 2000. The biology of Cryptosporidium. Contrib. Microbiol. 6:1-32. [DOI] [PubMed] [Google Scholar]
- 26.Tzipori, S., and H. Ward. 2002. Cryptosporidiosis: biology, pathogenesis and disease. Microb. Infect. 4:1047-1058. [DOI] [PubMed] [Google Scholar]
- 27.Tzipori, S., K. W. Angus, I. Campbell, and E. W. Gray. 1980. Cryptosporidium: evidence for a single-species genus. Infect. Immun. 30:884-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzipori, S., W. Rand, J. Griffiths, G. Widmer, and J. Crabb. 1994. Evaluation of an animal model system for cryptosporidiosis: therapeutic efficacy of paromomycin and hyperimmune bovine colostrum-immunoglobulin. Clin. Diagn. Lab. Immunol. 1:450-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widmer, G., S. Tzipori, C. J. Fichtenbaum, and J. K. Griffiths. 1998. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J. Infect. Dis. 178:834-840. [DOI] [PubMed] [Google Scholar]
- 30.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gillman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 31.Xiao, L., L. E. Escalante, C. Yang, I. M. Sulaiman, A. A. Escalante, R. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites on the small-subunit ribosomal RNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]