Abstract
VacA is a secreted toxin that plays a role in Helicobacter pylori colonization of the stomach and that contributes to the pathogenesis of peptic ulcer disease. Studies of VacA structure and function have been hindered by the lack of an efficient system for expression and genetic manipulation of this toxin. In this study, we developed methodology for expression of a functionally active VacA toxin in Escherichia coli. We then used a high-throughput screen to analyze a library of mutant toxins with pentapeptide insertions and identified six mutants that lacked the capacity to induce vacuolation of HeLa cells. The capacity to analyze VacA in this heterologous-expression system should greatly facilitate efforts to elucidate the structure and function of this toxin.
Colonization of the human stomach by Helicobacter pylori is associated with chronic gastritis and an increased risk of peptic ulceration and gastric cancer (4, 24). Numerous studies suggest that the vacuolating cytotoxin VacA is an important virulence factor secreted by H. pylori (1, 2, 20, 21, 23). When VacA is added to cultured cells, the most evident activity of the toxin is the induction of large cytoplasmic vacuoles (5, 16). VacA also has been reported to exhibit a number of other activities, including the formation of anion-selective channels, interference with antigen presentation, and induction of apoptosis (2, 20, 23). The mechanism of VacA activity is not yet completely understood.
Studies of VacA structure and function have been hampered by the lack of an efficient system for expression and genetic manipulation of this toxin. Previous attempts to express an active full-length VacA toxin in Escherichia coli have been unsuccessful (15, 17, 26, 27). Active toxin can be purified in small quantities from H. pylori broth culture supernatants (5, 17), and methodology for allelic-exchange mutagenesis of the H. pylori chromosomal vacA gene is available (3, 18, 25). However, this methodology is cumbersome, and mutagenesis of the portion of vacA encoding the carboxyl-terminal end of the mature toxin has been problematic due to a requirement for this region for proper toxin folding or secretion (25; M. S. McClain and T. L. Cover, unpublished observations). Several laboratories have successfully expressed functional VacA in transiently transfected tissue culture cells (9, 10, 12, 29, 30), but this approach does not allow the study of interactions between the toxin and the eukaryotic cell surface. Due to the limitations of the currently available systems, only a small number of inactive VacA mutant proteins have been characterized thus far.
The goal of the present study was to develop methodology that would permit a functionally active form of VacA to be expressed in E. coli. H. pylori VacA is the product of a single gene that encodes a 140-kDa precursor protein (5, 8). In H. pylori, this precursor protein is proteolytically processed to yield the mature secreted 88-kDa toxin (8, 22). Upon removal of the amino-terminal signal sequence, the mature VacA toxin contains an alanine as the amino-terminal amino acid. To express the mature VacA toxin in E. coli, a methionine must be present at the amino terminus in order to support initiation of translation. Previous studies have reported that several mutations near the VacA amino terminus result in ablation of toxin activity (10, 18, 25, 29, 30), and therefore the introduction of a methionine at this site could potentially abolish toxin activity. Based on these considerations, we first examined whether VacA expressed in H. pylori would be active if the first amino acid of the mature toxin was changed from alanine to methionine.
A 142-bp region from plasmid pA176 (containing an ∼3-kb portion of vacA in which a StuI site was introduced between codons 12 and 13) (18) was amplified by PCR using oligonucleotide primers AN5012 (5′-GGCTGCAGGAAAAGAAATGGAAATACAACAAAC) and AND3133 (5′-TGGAATGATCACGGTTGTAAAAAACATTGCATGACTTTGTTGCGG). The underlined nucleotides represent mutations relative to the wild-type vacA sequence to introduce a methionine codon and a BsrDI restriction site. The PCR product was digested with restriction enzymes StuI and BclI and ligated to StuI- and BclI-digested pA176 (18). The resulting plasmid, pMM589, was transformed into H. pylori strain VM022 (25). Transformants in which the sacB-kan cassette of VM022 was replaced by sequences containing the vacA-A1M mutation were selected by growth on plates containing 5.5% sucrose as described previously (3, 18, 25). The presence of the vacA-A1M mutation in one transformant, H. pylori VM085, was confirmed by PCR followed by digestion with the restriction enzyme BsrDI. H. pylori strain VM085 produced and secreted a VacA toxin (VacA-A1M) of the same apparent size as wild-type VacA as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, suggesting that the signal sequence was removed from the VacA-A1M protein as expected (data not shown). Addition of VacA-A1M to HeLa cells revealed that this toxin exhibited vacuolating activity indistinguishable from the activity of wild-type toxin (data not shown). This experiment indicated that the VacA-A1M substitution has no detectable effect on toxin activity.
To clone vacA in E. coli, a vacA fragment encoding the mature VacA toxin (amino acids 1 to 821, including the A1M substitution) was amplified by PCR from H. pylori strain 60190 by using oligonucleotide primers BA9146 (5′-CCCACTAGTAAGAGGAGACGCCATGTTTTTTACAACCGTG) andC7915 (5′-GGCTGCAGCTAAGCGTAGCTAGCGAAACG).The resulting PCR product was digested with SpeI and PstI (indicated by the underlined sequences) and ligated to XbaI- and PstI-digested pET-41b (conferring kanamycin resistance; Novagen) to create plasmid pMM592. Analysis of the vacA insert in pMM592 indicated that its nucleotide sequence was identical to that of vacA from H. pylori strain 60190, except for the A1M mutation. The plasmid was then transformed into E. coli strain ER2566 (New England Biolabs), which encodes an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible copy of the RNA polymerase gene from bacteriophage T7. Transformants were grown at 25°C overnight with shaking in Terrific broth (TB; 12 g of tryptone, 24 g of yeast extract, 4 ml of glycerol, 2.31 g of KH2PO4, and 12.54 g of K2HPO4 per liter) supplemented with 25 μg of kanamycin/ml (TB-KAN). Overnight cultures were diluted 1 to 100 in fresh TB-KAN and grown at 25°C for 4 h (to an optical density at 600 nm of 0.2 to 0.4). IPTG then was added to a final concentration of 250 μM, and growth continued for 20 h at 25°C.
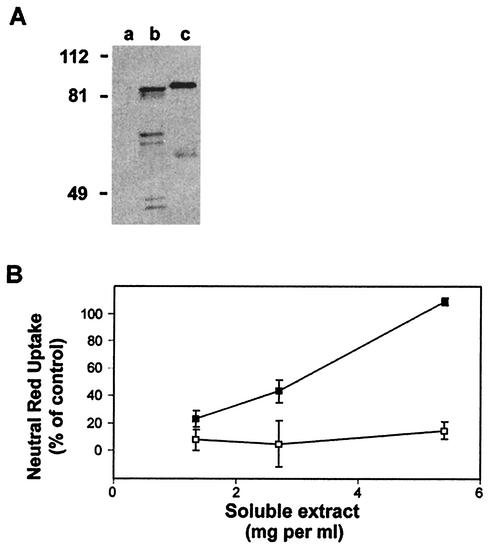
To prepare bacterial soluble extracts, the IPTG-induced cultures were pelleted and then resuspended in phosphate-buffered saline (PBS) containing 100 μg of egg white lysozyme per ml and a bacterial protease inhibitor cocktail (Sigma). The bacterial suspensions were incubated on ice for 15 min and then sonicated for two 10-s bursts at 4 W on ice. The sonicated suspensions were frozen in a methanol-dry ice bath and then thawed at 37°C. The sonication and freeze-thaw cycles were repeated two or three additional times as needed. Insoluble debris was pelleted, and the supernatant was filtered through a 0.22-μm-pore-size filter. Extracts were used immediately or stored at −20°C. Immunoblot analysis using anti-VacA antisera revealed the inducible expression of an immunoreactive protein (data not shown), and a significant portion of the immunoreactive protein was detected in the soluble fraction (Fig. 1A).
FIG. 1.
Expression of VacA-A1M in E. coli. (A) Immunoblot analysis, using anti-VacA rabbit antiserum, of soluble extract from IPTG-induced ER2566(pET-41b) (vector control; lane a), soluble extract from IPTG-induced ER2566(pMM592) (lane b), and VacA purified from H. pylori strain 60190 (lane c). (B) Equal amounts of protein from the soluble extract from IPTG-induced ER2566(pMM592) (VacA-A1M; ▪) and from the soluble extract from IPTG-induced ER2566(pET-41b) (vector control; □) were added to the medium overlying HeLa cells. Vacuolating activity was measured by a neutral red dye uptake assay. Results represent the means and standard deviations from quadruplicate samples and are expressed as the percentages of neutral red dye uptake relative to that for control cells treated with 10 μg of VacA purified from H. pylori strain 60190 per ml.
Interestingly, the immunoreactive VacA protein in E. coli soluble extracts appeared somewhat smaller than the 88-kDa VacA protein from H. pylori strain 60190 (Fig. 1A). Based on amino-terminal amino acid sequencing and matrix-assisted laser desorption ionization-time of flight mass-spectrometric analysis, the mature VacA toxin purified from H. pylori strain 60190 is predicted to consist of 821 amino acids (8, 22). DNA sequence analysis of the plasmid encoding recombinant VacA (pMM592) indicates that the cloned vacA gene also is expected to encode a protein with 821 amino acids. The only expected difference between the VacA protein expressed in H. pylori and the VacA protein expressed in E. coli is the substitution of methionine for alanine at the first amino acid position. This leads us to suggest that VacA may be posttranslationally modified in E. coli. One possibility is that there may be proteolytic truncation at the carboxyl-terminal end of the recombinant VacA produced in E. coli.
To determine whether the recombinant VacA exhibits biological activity, soluble extracts from induced cultures expressing VacA or from induced cultures transformed with the pET-41b vector alone were added to HeLa cells. HeLa cells were cultured in minimum essential medium (MEM; ICN Pharmaceuticals) containing 10% fetal bovine serum. Soluble extract from E. coli or VacA purified from H. pylori (6, 19) was added to HeLa cell monolayers in MEM containing 5 mM ammonium chloride. Medium overlying the cells was routinely removed after 1 h and replaced with fresh MEM containing 5 mM ammonium chloride to limit the development of an uncharacterized precipitate on the cell monolayers treated with recombinant VacA. Plates were incubated at 37°C in ambient air containing 5% CO2 for at least 2 h, until vacuoles developed. In contrast to VacA preparations purified from H. pylori broth culture supernatants, which were routinely acid activated prior to the addition of these preparations to HeLa cell monolayers (11), E. coli extracts containing recombinant VacA were not acid activated because acidification resulted in precipitation of proteins, including VacA.
The soluble extract from the VacA-producing E. coli, but not extract from the E. coli vector-only control, induced vacuole formation in HeLa cells. Vacuoles induced by recombinant VacA avidly sequestered neutral red dye, which allowed vacuolation to be quantified based on neutral red dye uptake (Fig. 1B) (7). Vacuole formation induced by the recombinant toxin was detectable about 2 h after addition of the recombinant toxin to cells and required the addition of a cell-permeant weak base such as ammonium chloride to the tissue culture medium (data not shown). These features of the recombinant VacA protein were identical to those of VacA isolated from H. pylori. In contrast to the VacA toxin purified from H. pylori, the recombinant VacA present in E. coli extracts exhibited vacuolating activity without requiring pH activation. Similarly, unpurified VacA toxin present in crude, unfractionated H. pylori broth culture supernatants also exhibits vacuolating activity without requiring pH activation. Further studies are needed to investigate possible differences in the quaternary structures of these different forms of VacA, which may account for differences in the requirement for pH activation.
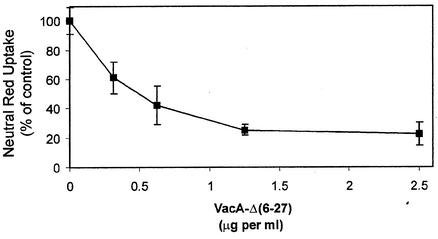
To further establish that the vacuolating activity in soluble extracts containing VacA-A1M was not due to some unidentified E. coli protein, we studied the ability of the mutant toxin VacA-Δ(6-27) to specifically inhibit the observed vacuolating activity. When isolated from H. pylori, VacA-Δ(6-27) lacks detectable vacuolating activity, and it exhibits a dominant-negative phenotype when added in combination with wild-type VacA to cells (25). Experimental evidence suggests that the dominant-negative effect is mediated by the formation of mixed oligomeric complexes containing both wild-type and mutant toxins (18, 25). We hypothesized, therefore, that if the vacuoles observed following addition of the VacA-A1M-containing extracts were due to the recombinant VacA, a mixture of recombinant VacA and VacA-Δ(6-27) purified from H. pylori would fail to induce vacuolation. Soluble extract from E. coli ER2566(pMM592) was mixed with purified, acid-activated VacA-Δ(6-27) from H. pylori strain AV452, and the mixed toxins were added to HeLa cell monolayers. The VacA-Δ(6-27) toxin purified from H. pylori was able to inhibit the vacuolating activity present in a soluble extract of E. coli ER2566(pMM592) (Fig. 2), indicating that the vacuolating activity observed in the extract was indeed due to recombinant VacA.
FIG. 2.
Recombinant VacA is inhibited by VacA-Δ(6-27) from H. pylori strain AV452. Soluble extract (5.5 mg per ml) from IPTG-induced ER2566(pMM592) was mixed with various amounts of acid-activated VacA-Δ(6-27) purified from H. pylori strain AV452 and was added to HeLa cells. Vacuolation was quantified by a neutral red dye uptake assay. Results represent the means and standard deviations from triplicate samples and are expressed as the percentages of neutral red dye uptake relative to that for cells treated with soluble extract from IPTG-induced ER2566(pMM592) alone.
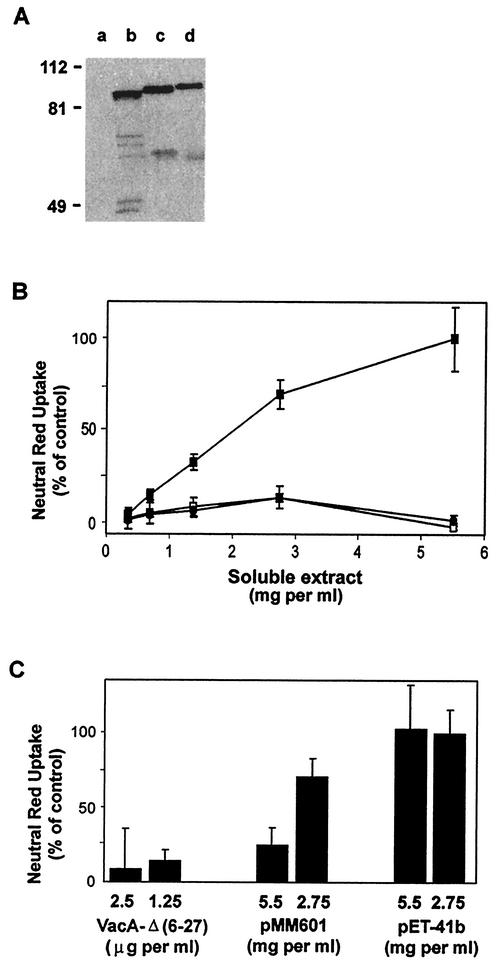
Expression of a functional recombinant VacA protein offers many opportunities for efficient construction and analysis of mutant toxins. As a first step in testing whether mutant toxins can be produced and analyzed in the recombinant expression system, we cloned, sequenced, and expressed in E. coli a recombinant form of the mutant VacA-Δ(6-27). We chose to express this mutant form specifically because of the dominant-negative phenotype associated with the VacA-Δ(6-27) toxin. Plasmid pMM601, containing DNA corresponding to the mature portion of the VacA-Δ(6-27) mutant toxin, was created by PCR amplification of the relevant vacA fragment from H. pylori strain AV452 (25) and subsequent cloning as described above for plasmid pMM592. E. coli containing plasmid pMM601 expressed an immunoreactive protein in the soluble fraction (Fig. 3A). Soluble extracts containing VacA-[A1M, Δ(6-27)] did not induce detectable vacuolation when added to HeLa cells (Fig. 3B). Importantly, E. coli extracts containing VacA-[A1M, Δ(6-27)] exhibited a dominant-negative phenotype when added to HeLa cells in combination with wild-type VacA purified from H. pylori 60190 (Fig. 3C) or VacA-A1M-containing E. coli extracts (data not shown). No inhibitory activity was detected in soluble extracts of E. coli transformed with the pET-41b vector control (Fig. 3C). These results demonstrate that a mutant VacA toxin can be produced recombinantly and that it exhibits properties very similar to those observed with the corresponding mutant toxin purified from H. pylori broth culture supernatant (25).
FIG. 3.
VacA-[A1M, Δ(6-27)] expressed in E. coli exhibits a dominant-negative phenotype. (A) Immunoblot analysis, using anti-VacA antiserum, of soluble extract from IPTG-induced ER2566(pET-41b) (vector control; lane a), soluble extract from IPTG-induced ER2566(pMM601) {expressing VacA-[A1M, Δ(6-27)]; lane b}, VacA-Δ(6-27) purified from H. pylori strain AV452 (lane c), and wild-type VacA purified from H. pylori strain 60190 (lane d). (B) Equal amounts of protein from the soluble extract from IPTG-induced ER2566(pMM592) (VacA-A1M; ▪), from the soluble extract from IPTG-induced ER2566(pMM601) {VacA-[A1M, Δ(6-27)]; □}, and from the soluble extract from IPTG-induced ER2566(pET-41b) (vector control; •) were added to the medium overlying HeLa cells. Vacuolating activity was measured by a neutral red dye uptake assay. Results represent the means and standard deviations from quadruplicate samples and are expressed as the percentages of neutral red dye uptake relative to that for cells treated with 5.5 mg of soluble extract from IPTG-induced ER2566(pMM592) per ml. (C) Acid-activated wild-type VacA purified from H. pylori strain 60190 (5 μg per ml, final concentration) was mixed with either acid-activated VacA-Δ(6-27) purified from H. pylori strain AV452, soluble extract from IPTG-induced ER2566(pMM601) {VacA-[A1M, Δ(6-27)]}, or soluble extract from IPTG-induced ER2566(pET-41b) (vector control). Mixtures of toxins were added to HeLa cells, and vacuolation was quantified by a neutral red dye uptake assay. Final concentrations of the indicated preparations are shown. Results represent the means and standard deviations of quadruplicate samples and are expressed as the percentages of neutral red dye uptake relative to that for control cells treated with 5 μg of acid-activated VacA purified from H. pylori strain 60190 per ml.
Having demonstrated the successful recombinant expression of a particular mutant toxin and having characterized it phenotypically, we proceeded to perform random mutagenesis of vacA. To do this, we subjected the VacA-A1M-encoding plasmid, pMM592, to pentapeptide scanning mutagenesis (13, 14). Plasmid pMM592 was subjected to random mutagenesis by using the transposon donor pGPS4 (GPS-LS linker-scanning system; New England Biolabs) according to the manufacturer's instructions. The transposon carries the cat gene (conferring chloramphenicol resistance) flanked by PmeI restriction sites. Plasmids containing transposon inserts were selected on Luria-Bertani agar plates supplemented with 25 μg of kanamycin and 20 μg of chloramphenicol per ml. This procedure generated a library of 195 transposon insertions. If the transposon insertions were random, this library would be predicted to contain approximately one transposon insertion for every 31 bp. The library was pooled, plasmid DNA was extracted, and the bulk of the transposon was excised by digesting the plasmid DNA with the restriction enzyme PmeI. Following digestion with PmeI and ligation, each plasmid was expected to contain a 15-bp insert specifying one of three possible amino acid sequences (CLNXX, XFKQX, or XFKHX, where the amino acid indicated by X depends on the sequence flanking the site of insertion) or a stop codon. The ligated plasmid pool then was transformed into the expression strain ER2566 to yield an expanded library of 371 colonies. Each colony was inoculated into 100 μl of TB-KAN in a well of a standard 96-well plate, and the plates were incubated with shaking at 25°C for 16 h.
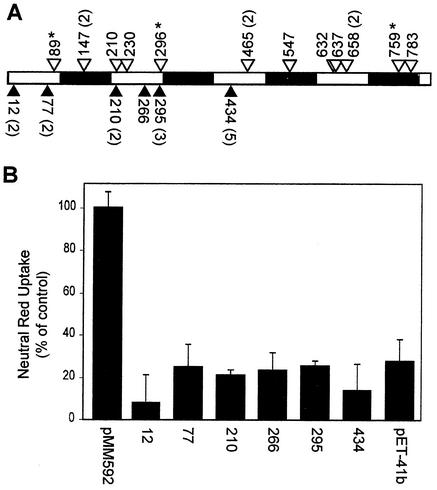
To assess the distribution of transposon insertions in vacA, 36 clones were selected at random from the expanded library in E. coli strain ER2566 and the sites of transposon insertion were mapped by PCR amplification of the vacA gene and digestion of the PCR products with PmeI. Fifteen of the 36 clones selected were determined to have a transposon insert within the vacA gene, whereas the other 21 clones had transposon inserts elsewhere in the plasmid. DNA sequence analysis of the PCR products revealed that 12 unique sites of insertion in vacA were represented (Fig. 4A). We conclude from this analysis that the transposon, as expected, inserted throughout the entire length of vacA. Three of the inserts (Fig. 4A) introduced nonsense mutations. Colonies representing the remaining nine inserts in vacA were cultured and lysed as described above to produce recombinant VacA proteins. Extracts from eight of the nine clones induced vacuolation when added to HeLa cells; extract from the clone containing an insertion after amino acid 210 did not induce vacuolation (data not shown).
FIG. 4.
Pentapeptide scanning mutagenesis of VacA. (A) Distribution of pentapeptide insertions throughout VacA. The 821-amino-acid sequence of VacA is represented by open and solid boxes, each corresponding to 100 amino acids. Open triangles, 12 pentapeptide insertions in VacA, identified by screening 36 randomly chosen mutants; solid triangles, 6 pentapeptide insertions that resulted in loss of VacA activity, identified in the high-throughput screen of 371 colonies. The amino acid of VacA preceding the site of each pentapeptide insertion is indicated. Numbers in parentheses, numbers of times clones with an identical insertion were isolated in each screen; asterisks, sites at which the insertion introduced a nonsense codon. The inserted amino acids are VFKHV (after amino acid 12), CLNID (after amino acid 77), LFKHT (after amino acid 210), VFKHQ (after amino acid 266), VFKHE (after amino acid 295), and CLNTS (after amino acid 434). (B) Analysis of six pentapeptide insertions that resulted in loss of VacA activity. Soluble extracts from IPTG-induced clones (designated as shown in panel A) were standardized so that they contained equal amounts of VacA and then were added to the medium overlying HeLa cells. As controls, extracts from ER2566(pMM592) (containing VacA-A1M with no mutations) or ER2566(pET-41b) were standardized by protein concentration and added to cells. Vacuolating activity was measured by a neutral red dye uptake assay. Results represent the means and standard deviations from quadruplicate samples and are expressed as the percentages of neutral red dye uptake relative to that for control cells treated with extract from ER2566 containing plasmid pMM592.
To identify additional pentapeptide insertion sites that inactivate the VacA toxin, all 371 individual colonies (including the 36 colonies analyzed as described above) from the cultured, expanded library were diluted 1 to 100 into 1 ml of TB-KAN in the wells of a 96-well deep-well dish. The deep-well dishes were incubated at 25°C with shaking for 4 h, and then VacA expression was induced by addition of IPTG. Induced cultures were pelleted and lysed directly in the deep-well dishes. IPTG-induced cultures were pelleted, washed in 0.9% NaCl, and resuspended in a solution (25 μl per ml of starting culture) that contained 10 mM Tris (pH 7.5), 100 mM NaCl, 1 mM EDTA, protease inhibitors (Complete Mini; Roche), and 20,000 U of ReadyLyse lysozyme (Epicentre) per ml. Bacteria were incubated at room temperature for 15 min with periodic mixing, after which a solution (75 μl per ml of starting culture) containing 50 mM Tris (pH 8.0), 2.67 mM MgCl2, and 67 U of Benzonase nuclease (Novagen) per ml was added. Samples were mixed briefly and subjected to four successive rounds of freezing (in a dry ice-methanol bath) and thawing at 37°C. The insoluble debris was pelleted, and the supernatants were used immediately or stored at −20°C.
To test the soluble extracts for vacuolating activity, an aliquot from each extract was added to the medium overlying HeLa cells. Vacuolation of HeLa cells was assessed by microscopic inspection. Based on this analysis, soluble extracts from 264 of the 371 colonies induced vacuolation of HeLa cells. In contrast, extracts from 107 of the 371 colonies induced either no detectable vacuolation or markedly less vacuolation than did extracts (prepared in a similar manner) from E. coli expressing wild-type VacA. Mutants that failed to induce HeLa cell vacuolation were studied further by immunoblotting to determine whether they each expressed a full-length toxin and whether these mutants expressed VacA at levels similar to the level of expression of wild-type VacA. The immunoblot analysis indicated that 92 of the 107 mutants expressed either markedly reduced amounts of VacA (75 mutants) or truncated forms of VacA (17 mutants). Presumably the insertions in these mutants reduced the stability of the recombinant VacA protein (making the protein more susceptible to degradation by E. coli), altered the folding of the recombinant protein (thereby reducing its abundance in the soluble fraction following cell lysis), or introduced nonsense mutations. We note that minimal efforts were made to control the growth state at which the cultures were induced in the high-throughput analysis, and it is possible that the growth state plays a role in the amount of VacA produced. Mutants that expressed either markedly reduced amounts of VacA or truncated forms of VacA were not studied further.
The remaining 15 mutants, producing very low or undetectable levels of vacuolating activity but expressing high levels of full-length VacA, were characterized in greater detail. Plasmids were isolated from these 15 mutants, and the mutations were mapped by restriction endonuclease and DNA sequence analyses. Among these 15 mutants, six different sites of transposon insertion were identified (Fig. 4A). Thus, a small subset of mutants was reproducibly identified in the screening process.
To study further the six unique insertions that inactivated VacA, a single mutant clone representing each site of insertion was cultured and fresh soluble extracts were prepared. An antigen detection enzyme-linked immunosorbent assay using anti-VacA antiserum was used to determine the relative amount of recombinant VacA in each extract (5, 25). VacA-containing samples were diluted in carbonate buffer (15 mM sodium carbonate and 35 mM sodium bicarbonate, pH 9.6) and adsorbed to the wells of a microtiter dish at 25°C overnight. Nonspecific protein binding sites were blocked by discarding the supernatant and filling the wells with PBS containing 0.05% Tween 20 and 0.1% bovine serum albumin. Plates were incubated for 3 h at 37°C and then washed three times with deionized water. Anti-VacA rabbit serum was added to each well in PBS containing 0.05% Tween 20, and the plates were incubated at 37°C for 1 h and then washed five times with deionized water. Secondary antibodies (goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase) were then added in PBS containing 0.05% Tween 20, and the plates were incubated at 37°C for 1 h and then washed five times with deionized water. Substrate (3,3′,5,5′-tetramethylbenzidine; Pierce) was added, and color development was quantified at 450 nm with an MRX microplate reader (Dynatech) after stopping development with 2 N H2SO4. Equivalent amounts of each recombinant VacA toxin were added to HeLa cell monolayers, and vacuolation was assessed microscopically and quantified by a neutral red dye uptake assay (Fig. 4B). As expected, each of the six mutant toxins exhibited significantly decreased vacuolating activity in comparison to wild-type recombinant VacA. In contrast to certain previously characterized mutant toxins that exhibited a dominant-negative phenotype (18, 25), none of the six inactive mutants exhibited a dominant-negative phenotype when mixed with soluble extracts expressing active wild-type VacA (data not shown). The reasons why certain mutants inhibit the vacuolating activity of wild-type VacA whereas other mutants do not exhibit an inhibitory phenotype remain to be determined.
This study demonstrates for the first time that a functionally active H. pylori vacuolating cytotoxin can be expressed in E. coli. Several previous studies have reported expression of full-length VacA in E. coli, but these recombinant toxins did not possess detectable vacuolating activity (15, 17, 26, 27). Notably, in each of these previous studies, the amino acid sequence at the amino-terminal end of the recombinant VacA was altered relative to that of VacA secreted by H. pylori. The amino-terminal addition of either a six-His tag (15, 17) or a glycine-serine dipeptide (26, 27) may account for the lack of activity observed in previous studies.
Despite many years of research in multiple laboratories, only a small number of inactive mutant VacA proteins have been characterized previously (10, 18, 25, 28-30). Some of the previously described inactive mutant VacA proteins contain large deletions that are expected to drastically alter VacA structure. In contrast, VacA can be inactivated by any of a variety of mutations, including single amino acid substitutions, near its amino terminus (10, 18, 25, 29, 30). The amino-terminal region of VacA (amino acids 1 to 32) is predicted to be highly hydrophobic and plays a role in the formation of anion-selective channels (18, 25). The pentapeptide insertion after amino acid 12 in the present study maps to this hydrophobic region, and we are therefore not surprised that this insertion inactivates VacA. VacA activity is likely to depend on the ability of the toxin to bind to cells, to form oligomeric complexes, to be internalized by cells, to form anion-selective channels, and perhaps to carry out enzymatic functions that have yet to be identified. The inactive mutants identified in the present study may be defective in one or more of these steps.
The successful expression of active, recombinant VacA and the ability to use this heterologous expression system to characterize vacA mutants should aid many new studies of this H. pylori virulence factor. Further investigations using this system are likely to provide new insights into the structure and mechanism of action of the VacA toxin.
Acknowledgments
We thank Ping Cao and Beverly Hosse for technical assistance and Victor Torres for helpful discussions. DNA oligonucleotides were synthesized by the Vanderbilt University DNA Chemistry Core Facility, and DNA sequence analysis was performed by the Vanderbilt University DNA Sequencing Laboratory.
This work was supported by NIH grants AI39657 and DK53623 and by the Medical Research Department of the Department of Veterans Affairs. The Vanderbilt University DNA Sequencing Laboratory is supported by the Vanderbilt-Ingram Cancer Center.
Editor: J. T. Barbieri
REFERENCES
- 1.Atherton, J. C. 1998. H. pylori virulence factors. Br. Med. Bull. 54:105-120. [DOI] [PubMed] [Google Scholar]
- 2.Atherton, J. C., T. L. Cover, E. Papini, and J. L. Telford. 2001. Vacuolating cytotoxin, p. 97-110. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C.
- 3.Copass, M., G. Grandi, and R. Rappuoli. 1997. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect. Immun 65:1949-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cover, T. L., D. E. Berg, M. J. Blaser, and H. L. T. Mobley. 2001. H. pylori pathogenesis, p. 510-558. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, San Diego, Calif.
- 5.Cover, T. L., and M. J. Blaser. 1992. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 267:10570-10575. [PubMed] [Google Scholar]
- 6.Cover, T. L., P. I. Hanson, and J. E. Heuser. 1997. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J. Cell Biol. 138:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cover, T. L., W. Puryear, G. I. Pérez-Pérez, and M. J. Blaser. 1991. Effect of urease on HeLa cell vacuolation induced by Helicobacter pylori cytotoxin. Infect. Immun. 59:1264-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover, T. L., M. K. R. Tummuru, P. Cao, S. A. Thompson, and M. J. Blaser. 1994. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 269:10566-10573. [PubMed] [Google Scholar]
- 9.de Bernard, M., B. Arico, E. Papini, R. Rizzuto, G. Grandi, R. Rappuoli, and C. Montecucco. 1997. Helicobacter pylori toxin VacA induces vacuole formation by acting in the cell cytosol. Mol. Microbiol. 26:665-674. [DOI] [PubMed] [Google Scholar]
- 10.de Bernard, M., D. Burroni, E. Papini, R. Rappuoli, J. Telford, and C. Montecucco. 1998. Identification of the Helicobacter pylori VacA toxin domain active in the cell cytosol. Infect. Immun. 66:6014-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bernard, M., E. Papini, V. de Filippis, E. Gottardi, J. Telford, R. Manetti, A. Fontana, R. Rappuoli, and C. Montecucco. 1995. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. J. Biol. Chem. 270:23937-23940. [DOI] [PubMed] [Google Scholar]
- 12.Galmiche, A., J. Rassow, A. Doye, S. Cagnol, J. C. Chambard, S. Contamin, V. de Thillot, I. Just, V. Ricci, E. Solcia, E. Van Obberghen, and P. Boquet. 2000. The N-terminal 34 kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J. 19:6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallet, B., D. J. Sherratt, and F. Hayes. 1997. Pentapeptide scanning mutagenesis: random insertion of a variable five amino acid cassette in a target protein. Nucleic Acids Res. 25:1866-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes, F., and B. Hallet. 2000. Pentapeptide scanning mutagenesis: encouraging old proteins to execute unusual tricks. Trends Microbiol. 8:571-577. [DOI] [PubMed] [Google Scholar]
- 15.Kuck, D., B. Kolmerer, C. Iking-Konert, P. H. Krammer, W. Stremmel, and J. Rudi. 2001. Vacuolating cytotoxin of Helicobacter pylori induces apoptosis in the human gastric epithelial cell line AGS. Infect. Immun. 69:5080-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leunk, R. D., P. T. Johnson, B. C. David, W. G. Kraft, and D. R. Morgan. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 26:93-99. [DOI] [PubMed] [Google Scholar]
- 17.Manetti, R., P. Massari, D. Burroni, M. de Bernard, A. Marchini, R. Olivieri, E. Papini, C. Montecucco, R. Rappuoli, and J. L. Telford. 1995. Helicobacter pylori cytotoxin: importance of native conformation for induction of neutralizing antibodies. Infect. Immun. 63:4476-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClain, M. S., P. Cao, H. Iwamoto, A. D. Vinion-Dubiel, G. Szabo, Z. Shao, and T. L. Cover. 2001. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J. Bacteriol. 183:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClain, M. S., W. Schraw, V. Ricci, P. Boquet, and T. L. Cover. 2000. Acid-activation of Helicobacter pylori vacuolating cytotoxin (VacA) results in toxin internalization by eukaryotic cells. Mol. Microbiol. 37:433-442. [DOI] [PubMed] [Google Scholar]
- 20.Montecucco, C., E. Papini, M. de Bernard, J. L. Telford, and R. Rappuoli. 1999. Helicobacter pylori vacuolating cytotoxin and associated pathogenic factors, p. 264-286. In J. E. Alouf and J. H. Freer (ed.), The comprehensive sourcebook of bacterial protein toxins. Academic Press, London, United Kingdom.
- 21.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell. Biol. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, V. Q., R. M. Caprioli, and T. L. Cover. 2001. Carboxy-terminal proteolytic processing of Helicobacter pylori vacuolating toxin. Infect. Immun. 69:543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papini, E., M. Zoratti, and T. L. Cover. 2001. In search of the Helicobacter pylori VacA mechanism of action. Toxicon 39:1757-1767. [DOI] [PubMed] [Google Scholar]
- 24.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 25.Vinion-Dubiel, A. D., M. S. McClain, D. M. Czajkowsky, H. Iwamoto, D. Ye, P. Cao, W. Schraw, G. Szabo, S. R. Blanke, Z. Shao, and T. L. Cover. 1999. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. J. Biol. Chem. 274:37736-37742. [DOI] [PubMed] [Google Scholar]
- 26.Wang, H. J., and W. C. Wang. 2000. Expression and binding analysis of GST-vacA fusions reveals that the C-terminal approximately 100-residue segment of exotoxin is crucial for binding in HeLa cells. Biochem. Biophys. Res. Commun. 278:449-454. [DOI] [PubMed] [Google Scholar]
- 27.Wang, W. C., H. J. Wang, and C. H. Kuo. 2001. Two distinctive cell binding patterns by vacuolating toxin fused with glutathione S-transferase: one high-affinity m1-specific binding and the other lower-affinity binding for variant m forms. Biochemistry 40:11887-11896. [DOI] [PubMed] [Google Scholar]
- 28.Ye, D., and S. R. Blanke. 2002. Functional complementation reveals the importance of intermolecular monomer interactions for Helicobacter pylori VacA vacuolating activity. Mol. Microbiol 43:1243-1253. [DOI] [PubMed] [Google Scholar]
- 29.Ye, D., and S. R. Blanke. 2000. Mutational analysis of the Helicobacter pylori vacuolating toxin amino terminus: identification of amino acids essential for cellular vacuolation. Infect. Immun 68:4354-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye, D., D. C. Willhite, and S. R. Blanke. 1999. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. J. Biol. Chem. 274:9277-9282. [DOI] [PubMed] [Google Scholar]