Abstract
Moraxella catarrhalis is a strict human pathogen and a significant cause of respiratory disease and otitis media. In direct response to these infections, research efforts have focused primarily on the identification of potential vaccine targets. The general biology of M. catarrhalis, however, including the mechanisms utilized to survive in the human host, remains poorly understood. Previous work has demonstrated that M. catarrhalis expresses iron-repressible proteins, suggesting the presence of iron acquisition systems under the control of a ferric uptake regulator (Fur). In this study M. catarrhalis fur has been cloned and sequenced from strain 7169. A deletion-insertion mutation of 7169 fur resulted in upregulation of iron-repressible outer membrane proteins in the absence and presence of iron. This mutant strain, 7169fur1, was significantly more sensitive to the bactericidal activity of normal human serum than the resistant wild-type strain. These data suggest that constitutive expression of iron-regulated proteins may provide multiple targets for human antibodies. In addition, the 7169 fur mutant provides an important tool for further investigation of the iron acquisition mechanisms utilized by M. catarrhalis.
Moraxella catarrhalis is a gram-negative diplococcus that has emerged as the third leading bacterial cause of acute otitis media (AOM) in young children (1). This organism, frequently identified as a commensal of the upper and lower respiratory tracts of healthy individuals, is also responsible for sinusitis, pulmonary infections in adults with underlying lung disease (chronic obstructive pulmonary disease), nosocomial disease, and a variety of severe infections in immunocompromised hosts (9, 13, 17, 24). An estimated 24 million cases of AOM occur each year in American children under the age of 2 years (23). Otitis media is the leading cause of pediatric office visits, resulting in substantial direct and indirect costs for diagnosis and treatment (17).
AOM often recurs despite antibiotic treatment, resulting in substantial morbidity and an increased rate of learning disabilities that are recognized when children enter the formal schooling environment (23). In addition, antibiotics have been overprescribed, and as a result more than 90% of M. catarrhalis clinical isolates are β-lactamase positive (4). Due to the limited success of antibiotic treatment of this disease and the significant burden on the health care system, there is a compelling need for more research efforts aimed at identifying effective vaccine antigens against M. catarrhalis infections (4).
Recent studies have focused on identifying and evaluating potential M. catarrhalis outer membrane components as possible vaccine antigens. These include outer membrane proteins (OMPs) C, D, and E, M. catarrhalis outer membrane protein B (CopB), ubiquitous surface proteins A1 and A2 (UspA1 and UspA2), lipooligosaccharide (LOS), transferrin binding protein B (TbpB), and lactoferrin binding protein B (LbpB) (1, 11, 14, 18). Although these components show promise, their intermittent expression, the variability in epitopes, the risk of phase variation, and serotype heterogeneity clearly limit the use of these structures as effective antigens capable of inducing immunologic memory (14).
In the present studies, the ferric uptake regulator (fur) gene from M. catarrhalis strain 7169 was identified and cloned. Recombinant 7169 Fur was capable of complementing an E. coli fur-deficient mutant, demonstrating that M. catarrhalis Fur was expressed as a functional protein in this species. An isogenic mutant was constructed by allelic exchange, and the resulting fur mutant, 7169fur1, exhibited constitutive expression of iron-repressible proteins. More importantly, this construct was significantly more sensitive to the bactericidal activity of normal human serum (NHS) than the wild-type strain. These data suggest that 7169fur1 expression of iron-repressible proteins may provide more effective targets for human antibodies.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. catarrhalis 7169, a middle-ear isolate from a child with otitis media, was used to construct the fur homologue-deficient mutant 7169fur1 (11). Culture conditions for analyzing M. catarrhalis iron requirements by using Chelex-treated chemically defined medium (CDM) have been described previously (2, 11, 12). M. catarrhalis strains were cultured routinely at 35°C in 5% CO2 on brain heart infusion (BHI) agar plates or at 37°C with rotary shaking at 225 rpm in liquid BHI medium. The mutant strain 7169fur1 was cultured in the presence of 20 μg of kanamycin per ml. Escherichia coli XL1-Blue, E. coli H1780 (fiu::λplacMu53 and inactivated fur), and E. coli MC4100 (parent strain of H1780, obtained from the E. coli Genetic Stock Center at Yale University) were cultured on Luria-Bertani (LB) agar plates or in liquid LB medium in the presence of the appropriate antibiotic (ampicillin [100 μg/ml] and/or kanamycin [20 μg/ml]) under the conditions described above.
General DNA manipulations.
Restriction endonucleases, T4 ligase, and all other standard molecular biology reagents were purchased from New England Biolabs, Inc. (Beverly, Mass.), or Promega (Madison, Wis.) and were used according to standard methods. M. catarrhalis chromosomal DNA was isolated as described previously (20). Platinum Taq High Fidelity Polymerase, used for PCR analysis, was obtained from Invitrogen Life Technologies Corp. (Carlsbad, Calif.). PCR products and plasmid DNA samples were purified by using the MinElute kit and the QIAprep spin kit, respectively (Qiagen, Santa Clarita, Calif.). PCR amplifications were performed using genomic M. catarrhalis 7169 DNA for 25 cycles at annealing temperatures specific for the primer pairs utilized (described below). Automated DNA sequencing was performed (at the RPCI Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, N.Y.) and analyzed (with MacVector, version 6.5.3; Genetics Computer Group, Madison, Wis.) for all constructs.
Cloning and mutagenesis of M. catarrhalis 7169 fur.
PCR primers for cloning the 7169 fur homologue were designed based on the M. catarrhalis Fur homologue sequence (located in the National Center for Biotechnology Information [NCBI] data bank under Incyte Genomics sequence 38, patent WO0078968, accession no. AX067463) found through BLAST searches with other Fur homologues. The 7169 fur open reading frame (ORF), plus 92 nucleotides upstream of the putative 5′ transcription start site and 96 nucleotides downstream of the 3′ stop site, was amplified using primers 391 (5′-GTGATGATTTTGGGGTGC-3′) (sense) and 375 (5′-GGATTTTTTGACCTTTGGGTGG-3′) (antisense). The resulting 629-nucleotide PCR product was ligated into pGEM-T Easy (TA-cloning vector; Promega), resulting in pFUR-3KF. PCR analysis, restriction digestion, and sequence analysis were performed using this plasmid to confirm the cloning of this gene and to determine the nucleotide organization of the 7169 fur gene.
An inverse PCR strategy was employed to construct a deletion-insertion isogenic mutant of 7169 fur by using pFUR-3KF as the template and primers 415 (5′-TATAGGATCCTTGAGATTGTCCAAGATGAAC-3′) (sense) and 416 (5′-CGCGAGATCTAAAACACGATACACCGTTG-3′) (antisense), with engineered BamHI and BglII sites, respectively (underlined). This resulted in a deletion of 62 nucleotides internal to the 7169 fur coding region. The nonpolar kanamycin resistance cassette aphA-3 was amplified from pUC18K by using primers 417 (5′-TATAAGATCTGGGTGACTAACTAGGAGGAATAAATGGCTA-3′) (sense) and 342 (5′-GCGGGATCCGTCGACTCTAGAGGATCCCCGGGTCATTA-3′) (antisense), with corresponding engineered restriction sites for directional cloning (15). PCR products obtained from the reactions described were subjected to restriction digestion and ligation, resulting in pFURK-3. Sequence analysis was performed to confirm the proper insertion of the cassette.
Primers 391 and 375 were used to amplify a 1.4-kb fragment from pFURK-3 (fur plus aphA-3) which was used to naturally transform wild-type M. catarrhalis 7169. In brief, a 100-μl aliquot of an early-log-phase 7169 culture (optical density at 600 nm, 0.2) was plated onto BHI agar, and 20 ng of the purified amplicon DNA was spotted onto a portion of the bacterial lawn. After incubation for 5 h under standard growth conditions, the area of the bacterial lawn that had been inoculated with the mutagenesis construct was swabbed onto selective plates containing kanamycin. Chromosomal DNA from 7169fur1 was isolated and subjected to PCR analysis (with primers 391 and 375) as well as sequence analysis to confirm the integration of the inactivated fur into the genome.
β-Galactosidase enzyme assay.
E. coli H1780 (fur inactivated), H1780 plus pFUR-3KF, and H1780 plus pFURK-3 were analyzed for β-galactosidase activity essentially as described previously, with the exception that isopropyl-β-d-thiogalactopyranoside (IPTG) was not used. This assay was performed in triplicate for each sample (6, 16).
Siderophore production in E. coli.
Chrome azurol S (CAS) agar plates were prepared according to the method of Schwyn and Neilands and were used to detect siderophore production in E. coli MC4100, H1780, H1780 plus pFUR-3KF, and H1780 plus pFURK-3 (21). This is a well-defined colorimetric assay for E. coli based on the affinity of siderophores for Fe(III) (21). Data shown are results of three separate experiments.
OMP analysis.
M. catarrhalis strains 7169 and 7169fur1 were cultured under chemically defined iron-stressed (CDM 0) and iron-replete (CDM 100) conditions. After 9 h, cultures were harvested and OMPs were prepared. OMP preparations were isolated by Zwittergent extraction and analyzed by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-7.5% PAGE). Western blotting with monoclonal antibody 11C6 was performed as described previously (12).
Bactericidal assays.
A 200-μl microscale serum bactericidal assay was performed in a 24-well plate essentially as described previously with the following modifications (25). Briefly, NHS (pooled from 10 healthy adult donors) was diluted to 10 and 25% in phosphate-buffered salt solution (PBSS). Bacteria (10 μl containing 106 cells) were inoculated either into 190-μl reaction wells containing the diluted NHS or into control wells containing heat-inactivated (HI) NHS or PBSS alone and were incubated at 37°C with shaking for 30 min. Serial dilutions (1:10) of each reaction well were plated onto BHI agar, and following a 24-h incubation, the resulting colonies were counted. Percent survival was calculated by comparing the number of CFU after the 30-min incubation in serum with the CFU of the inoculum incubated in PBSS alone. Results presented are from three independent assays performed on separate days.
Nucleotide sequence accession number.
The nucleotide sequence of M. catarrhalis 7169 fur has been deposited in GenBank under accession no. AY172972.
RESULTS AND DISCUSSION
Identification, cloning, and analysis of M. catarrhalis 7169 fur.
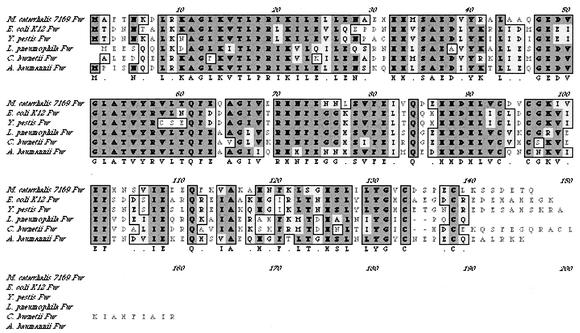
The putative M. catarrhalis fur gene was identified by BLAST searches of the patented M. catarrhalis genome through the NCBI. The M. catarrhalis 7169 fur homologue was cloned as described in Materials and Methods, resulting in pFUR-3KF. Nucleotide sequence analysis indicated that the cloned DNA fragment contained a single ORF of 441 bp with a predicted gene product of 146 amino acids. Database searches with the deduced polypeptide sequence revealed high similarities to other Fur proteins (70 to 84%). A ClustalW alignment of known Fur proteins is shown in Fig. 1. There is also a potential stem-loop structure encoded in the 3′ untranslated region of 7169 fur. This phenomenon has also been reported for the fur homologue of Acinetobacter baumannii (5). Analysis of sequence upstream of fur identified a putative ORF predicted to be an omlA homologue, as is seen for a number of other bacterial species (10).
FIG. 1.
ClustalW alignment of the amino acid sequence of M. catarrhalis 7169 Fur with Fur proteins from other gram-negative species. Dark shading, identical amino acids; light shading, similar amino acids.
Expression of 7169 fur in E. coli and complementation of an E. coli fur mutant.
In order to determine if the cloned 7169 fur encoded a functional protein, pFUR-3KF was used to transform the E. coli fur mutant H1780 by electroporation (6). H1780 was engineered to be fur deficient and to include the Fur-regulated gene fiu fused to a promoterless lacZ gene. This reporter gene, fiu-lacZ, cannot be repressed in this strain due to the fur mutation, and therefore the gene encoding the enzyme β-galactosidase is constitutively expressed (6). A deletion-insertion mutation of 7169 fur, pFURK-3, was also constructed and used to transform H1780 as a negative control.
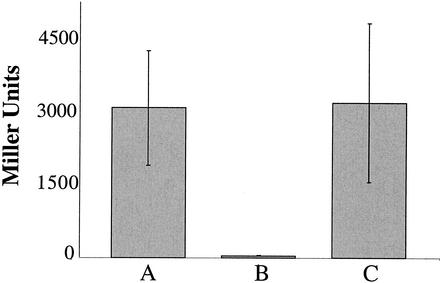
All strains evaluated for β-galactosidase activity were grown in BHI liquid medium (naturally iron replete), since iron is required to ensure that Fur is functional (19). In these studies, H1780, H1780 plus pFUR-3KF, and H1780 plus pFURK-3 were compared. The results presented in Fig. 2 clearly demonstrate that the recombinant 7169 Fur is in fact expressed in a functional form in E. coli. High β-galactosidase activity was observed for E. coli H1780 (Fig. 2, bar A). Complementation of H1780 with pFUR-3KF rescued the Fur defect of this strain and resulted in the repression of transcription of the fiu-lacZ reporter gene, as shown by the sharp decrease in detectable β-galactosidase activity (Fig. 2, bar B). When 7169 pFURK-3, containing the disrupted fur gene, was transformed into the E. coli mutant, β-galactosidase activity was restored as predicted (Fig. 2, bar C).
FIG. 2.
β-Galactosidase enzyme activity assay. The three bacterial strains were used to test for repression of transcription of the fiu-lacZ reporter gene. (A) H1780 (fur inactive); (B) H1780 plus pFUR-3KF (with the fur defect complemented); (C) H1780 plus pFURK-3 (7169 fur disrupted). Means from three independent experiments are shown. Error bars, standard deviations.
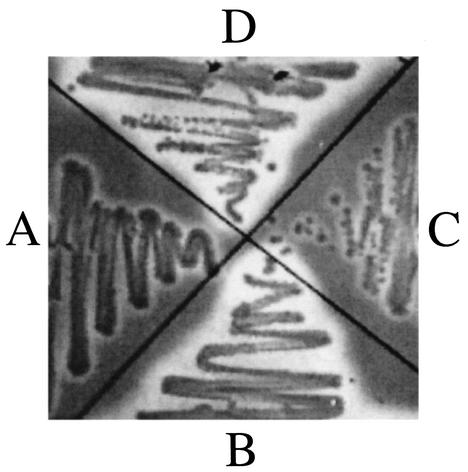
An E. coli system for Fur analysis was utilized as a second method to confirm that the cloned 7169 fur encodes a functional protein. Under iron-replete conditions, Fur traditionally acts as a transcriptional repressor of genes encoding iron acquisition system components, including those controlling siderophore production. Based on this, recombinant 7169 Fur was evaluated for the ability to repress the constitutive siderophore production in H1780 by using the CAS blue agar plate assay described by Schwyn and Nielands (21). Siderophore production is detected by the release of free dye from the iron-dye complex, resulting in a color change from blue to orange (appearing as halos in a black-and-white reproduction). E. coli strain MC4100 was used as a negative control (Fig. 3, quadrant A), and H1780 was used as a positive control (Fig. 3, quadrant B), for this experiment. H1780 complemented with pFUR-3KF resulted in repression of siderophore production (Fig. 3, quadrant C). In contrast, H1780 complemented with the disrupted fur construct, pFURK-3, continued siderophore production at a level comparable to that of H1780 alone (Fig. 3, quadrant D). These data confirm that the 7169 Fur is expressed and functional in E. coli. In addition, these data suggest that 7169 fur is important in iron-regulated pathways utilized by M. catarrhalis.
FIG. 3.
CAS plate assay used for the detection of siderophore production. (A) E. coli strain MC4100 (parental strain); (B) E. coli strain H1780; (C) H1780 plus pFUR-3KF; (D) H1780 plus pFURK-3. The large halos in quadrants B and D represent siderophore production. This is not apparent in quadrants A and C, although there are minor halos that could be the result of heavy bacterial growth or low-level siderophore production.
Construction of an isogenic 7169 fur mutant, 7169fur1.
To begin to determine the role of Fur in M. catarrhalis 7169, an isogenic fur mutant was constructed by using inverse PCR combined with insertional inactivation. Briefly, the 850-bp nonpolar kanamycin resistance cassette (aphA-3) from pUC18K was inserted into a 62-bp internal deletion within the 7169 fur ORF by using pFUR-3KF as a template, as described in Materials and Methods. This construct, termed pFURK-3, was used as a negative control in the complementation studies described above. PCR was used to amplify this entire insert containing the insertionally inactivated fur gene, and the resulting 1.4-kb amplicon was used to naturally transform 7169. PCR and sequence analysis of chromosomal DNA from several kanamycin-resistant transformants demonstrated that proper allelic exchange had occurred, and one of the resulting isogenic fur mutants, termed 7169fur1, was selected for further evaluation.
Growth analysis and OMP profile of 7169fur1.
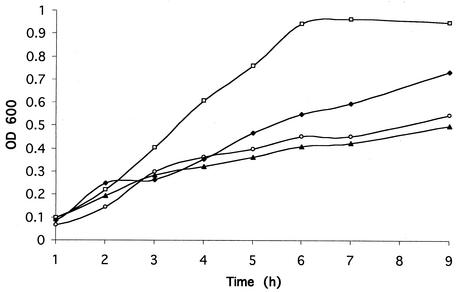
In order to initially investigate the effect of an inactive fur gene on 7169, growth analyses were performed (Fig. 4). These data show that under the iron-limiting conditions of CDM 0, wild-type 7169 and 7169fur1 showed similar growth patterns. In contrast, under iron-replete conditions using CDM 100, 7169fur1 showed a significant lag in growth compared to wild-type 7169. We hypothesize that this growth deficiency is the direct result of the inability of 7169fur1 to regulate iron acquisition systems, resulting in toxic levels of iron within these organisms (3). These data are consistent with previous studies describing fur mutations in other bacterial species (7, 22).
FIG. 4.
Growth analysis of 7169 and 7169fur1 in CDM 0 and CDM 100. □, 7169 in CDM 100; ⧫, 7169fur1 in CDM 100; ○, 7169 in CDM 0; ▴, 7169fur1 in CDM 0. Plot shown is representative of all growth analyses performed with time points ranging from 1 to 9 h.
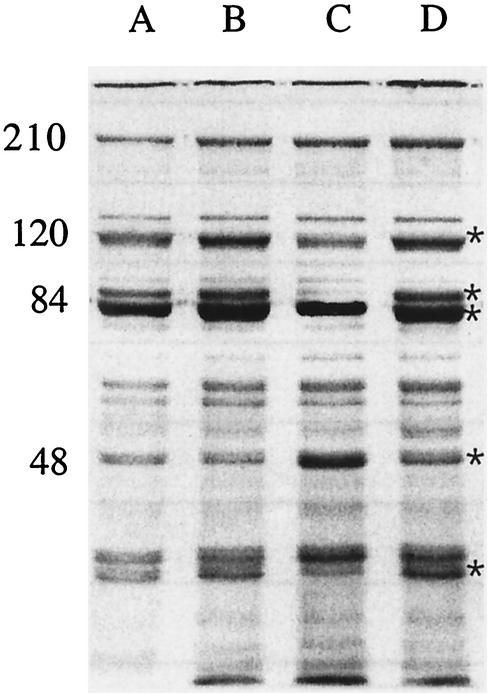
After 9 hours of growth in CDM 0 and CDM 100, cultures were harvested and OMP profiles were analyzed as described. Both 7169 and 7169fur1, grown without iron, revealed iron-stressed phenotypes (Fig. 5, lanes A and B). However, in the presence of iron, 7169fur1 retained an iron-stressed phenotype, whereas 7169 showed repression of iron-regulated protein expression (Fig. 5, lanes C and D, respectively) (2, 10).
FIG. 5.
SDS-7.5% PAGE analysis of OMPs purified from 7169 and 7169fur1. Lanes: A, 7169 grown in CDM 0; B, 7169fur1 grown in CDM 0; C, 7169 grown in CDM 100; D, 7169fur1 grown in CDM 100. Molecular weights (in thousands) are shown. Asterisks indicate iron stress proteins for M. catarrhalis as previously described (2, 10).
7169fur1 is highly susceptible to the bactericidal action of NHS.
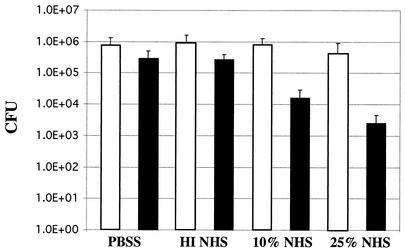
Previous studies have suggested that iron-regulated OMPs from various bacterial species have potential as vaccine antigens (9, 12, 14). While the data demonstrate that these proteins may be useful targets under iron-limited conditions, it is likely that these proteins will not be expressed once these bacteria acquire iron. In the latter scenario, those iron-regulated proteins would no longer be available for interaction with elements of the immune system whether in vitro or in vivo. The data presented above demonstrate that 7169fur1 expresses known iron-repressible target OMPs under both iron-limited and iron-replete conditions. Comparative studies were performed to determine if 7169fur1 was more sensitive than the parental strain 7169 to the bactericidal activity of NHS. In this analysis, comparable inocula of 7169 and 7169fur1 were incubated for 30 min in 10 and 25% pooled NHS. The controls included incubation with HI NHS and with buffer alone. 7169 demonstrated resistance to pooled NHS, which is consistent with the findings of previous studies (Fig. 6) (12). In contrast, 7169fur1 exhibited a dramatic 2-log10-unit decrease over the 30-min incubation period at both serum concentrations (Fig. 6). This represents a >99% rate of killing, suggesting that the unregulated expression of outer membrane iron-repressible proteins in 7169fur1 leads to a highly serum sensitive phenotype.
FIG. 6.
Susceptibilities of 7169 (open bars) and 7169fur1 (solid bars) to the bactericidal activity of NHS. 7169 was compared to 7169fur1 in 10 and 25% NHS. Negative controls included buffer alone (PBSS) and HI NHS (50% pooled NHS inactivated by incubation at 56°C for 30 min). Means from three separate experiments are shown. Error bars, standard deviations. By use of Student's t test, the differences between results for 7169fur1 in 10 and 25% NHS and results for the wild type are statistically significant (P < 0.001).
This is an extremely interesting observation, and these are the first data comparing the serum sensitivity of a fur mutant with that of the respective wild-type strain. Although the mechanism(s) involved in this bactericidal activity is currently undefined, there are various possible explanations. One possibility is that under the iron-replete conditions used for these studies, it is likely that 7169 exhibited limited to no expression of iron-repressible proteins whereas the unregulated 7169fur1 strain continued to express these proteins. This suggests that antibodies directed to these iron-regulated proteins may be involved in this mechanism of killing.
To date, most studies involving the analysis of fur from various bacteria have centered around the overall impact of iron acquisition. It is well established for other organisms that Fur, in the presence of ferrous iron, can bind to response elements in the promoter regions of iron-regulated genes, subsequently repressing their transcription (19). The studies presented above demonstrate that 7169 Fur complements an E. coli fur mutant and thus functions in a manner similar to that described for other organisms. Although various M. catarrhalis iron-repressible proteins have been described, a comprehensive analysis of the mechanisms of iron acquisition utilized by this pathogen remains elusive. The M. catarrhalis 7169 fur mutant strain will be indispensable for future studies aimed at addressing this important biologic issue.
Previous studies have demonstrated that antibodies to surface-exposed iron-regulated proteins of M. catarrhalis, i.e., TbpB, LbpB, and CopB, have been detected in human serum (9, 13, 14, 24). These proteins, although immunogenic, are variably repressed in the presence of iron. In addition, there have been multiple studies investigating the vaccine potential of TbpB for the pathogenic Neisseriaceae (8). Strain 7169fur1 is clearly more serum sensitive than the wild-type strain, yet despite the encouraging data to date, Fur mutants have not been investigated as vaccine antigens.
In general, the surface-exposed nature of these outer membrane receptors, their likely expression in vivo, and their ability to elicit functional antibodies are favorable characteristics of effective vaccine candidates. However, these proteins can be highly heterogeneous in nature, and their expression on the bacterial surface can be rapidly repressed. A further complication involves the procedures used to isolate and purify these individual proteins, as these processes can destroy conformational epitopes and thus result in induction of an ineffective immune response.
Recent studies involving vaccine antigens have shown that multiple bacterial components are preferred based on the ability of the bacteria to vary surface components in response to immunologic pressures. Thus, single, isolated proteins may not be ideal candidates, as the generation of a polyclonal response against multiple surface antigens has the potential to be more effective. The 7169fur1 construct, described in this study, constitutively expresses iron-regulated proteins on the surface of the bacterium, and these proteins are likely in their native confirmation. This characteristic circumvents the problems associated with iron-repressible protein expression and the potential loss of conformational epitopes during protein purification procedures. Another important aspect of this isogenic fur mutant is the fact that this construct appears to maintain expression of other potential vaccine antigens including LOS, OMPs C, D, and E, and UspA1 and -A2. Despite these favorable characteristics, far more detailed studies are warranted due to concerns regarding the safety of vaccines. Future analyses will not only include studies on iron-acquisition but will focus on evaluating the ability of 7169fur1 to elicit cross-reactive antibodies to surface components that are protective against other strains of M. catarrhalis.
Acknowledgments
We thank Klaus Hantke and Erin R. Murphy for providing us with E. coli strain H1780, Eric J. Hansen for his kind advice on cloning the 7169 fur gene, Alan Lesse for assistance with statistical analyses, and Nicole R. Luke for helpful suggestions while reviewing the manuscript.
This research was supported by grant AI46469 from the National Institutes of Health (NIAID) to A.A.C.
Editor: D. L. Burns
REFERENCES
- 1.Aebi, C., L. D. Cope, J. L. Latimer, S. E. Thomas, C. A. Slaughter, G. H. McCracken, Jr., and E. J. Hansen. 1998. Mapping of a protective epitope of the CopB outer membrane protein of Moraxella catarrhalis. Infect. Immun. 66:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campagnari, A. A., K. L. Shanks, and D. W. Dyer. 1994. Growth of Moraxella catarrhalis with human transferrin and lactoferrin: expression of iron-repressible proteins without siderophore production. Infect. Immun. 62:4909-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamnongpol, S., W. Dodson, M. J. Cromie, Z. L. Harris, and E. A. Groisman. 2002. Fe(III)-mediated cellular toxicity. Mol. Microbiol. 45:711-719. [DOI] [PubMed] [Google Scholar]
- 4.Dagan, R. 2000. Treatment of acute otitis media—challenges in the era of antibiotic resistance. Vaccine 19(Suppl. 1):S9-S16. [DOI] [PubMed] [Google Scholar]
- 5.Daniel, C., S. Haentjens, M. Bissinger, and R. J. Courcol. 1999. Characterization of the Acinetobacter baumannii Fur regulator: cloning and sequencing of the fur homolog gene. FEMS Microbiol. Lett. 170:199-209. [DOI] [PubMed] [Google Scholar]
- 6.Hantke, K. 1987. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol. Gen. Genet. 210:135-139. [DOI] [PubMed] [Google Scholar]
- 7.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jodar, L., I. M. Feavers, D. Salisbury, and D. M. Granoff. 2002. Development of vaccines against meningococcal disease. Lancet 359:1499-1508. [DOI] [PubMed] [Google Scholar]
- 9.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 10.Lowe, C. A., A. H. Asghar, G. Shalom, J. G. Shaw, and M. S. Thomas. 2001. The Burkholderia cepacia fur gene: co-localization with omlA and absence of regulation by iron. Microbiology 147:1303-1314. [DOI] [PubMed] [Google Scholar]
- 11.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 67:5815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luke, N. R., T. A. Russo, N. Luther, and A. A. Campagnari. 1999. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane protein B1 defined by monoclonal antibody 11C6. Infect. Immun. 67:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMichael, J. C. 2000. Progress toward the development of a vaccine to prevent Moraxella (Branhamella) catarrhalis infections. Microbes Infect. 2:561-568. [DOI] [PubMed] [Google Scholar]
- 14.McMichael, J. C. 2000. Vaccines for Moraxella catarrhalis. Vaccine 19(Suppl. 1):S101-S107. [DOI] [PubMed] [Google Scholar]
- 15.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy, E. R., A. Dickenson, K. T. Militello, and T. D. Connell. 1999. Genetic characterization of wild-type and mutant fur genes of Bordetella avium. Infect. Immun. 67:3160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers, L. E., Y. P. Yang, R. P. Du, Q. Wang, R. E. Harkness, A. B. Schryvers, M. H. Klein, and S. M. Loosmore. 1998. The transferrin binding protein B of Moraxella catarrhalis elicits bactericidal antibodies and is a potential vaccine antigen. Infect. Immun. 66:4183-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 20.Russo, T. A., J. E. Guenther, S. Wenderoth, and M. M. Frank. 1993. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol. Microbiol. 9:357-364. [DOI] [PubMed] [Google Scholar]
- 21.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 22.Staggs, T. M., J. D. Fetherston, and R. D. Perry. 1994. Pleitropic effects of a Yersinia pestis fur mutation. J. Bacteriol. 176:7614-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teele, D. W., J. O. Klein, B. M. Word, B. A. Rosner, S. Starobin, R. Earle, Jr., C. S. Ertel, G. Fisch, R. Michales, R. Heppen, and N. P. Strause. 2001. Antimicrobial prophylaxis for infants at risk for recurrent acute otitis media. Vaccine 19(Suppl. 1):S140-S143. [DOI] [PubMed] [Google Scholar]
- 24.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaleski, A., N. K. Scheffler, P. Densen, F. K. Lee, A. A. Campagnari, B. W. Gibson, and M. A. Apicella. 2000. Lipooligosaccharide P(k) (Galα1-4Galβ1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect. Immun. 68:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]