Abstract
Adeno-associated virus type 2 (AAV-2) establishes latency by site-specific integration into a unique locus on human chromosome 19, called AAVS1. During the development of a sensitive real-time PCR assay for site-specific integration, AAV-AAVS1 junctions were reproducibly detected in highly purified AAV wild-type and recombinant AAV vector stocks. A series of controls documented that the junctions were packaged in AAV capsids and were newly generated during a single round of AAV production. Cloned junctions displayed variable AAV sequences fused to AAVS1. These data suggest that packaged junctions represent footprints of AAV integration during productive infection. Apparently, AAV latency established by site-specific integration and the helper virus-dependent, productive AAV cycle are more closely related than previously thought.
Adeno-associated virus (AAV) has evolved a biphasic life cycle to ensure persistence in its primate host. It needs an unrelated helper virus, adenovirus (Ad), or herpesvirus for productive infection (16). In the absence of a helper, AAV establishes latency by preferential integration into a specific site on human chromosome 19 (chr.19), 19q13.3q-ter, called AAVS1 (10). AAV type 2 (AAV-2) contains a linear single-stranded DNA genome of 4.7 kb that covers the genes rep and cap, flanked by 145-bp inverted terminal repeats (ITRs) (27) that serve as origins of replication and as cis elements for chromosomal integration. The AAV regulatory proteins Rep78 and/or Rep68 (Rep78/68) are necessary for AAV DNA replication both in vivo and in vitro. Rep78/68 binds to the Rep-binding site (RBS) on the AAV-ITR (26) and nicks and unwinds the ITR at the terminal resolution site (trs) (7). Rep78/68 also mediates site-specific AAV integration at AAVS1 on chr.19 that comprises DNA sequences homologous to the RBS and the trs of the AAV-ITR (9, 23). In vitro studies showed ternary complex formation of Rep68 with the AAV-ITR and AAVS1 (31) and Rep68-mediated initiation of DNA replication starting from an origin sequence comprised of AAVS1 (29). A 33-bp sequence spanning the chr.19 RBS, a spacer sequence, and a trs homology element are sufficient to mediate site-specific AAV integration in vivo (12, 14). chr.19 integration sites have been analyzed either in selected cell clones or in 293 cells carrying AAVS1 on an Epstein-Barr virus-based episome (1, 2, 9, 17, 20, 23, 28, 32). To analyze integration into the authentic chr.19 preintegration locus early after AAV-2 infection, we have recently developed a sensitive and quantitative real-time PCR assay for AAV-ITR/AAVS1 junctions. Within 4 days postinfection (p.i.) site-specific integration frequency reached 10 to 20% of unselected HeLa cells. Further analysis showed that at least 1 in 1,000 infectious AAV-2 integrated site specifically (5, 6).
MATERIALS AND METHODS
Plasmids.
The following plasmids have been described: pTAV2-0 (4), pDG (3), pAAVS1-TR (6), and psub201 (22). Plasmid psubgfpneo was generated by inserting the gfpneo cassette derived from pTR-UF5 (36) between the 190-bp, XbaI-flanked AAV-ITRs of psub201.
Preparation of highly purified AAV wild-type stocks.
Conventional AAV-2 stocks were produced and titrated as described previously (6). Cells were infected at a multiplicity of infection (MOI) of 500 unless otherwise stated. Highly purified AAV stocks were generated on pTAV2-0 transfected and Ad type 2 (Ad2)-infected HeLa cells. The corresponding AAV-negative control preparations were generated in parallel without pTAV2-0 transfection. Virus purification was done according to a protocol published previously (35). In brief, phosphate-buffered saline (PBS)-washed cells were pelleted by low-speed centrifugation, resuspended in PBS, and adjusted to 0.5% deoxycholic acid. Unencapsidated nucleic acids were degraded by treatment with 5,000 U of benzonase (Purity Grade II; Merck) for 90 min at 37°C. Cell debris was pelleted at 8,100 × g for 30 min. Supernatants were frozen, thawed, centrifuged again, and then purified on iodixanol discontinuous density gradients. The 40% fraction containing AAV was aspirated and subjected to heparin affinity chromatography. Eluted virions were dialyzed against PBS and stored at −80°C.
Preparation of highly purified rAAV stocks.
Recombinant AAV-2 (rAAV-2) stocks were prepared on 293 cells by cotransfection of pDG and psubgfpneo as described by Huser et al. (6). Highly purified rAAV was prepared according to the protocol described above for the AAV wild type.
Quantitative, real-time PCR for the determination of site-specific integration.
Targeted integration of AAV-2 into AAVS1 was detected by real-time PCR of virus-cell junctions and calculated with the aid of known concentrations of a standard plasmid, as described previously (6). For calculations of AAV-ITR/chr.19 junction copy numbers per cell, a DNA content of 10 pg per cell was assumed, reflecting the hypertriploid (3n+) karyotype of HeLa cells (13).
Quantitative, real-time PCR for the determination of virion genome copies.
Genome copy numbers of AAV-2 wild-type or rAAV vector stocks were quantified by by real-time PCR as follows. Aliquots of highly purified (benzonase-treated) virion stocks (see above) were digested with proteinase K, extracted with phenol, precipitated, and resuspended in Tris-EDTA. DNAs were purified by adsorption chromatography on QIAquick spin columns (Qiagen, Hilden, Germany). Real-time LightCycler PCR was performed with aliquots diluted to a final volume of 20 μl. The reaction mixture included polymerase (LightCycler Kit-Fast Start DNA Master SYBR Green I; Roche) and 5 mM MgCl2. For the determination of AAV-2 wild-type genomes, a 500 nM concentration of primer PAAV-F1 (GCCAACTCCATCACTAGGGG; AAV-2 nucleotides 121 to 140) and a 500 nM concentration of primer PAAV-W1 (CCCGCTTCAAAATGGAGACC; AAV-2 nucleotides 299 to 280) were used. For the determination of rAAV-gfp, a primer pair specific for the cytomegalovirus promoter, PAAV-F3 (TGGGCGTGGATAGCGGTTTG) and PAAV-B3 (AACAGCGTGGATGGCGTCTC), were used. Amplification was done at 95°C for 10 min to activate the polymerase, followed by 40 cycles at 95°C for 0 s, 60°C for 4 s, and 72°C for 15 s.
Detection of virus-cell junctions in purified AAV preparations.
Samples of the highly purified AAV stock (1 ml) or of the purified negative-control preparation (1 ml) spiked with 4 × 107 copies of pAAVS1-TR were digested for 30 min at 37°C with 250 U of benzonase as indicated above. Controls without benzonase were run in parallel. Samples were pelleted by using the S55-S rotor and the RC-M120 GX ultracentrifuge (Sorvall) at 120,000 × g and 4°C for 2 h. Each pellet was resuspended in 2 ml of PBS, centrifuged again, and resuspended in 300 μl of Tris-EDTA. Samples were adjusted to 1% sodium lauryl sarcosinate, digested with proteinase K for 2 h at 56°C, and then extracted with phenol-chloroform. DNA samples were precipitated with ethanol, purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany), and eluted with 30 μl of Tris-Cl. Then, 5-μl aliquots were used for the determination of junction copy numbers by real-time PCR as described previously (6).
Analysis of PCR products.
Aliquots of the PCR products were cloned into pCR4-TOPO (Invitrogen) and subjected to DNA sequencing as described previously (6).
RESULTS
Detection of AAV-ITR/AAVS1 junctions in highly purified AAV preparations.
The recently established real-time PCR assay for site-specific AAV-2 integration (Fig. 1) regularly detects targeted integration by 8 h p.i. (6). Repeated analyses of earlier time points led to the consistent finding that in AAV-2-infected cells (MOI, 500) harvested immediately after the 45-min adsorption period, low but specific PCR signals of AAV-ITR/AAVS1 junctions were detected, whereas mock-infected parallel cultures were negative. To exclude the possibility that the PCR signals resulted from fragmented genomic DNA possibly contaminating AAV freeze-thaw stocks, AAV stocks were purified to the high purification grade developed for rAAV gene therapy vectors as outlined in Materials and Methods (35). As seen in Table 1 no (0) junctions were detected in 1 μg of uninfected HeLa cell DNA. AAV-infected HeLa cell DNA extracted immediately after removal of the virus (0 h p.i.) gave rise to 173 junction copies that increased to 2,150 junction copies at 72 h p.i. The data are similar to the results obtained with conventional AAV preparations. Analysis of the highly purified AAV preparation alone yielded 124 junction copies per 2 × 108 virion genomes, whereas the controls remained negative (Table 1). Thus, highly purified AAV-2 preparations contain low levels of AAV-ITR/AAVS1 junction DNAs that survive a rigorous virus purification scheme. Efficient protection of the AAV-ITR/AAVS1 junctions by AAV capsids appears to be the most likely explanation for this finding.
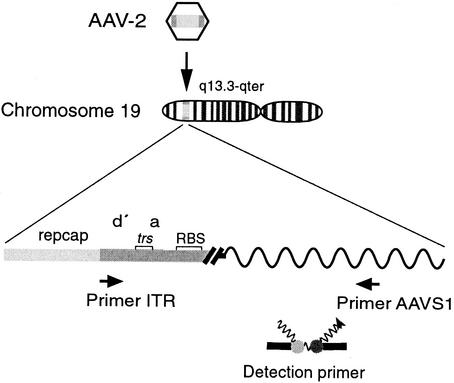
FIG. 1.
Quantitative real-time PCR for site-specific integration of AAV-2 into AAVS1 on chr.19. Shown is a schematic representation of the real-time PCR assay as described by Hüser et al. (6). Primers are indicated by arrows; primer ITR hybridizes to AAV-2 sequences at positions 4526 to 4545 (27), and primer AAVS1 hybridizes to AAVS1 on chr.19 at positions 1609 to 1593 (9). Sequence elements within the ITR are as outlined in Fig. 3B. trs, terminal resolution site. Fluorescent dye-labeled probes hybridize to the amplified DNA fragment (donor probe, nucleotides 1541 to 1560; acceptor probe, nucleotides 1562 to 1583). Close proximity of the attached dyes elicits fluorescence resonance energy transfer.
TABLE 1.
Real-time PCR analysis of AAV-ITR/AAVS1 junctions
| Sample | Amt of DNA in PCR analysis | No. of junction copiesa
|
|
|---|---|---|---|
| Per reaction (PCR raw data) | Per 109 virion genomes | ||
| Mock-infected HeLa cells | 1 μg | 0 | NA |
| AAV-2-infected cells at 0 h p.i.b | 1 μg | 173 | NA |
| AAV-2-infected cells at 72 h p.i.b | 1 μg | 2,150 | NA |
| AAV-2 mixed with HeLa cellsc | 1 μg of HeLa DNA and AAV DNA extracted from 5 × 108 virion genomes | 69 | 144 |
| AAV-2 prepn 1d | Extracted from 2 × 108 virion genomes | 124 | 729 |
| AAV-2 prepn 2d | Extracted from 1 × 109 virion genomes | 37 | 34 |
| AAV-2 prepn 3d | Extracted from 2 × 109 virion genomes | 24 | 13 |
| Plasmid pTAV2-0e | 5 × 108 copies | 0 | NA |
| Ad2-infected cells at 72 h p.i. | 1 μg | 0 | NA |
| rAAV (psubgfpneo) prepn 1f | Extracted from 2 × 1011 virion genomes | 570 | 6 |
| rAAV (psubgfpneo) prepn 2f | Extracted from 2 × 1011 virion genomes | 3,826 | 32 |
| rAAV (psubgfpneo) prepn 3f | Extracted from 1 × 109 virion genomes | 154 | 154 |
| Plasmid psubgfpneog | 6 × 1010 copies | 0 | NA |
| Plasmid pDGg | 1 × 1011 copies | 0 | NA |
Junction copy numbers per reaction were determined and quantified with the aid of standards run in parallel. LightCycler analysis was performed as represented in Fig. 2. Note that a reliable quantification of positive results is possible only for values of >200 copies per reaction. Virion genomes were determined in AAV-2 wild-type or rAAV stocks by quantitative real-time PCR as outlined in Materials and Methods. NA, not applicable.
Cells were infected with a highly purified AAV-2 stock at an MOI of 500 (4 × 108 virion genomes at a maximum). Total genomic DNA was extracted either immediately after the 45-min adsorption period (0 h p.i.) and removal of the virus or at 72 h p.i.
Cells were mixed with highly purified AAV-2 and were extracted immediately.
Total DNA was isolated from independent, highly purified AAV preparations.
A 2.5-fold excess of plasmid pTAV2-0 (AAV-2 wild type) over the estimated number of viral particles in the AAV-2 preparation 1 sample was subjected to PCR.
Equal aliquots of three separately prepared AAV vector stocks were analyzed.
Plasmids psubgfpneo and pDG were used for the production of rAAV vector stocks and served as negative controls.
To verify this assumption, the efficiency of the benzonase digestion step during AAV preparation had to be evaluated, ideally by the demonstration of efficient enzymatic degradation of an excess of added junction DNA. To address this point, a highly purified AAV-2 preparation and an AAV-negative control preparation were generated and purified step by step in parallel, as outlined in Materials and Methods. No AAV-ITR/AAVS1 junction copies were detected in the AAV-negative control preparation, whereas the purified AAV-2 preparation (AAV titer, 5 × 108 infectious units/ml) was positive. This finding excluded DNA or virus contamination during the preparation procedure. To demonstrate that an excess of added unprotected junction DNA is efficiently degraded during benzonase digestion, the AAV-negative control preparation was spiked with the plasmid pAAVS1-TR that covers a cloned AAV-ITR/AAVS1 junction (Fig. 2). After benzonase digestion (250 U for 30 min) real-time PCR detected 103 junction copies/ml, whereas the undigested sample had 6 × 106 junction copies/ml. The efficient, >5,000-fold reduction of unprotected DNA was in contrast to an only 2-fold reduction of junction copy number in the highly purified AAV preparation (Fig. 2). This minor reduction may result from junctions packaged in leaky or otherwise defective capsids. Benzonase accessibility of one primer-binding site is already sufficient to render the template undetectable by the PCR assay. In summary, the obvious explanation for the benzonase resistance of AAV-ITR/AAVS1 junctions in highly purified AAV preparations is efficient protection by packaging in AAV capsids.
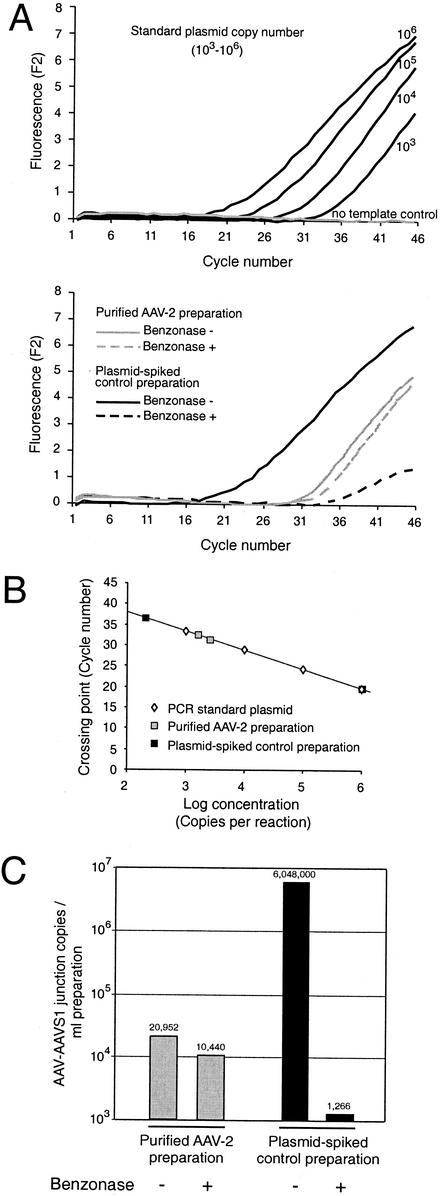
FIG. 2.
PCR analysis of AAV-AAVS1 junctions packaged in AAV capsids. A highly purified wild-type AAV-2 preparation (1 ml) and the equivalent volume of a parallel mock purification of an AAV-negative control preparation, spiked with 4 × 107 copies of pAAVS1-TR, were treated with 250 U of benzonase for 30 min as indicated. (A) Raw data of real-time PCR of AAV-ITR/AAVS1 junctions in the AAV and control preparations. Known copy numbers (103 to 106) of standard plasmid pAAVS1-TR were analyzed in parallel for the generation of a standard curve as described previously (6). Note that samples and controls, although analyzed simultaneously, are represented in separate panels to simplify the visual discrimination of the curves. (B) A standard curve was calculated from the raw data of known standards displayed in panel A. Sample concentrations were calculated with the aid of the standard curve. (C) Copy numbers of AAV-ITR/AAVS1 junctions per ml of the highly purified AAV or plasmid-spiked negative-control preparations are derived from the values given in panel B. Only one of the six samples was analyzed, so the copy numbers in panel B had to be adjusted accordingly. The values are indicated above the respective columns.
Additional AAV wild-type preparations were produced on PCR-tested HeLa or 293 cells by (i) coinfection of AAV-2 and Ad2, (ii) transfection of pTAV2-0 followed by Ad2 infection, or (iii) cotransfection of pTAV2-0 and pDG, a plasmid covering the Ad helper genes. In seven of nine virus preparations packaged AAV-ITR/AAVS1 junctions were detected (Table 1 displays the quantification for three independent preparations). Junction copy numbers per 109 virion genomes were highly variable, possibly reflecting differences in the infection kinetics. Furthermore, rAAV vector stocks were analyzed that were prepared as described in Materials and Methods. Three out of three independently generated rAAV preparations contained junctions detected by real-time PCR (Table 1). The ratios of packaged junctions compared to virion genome titers were similar to those of AAV-2 wild-type preparations. The results, taken together, show that both AAV wild-type and rAAV vector stocks regularly contain packaged AAV-ITR/AAVS1 junctions at variable copy numbers. Since virus stocks were produced by plasmid transfections with tested reagents, packaged junctions were newly generated during a single AAV production round.
DNA sequence analysis of packaged junctions.
To analyze the packaged DNA sequences, PCR products were cloned into pCR4-TOPO as described previously (6). DNAs of randomly selected clones were subjected to DNA sequence analysis (Fig. 3). AAV-ITR sequences fused to variable integration sites of AAVS1 were found. Although the PCR assay detects only the right-hand AAV-ITR (6), AAV p5 promoter sequences were detected in one clone and parts of AAV cap and the polyadenylation signal in two others. These findings point to extensive rearrangements of the entire AAV genome. Although the cloning procedure and sequence analysis were identical to those in the study of AAV site-specific integration at AAVS1 on chr.19, the DNA structures of the packaged junctions were more rearranged than the ones derived from latently infected HeLa cells after AAV infection (5, 6). We therefore favor the interpretation that the detected sequence rearrangements indicate that the integration, rescue, replication, and/or recombination events took place before packaging rather than during PCR amplification and subcloning of the junctions. Since we were able to reisolate junctions with identical rearrangements from the same AAV preparation, this notion is further underlined. Either of the rearranged sequences in Fig. 3 can be explained as an intermediate of AAV integration into AAVS1. None of the clones analyzed contained unrelated DNA sequences as described recently for DNAs cloned from rAAV-transduced cells (15).
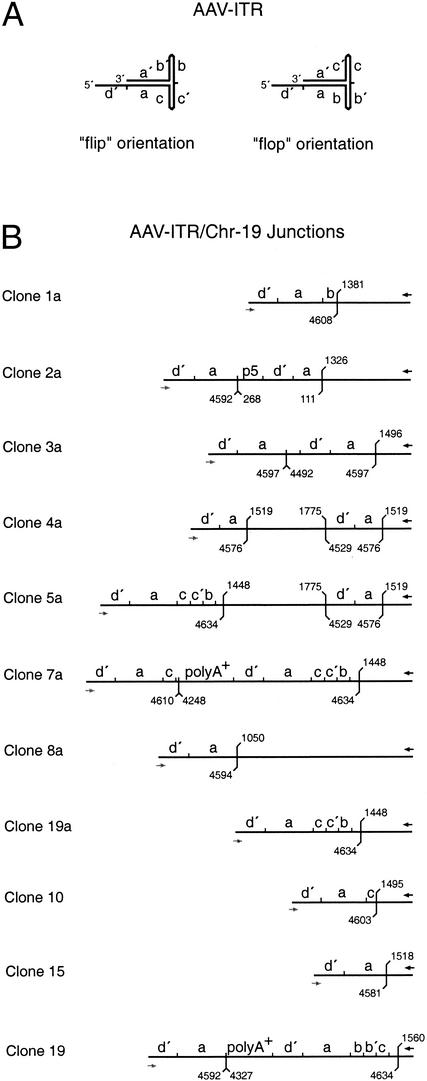
FIG. 3.
Sequence analysis of junctions packaged into AAV-2 virions. Junctions amplified from highly purified AAV-2 stocks were cloned into pCR4-TOPO, and the DNA sequence was determined. (A) Structure of the AAV-ITR sequence elements in either the “flip” or the “flop” orientation. (B) Structural maps deduced from DNA sequence analysis of cloned junctions. The hybridization sites of the primers are indicated either by black arrows (PAAVS1) or gray arrows (PITR). In some sequences a second internal primer-binding site exists. The positions of the last AAVS1-specific and of the last AAV-ITR-specific nucleotides are indicated. The numbering is according to published DNA sequences (9, 27). Within the AAV-ITR small letters (d′, a, b, c, and c′) indicate palindromic sequence elements. p5, AAV p5 promoter sequence; polyA+, 90 to 150 bp of AAV cap and the common AAV polyadenylation signal sequence.
DISCUSSION
The current concept of site-specific AAV integration.
Targeted AAV integration requires three essential components: Rep78/68 in trans and both the AAV-ITR and the AAVS1 of chr.19 in cis, which together form a ternary complex (31). A recent report suggests that that the AAV p5 promoter elements add to the effect (19). Typical AAV DNA replication intermediates have a head-to-head or tail-to-tail genome conformation (16). This is also true for AAV wild-type genomes (8). Other studies have demonstrated integrated AAV genomes preferentially displaying head-to-tail concatemeric structures (1, 32). These findings led Linden et al. (12) to propose a circular AAV intermediate as a template for limited rolling circle replication, thereby explaining the generation of head-to-tail concatemers. After ternary complex formation of Rep78/68, the AAV RBS and the chr.19 RBS Rep-mediated nicking of the chr.19 trs is assumed to initiate targeting. Rep is then assumed to attach to the free 5′ end of the chr.19 single-stranded nick, in analogy to the single-stranded AAV genome in which covalent Rep binding to the free 5′ end has been described (25). The free 3′ end of the nicked chr.19 sequence may serve as primer for the polymerase that replicates along the gap. To explain AAV integration, Rep is postulated to switch templates between the circular AAV genome and the chr.19 preintegration site (11), a critical component of the model that remains to be proven. Young and Samulski (34) recently showed that in cell culture nicking of the AAV-ITR, trs was not required for targeted integration and that overexpression of Rep alone was sufficient to induce rearrangements at AAVS1 (33). Based on these findings, Rep78/68 is assumed to initiate replication on chr.19 by interaction with RBS and trs. The AAV genome will be attracted to enter the Rep-AAVS1 complex. Integration of the AAV genome is assumed to rely on cellular recombination mechanisms also active in gene amplification (34). This assumption is backed by previous in vitro studies that not only demonstrated ternary complex formation of Rep with the AAV-ITR and AAVS1 (31) but also demonstrated Rep-dependent initiation of DNA synthesis starting off a DNA sequence within AAVS1 that serves as the origin sequence (29). Thus, the described ability of Rep not only to site specifically nick DNA but also to ligate free single-stranded DNAs (24) in conjunction with ternary complex formation could explain our finding of covalently attached AAV-ITRs and AAVS1 of chr.19. Irrespective of the mode of AAV integration, free AAV-ITR/AAVS1 junction sequences can be viewed as episomal integration intermediates that, due to the presence of packaging signals within the AAV-ITR, can be encapsidated in AAV virions. The recent description of chr.19 containing AAV particles rescued from a latently infected cell clone established by a trs-defective AAV genome (34) further supports the notion of AAV/AAVS1 junctions being aberrant replication intermediates. Thus far, we could only analyze the DNA sequences between the defined primers on chr.19 and on the AAV-ITR. At present, we have no experimental data demonstrating whether other covalently attached genomic DNAs can be packaged. Due to the low percentage of packaged junctions in the virion stocks, cloning of complete packaged genomes and detection of “unknown” DNAs, e.g., with some combinations of degenerate primers, is beyond the scope of this study.
Potential mechanisms of AAV-ITR/AAVS1 generation during productive AAV infection.
The generally accepted concept of the bipartite AAV life cycle is represented in Fig. 4. Targeted AAV integration leads to the establishment of latency in AAV-infected cells. The helper virus-dependent, productive AAV cycle is viewed as separate in the sense that an unrelated helper virus, typically Ad or herpesvirus, induces high-level AAV replication of the episomal AAV template (16). In the case of a latently infected cell line, Ad will rescue AAV from the integrated state. It is unclear whether this involves excision of the AAV copy, followed by replication of the AAV episome, or whether DNA replication uses the integrated AAV copy as a template.
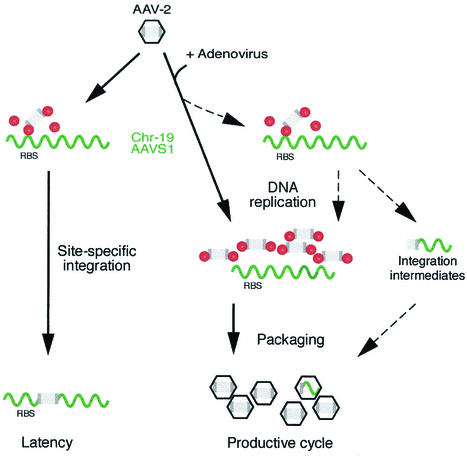
FIG. 4.
Rep-dependent generation of packaged AAV-2/chr.19 integration intermediates during productive AAV infection. Schematic representation of the bipartite AAV life cycle. The AAV genome is drawn in gray with the ITRs shaded darker. Rep molecules are represented as red bullets. The human chr.19 preintegration site called AAVS1 is represented as wavy green line. Black arrows indicate the commonly accepted steps of the AAV life cycle. Dashed arrows indicate additional, bypassing steps postulated to explain the generation of packaged junctions. Latency is established upon infection of AAV-2 in the absence of a helper virus, leading to limited and autoregulated Rep expression. AAV integration at AAVS1 of chr.19 is mediated by ternary complex formation of Rep with the RBS of AAV-ITR and of AAVS1. The productive cycle is initiated by Ad coinfection, when Rep expression is derepressed and episomal AAV DNA replication is initiated. Under these conditions, most of Rep is bound to the RBS on the replicating AAV genomes. AAV capsid proteins package the replicated AAV genomes. This concept is extended (dashed arrows) by the assumption that limited AAV integration can take place in the presence of Ad, leading to the generation of AAV-ITR/chr.19 integration intermediates, which may serve as templates for further replication. Since the AAV-ITR comprises the AAV packaging signals, these integration intermediates can be packaged into AAV capsids.
The data presented here point to a closer relationship between AAV latency and productive AAV replication as outlined in Fig. 4. Packaged AAV-ITR/AAVS1 junctions can be viewed as aberrant integration intermediates of productive infection. AAV appears to have an inherent potential to integrate at AAVS1 irrespective of the presence of a helper virus. We assume that after AAV infection the p5 promoter is able to express enough Rep to initiate site-specific integration before the cascade of Ad helper functions has been fully expressed and the AAV genome starts replication. Ad-induced Rep78 expression and AAV DNA replication were detected at 10 to 12 h p.i. (21). In our study of Rep expression in the absence of a helper virus, we were able to detect Rep78 on Western blots as early as 8 h p.i. In addition, by using sensitive immunofluorescence protocols, an occasional Rep-positive cell could be detected as early as 5 h p.i. (R. Heilbronn et al., unpublished data). In latently infected cell clones under conditions of autoregulated Rep expression, a few thousand Rep molecules per cell have been measured (33). In addition, a low level of targeted AAV integration has been detected by 8 h p.i. (6). Thus, limited targeted integration may occur very early after AAV infection, before the onset of Ad-stimulated AAV replication. A recent report describing site-specific integration of an AAV-ITR-containing Ad vector expressing Rep78 (18) and our preliminary data on limited AAV integration in the presence of replicating wild-type Ad (D. Hüser and R. Heilbronn, unpublished data) support this notion. However, upon AAV replication, the increasing copy number of the AAV-ITR is assumed to compete for binding with the chr.19 RBS, which has been shown to bind to Rep in vitro with reduced affinity (33). In addition, the nuclear compartmentalization of the AAV genome and of Rep in Ad replication centers at later time points postinfection (30) presumedly limits chr.19 targeting. Based on these findings, we assume that chr.19-specific AAV targeting is limited to the first few hours after AAV and Ad coinfection. In addition, due to cell lysis AAV integration in the presence of Ad will remain without consequences for the infected cell.
In summary, the packaged AAV-ITR/AAVS1 junctions detected in AAV virion stocks can be viewed as footprints of integration events during productive AAV replication. Interestingly, packaged junctions were detected irrespective of the AAV production method. Ad2 infection, as well as transfection of the defined Ad5 helper genes, into 293 cells led to formation of these junctions. Obviously, the latent and the productive AAV replication cycles are more closely related than previously thought.
Implications for gene therapy and rAAV production protocols.
Packaged AAV-ITR/AAVS1 junctions were not only found in AAV wild-type preparations but rAAV vector stocks were also found to harbor comparable amounts of packaged junctions. Typical inoculation doses in mice and rats range between 1011 and 1013 genome equivalents. With this dose it is expected that 104 to 106 packaged junctions will be transferred. Due to restrictions in primer design, we only analyzed vectors based on psub201 (22). It will be interesting to see how optimized rAAV vectors such as the pUF2 series (36) and the rAAVs derived from them will perform. We have to envision that many, if not most, rAAV preparations harbor low and as-yet-undetected levels of packaged junctions. This point has to be reevaluated critically. However, to our knowledge unwanted side effects attributable to the presence of ITR/chr.19 junctions have not been documented so far.
Acknowledgments
We thank M. Gere for skilled technical assistance, R. Joncker for secretarial support, and M. Boshart for helpful discussions and critical reading of the manuscript. We also thank J. Kleinschmidt, N. Muzyczka, and R. J. Samulski for plasmids.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 506).
REFERENCES
- 1.Giraud, C., E. Winocour, and K. I. Berns. 1995. Recombinant junctions formed by site-specific integration of adeno-associated virus into an episome. J. Virol. 69:6917-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giraud, C., E. Winocour, and K. I. Berns. 1994. Site-specific integration by adeno-associated virus is directed by a cellular DNA sequence. Proc. Natl. Acad. Sci. USA 91:10039-10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 4.Heilbronn, R., A. Bürkle, S. Stephan, and H. zur Hausen. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hüser, D., and R. Heilbronn. 2003. Adeno-associated virus integrates site-specifically into human chromosome 19 in either orientation and with equal kinetics and frequency. J. Gen. Virol. 84:133-137. [DOI] [PubMed] [Google Scholar]
- 6.Hüser, D., S. Weger, and R. Heilbronn. 2002. Kinetics and frequency of adeno-associated virus site-specific integration into human chromosome 19 monitored by quantitative real-time PCR. J. Virol. 76:7554-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Im, D.-S., and N. Muzyczka. 1990. The AAV origin-binding protein Rep68 is an ATP-dependent site-specific endonuclease with helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 8.Kotin, R. M., and K. I. Berns. 1989. Organization of adeno-associated virus DNA in latently infected Detroit 6 cells. Virology 170:460-467. [DOI] [PubMed] [Google Scholar]
- 9.Kotin, R. M., R. M. Linden, and K. I. Berns. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotin, R. M., J. C. Menninger, D. C. Ward, and K. I. Berns. 1991. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics 10:831-834. [DOI] [PubMed] [Google Scholar]
- 11.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. I. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 93:11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linden, R. M., E. Winocour, and K. I. Berns. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA 93:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macville, M., E. Schrock, H. Padilla-Nash, C. Keck, B. M. Ghadimi, D. Zimonjic, N. Popescu, and T. Ried. 1999. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59:141-150. [PubMed] [Google Scholar]
- 14.Meneses, P., K. I. Berns, and E. Winocour. 2000. DNA sequence motifs which direct adeno-associated virus site-specific integration in a model system. J. Virol. 74:6213-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, D. G., E. A. Rutledge, and D. W. Russell. 2002. Chromosomal effects of adeno-associated virus vector integration. Nat. Genet. 30:147-148. [DOI] [PubMed] [Google Scholar]
- 16.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2359. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott, Philadelphia, Pa.
- 17.Palombo, F., A. Monciotti, A. Recchia, R. Cortese, G. Ciliberto, and N. La Monica. 1998. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus-adeno-associated virus vector. J. Virol. 72:5025-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philpott, N. J., C. Giraud-Wali, C. Dupuis, J. Gomos, H. Hamilton, K. I. Berns, and E. Falck-Pedersen. 2002. Efficient integration of recombinant adeno-associated virus DNA vectors requires a p5-rep sequence in cis. J. Virol. 76:5411-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philpott, N. J., J. Gomos, K. I. Berns, and E. Falck-Pedersen. 2002. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl. Acad. Sci. USA 99:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pieroni, L., C. Fipaldini, A. Monciotti, D. Cimini, A. Sgura, E. Fattori, O. Epifano, R. Cortese, F. Palombo, and N. La Monica. 1998. Targeted integration of adeno-associated virus-derived plasmids in transfected human cells. Virology 249:249-259. [DOI] [PubMed] [Google Scholar]
- 21.Redemann, B. E., E. Mendelson, and B. J. Carter. 1989. Adeno-associated virus rep protein synthesis during productive infection. J. Virol. 63:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samulski, R. J., L.-S. Chang, and T. Shenk. 1987. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J. Virol. 61:3096-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941-3950. (Erratum, 11:1228, 1992.) [DOI] [PMC free article] [PubMed]
- 24.Smith, R. H., and R. M. Kotin. 2000. An adeno-associated virus (AAV) initiator protein, Rep78, catalyzes the cleavage and ligation of single-stranded AAV ori DNA. J. Virol. 74:3122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder, R. O., D.-S. Im, and N. Muzyczka. 1990. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J. Virol. 64:6204-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder, R. O., D.-S. Im, T. Ni, X. Xiao, R. J. Samulski, and N. Muzyczka. 1993. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J. Virol. 67:6096-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsunoda, H., T. Hayakawa, N. Sakuragawa, and H. Koyama. 2000. Site-specific integration of adeno-associated virus-based plasmid vectors in lipofected HeLa cells. Virology 268:391-401. [DOI] [PubMed] [Google Scholar]
- 29.Urcelay, E., P. Ward, S. M. Wiener, B. Safer, and R. M. Kotin. 1995. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J. Virol. 69:2038-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weitzman, M. D., K. J. Fisher, and J. M. Wilson. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weitzman, M. D., S. R. M. Kyöstiö, R. M. Kotin, and R. A. Owens. 1994. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. USA 91:5808-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, C. C., X. Xiao, X. Zhu, D. C. Ansardi, N. D. Epstein, M. R. Frey, A. G. Matera, and R. J. Samulski. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71:9231-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young, S. M., Jr., D. M. McCarty, N. Degtyareva, and R. J. Samulski. 2000. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 74:3953-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young, S. M., Jr., and R. J. Samulski. 2001. Adeno-associated virus (AAV) site-specific recombination does not require a Rep-dependent origin of replication within the AAV terminal repeat. Proc. Natl. Acad. Sci. USA 98:13525-13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zolotukhin, S., B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973-985. [DOI] [PubMed] [Google Scholar]
- 36.Zolotukhin, S., M. Potter, W. W. Hauswirth, J. Guy, and N. Muzyczka. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]