Abstract
Several picornaviruses shut down host cellular protein synthesis by proteolytic cleavage of the eukaryotic initiation factor (eIF) 4GI and eIF4GII isoforms. Viral RNA translation is maintained by a cap-independent mechanism. Here, we identify the human rhinovirus 2 2Apro cleavage site in eIF4GII in vitro as PLLNV699*GSR; this sequence lies seven amino acids C-terminal to the cleavage site previously identified in eIF4GI (LSTR681*GPP).
Picornaviruses, which include human rhinovirus (HRV), poliovirus (PV), and coxsackievirus (CV) as well as the animal pathogen foot-and-mouth disease virus, contain a single-stranded RNA of positive polarity. Eukaryotic cellular mRNAs possess at their 5′ ends a cap structure (m7GpppX, where X is any nucleotide) (27), which is important for ribosomal recruitment mediated by a cap-binding protein complex, eukaryotic initiation factor (eIF) 4F. The genomic viral RNA lacks a 5′-terminal cap structure (9, 22), and virus translation proceeds by a cap-independent mechanism, whereby ribosomes bind directly to an internal ribosome entry site of the viral RNAs (11, 23). The cap-binding protein complex eIF4F is a three-subunit complex. The three subunits are eIF4E, which interacts directly with the cap (19, 21, 29), eIF4A, an ATPase which in conjunction with eIF4B exhibits RNA helicase activity (25), and eIF4G (formerly p220 or eIF4γ [31]), which is a large scaffolding protein that plays a key role in the assembly of the mRNA-ribosome initiation complex. eIF4G binds directly to the ribosome-associated eIF3, thus delivering the small ribosomal subunit to the mRNA (12, 18). We have cloned and characterized a homologue of eIF4G, which we have termed eIF4GII (4), while the original eIF4G (31) was renamed eIF4GI. eIF4GII is 41% identical to eIF4GI, binds eIF4E, eIF3, and eIF4A, and functionally complements eIF4GI (4, 10).
Different picornavirus proteinases can cleave both isoforms of eIF4G, generating in each case their respective N- and C-terminal cleavage products, cpN and cpC (reviewed in references 2 and 28). The cpC of eIF4G retains the capacity to interact with internal ribosome entry sites as well as with eIF3 and eIF4A (15-17, 20, 24) and can therefore support cap-independent translation. Indeed, initiation of translation on HRV and PV RNA is stimulated under these conditions (7, 14, 32). However, as the eIF4G cpC lacks the eIF4E-binding site, it is unable to support cap-dependent translation of cellular mRNAs (reviewed in reference 8) or does this inefficiently (1).
We have previously shown, using PV1 and HRV14 as model systems (5, 30), that eIF4GII cleavage precisely coincides with the inhibition of host cellular protein synthesis, whereas the cleavage of eIF4GI occurs earlier. However, in HRV2-infected cells, eIF4GI and eIF4GII are cleaved at similar rates, coincident with the shutoff of host cell protein synthesis (26).
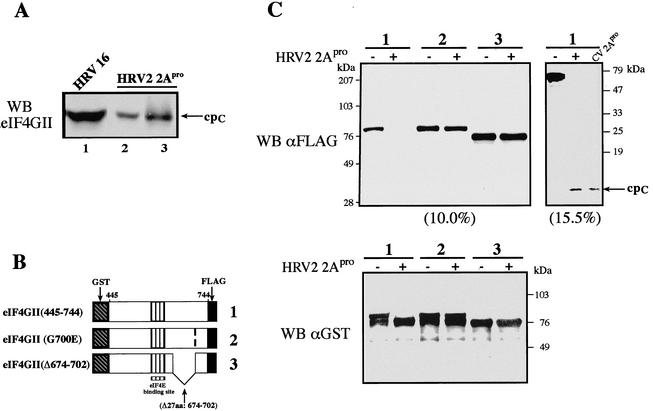
To begin to understand the differences in the kinetics of eIF4GI and eIF4GII cleavage, we set out to determine the in vitro HRV2 2Apro cleavage site in human eIF4GII and compare it to that previously determined for HRV2, CVB4, and PV1 2Apro on eIF4GI. First, we used HRV2 2Apro to cleave recombinant eIF4GII (4). After incubation of recombinant eIF4GII (20 μg for 30 h at 30°C) with the purified enzyme in vitro, we examined the status of eIF4GII by using antibodies against N- and C-terminal regions of the protein. The C-terminal fragment of endogenous or purified recombinant eIF4GII generated by in vitro cleavage ran at about 90 kDa, with mobility identical to that found in vivo in HRV16-infected cells (Fig. 1A, compare lanes 2 and 3 to lane 1) or in HRV2-infected cells (data not shown). Five cycles of N-terminal sequencing by automated Edman degradation of the recombinant eIF4GII cpC (Fig. 1A, lane 3) generated the following amino acids: glycine, serine, arginine, arginine, and serine. These amino acids correspond to the sequence V700GSRR704 on eIF4GII, indicating that HRV2 2Apro must cleave eIF4GII at Val699*Gly700 (Fig. 2). We attempted to determine the N-terminal sequence of the endogenous eIF4GII cpC isolated by immunoprecipitation from HRV16-infected cells but were unsuccessful, as the preparation contained a mixture of polypeptides.
FIG. 1.
(A) Identification of the 2Apro cleavage site in eIF4GII. HeLa-I cells were infected with HRV16 (100 50% tissue culture infective doses per cell) as described previously (30). Cell extract (60 μg of protein; lane 1) was prepared 6 h postinfection and loaded on a gel in parallel with 40 μg of HeLa S10 (lane 2) or purified recombinant eIF4GII (1/40 of the reaction mixture, ∼0.5 μg; lane 3) that was incubated in the presence of purified HRV2 2Apro. Proteins were resolved by SDS-8% PAGE and blotted onto nitrocellulose. The blot was treated with polyclonal antibodies against the cpC of eIF4GII. WB, Western blot. (B and C) eIF4GII mutants that are resistant to HRV2 2Apro cleavage. (B) Scheme of wild-type eIF4GII (1) and mutant (2 and 3) fragments. GST-eIF4GII (aa 445 to 744)-FLAG fragments contained a single point mutation (G700E) or a deletion (Δ674-702). (C) eIF4GII proteins were expressed in E. coli and purified on a glutathione-Sepharose column (4). Purified proteins (0.5 μg) were incubated in the presence or absence of recombinant 2Apro (2.8 μg) in buffer containing 100 mM potassium acetate, 20 mM Tris-HCl (pH 7.6), 2.5 mM magnesium acetate, and 10% glycerol for 1 h at 30°C. Laemmli buffer was added to stop the reaction. Samples were resolved by SDS-PAGE (10.0, 15.5, or 12.5% [bottom] acrylamide). Proteins were transferred onto a nitrocellulose membrane, which was subsequently incubated with anti-GST or anti-FLAG antibodies as indicated. The small (62-aa) C-terminal cleavage product containing the FLAG epitope was resolved only in a 15.5% gel (cpC). The wild-type eIF4GII fragment was cleaved by HRV2 or CVB4 2Apro, as indicated. Positions of molecular mass standards are shown on the right.
FIG. 2.
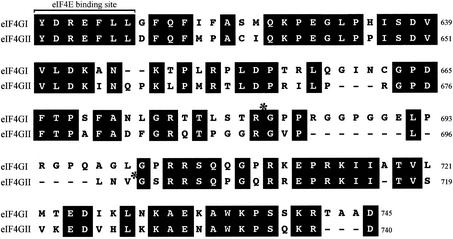
Sequence alignment of human eIF4GI and eIF4GII proteins (sequence comparison starts from the eIF4E-binding site) (4, 18). Asterisks, positions of 2Apro cleavage sites in eIF4GI and eIF4GII. The full-length eIF4GI and eIF4GII sequence accession numbers are AY082886 (3) and AF012072 (4), respectively.
To further substantiate our results, we introduced mutations at the HRV2 2Apro cleavage site in eIF4GII. Glutathione S-transferase (GST)-eIF4GII (amino acids [445 to 744])-FLAG fragments, containing either a single point mutation (G700E) in or a 27-aa deletion (Δ674-702) of the putative hinge region (Fig. 1B) were generated, expressed in Escherichia coli, and purified by glutathione-Sepharose affinity chromatography. The purity and integrity of the proteins were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining (data not shown). Equal amounts of the fusion proteins (wild type and two mutants) were incubated with the purified HRV2 2Apro or buffer alone at 30°C for 1 h. Protein samples were resolved by SDS-12.5% PAGE and analyzed by Western blotting using antisera against either the FLAG or GST tag (Fig. 1C). Both mutants were resistant to 2Apro cleavage (lanes 2 and 3), strongly supporting the conclusion that HRV2 2Apro directly cleaves eIF4GII between V699 and G700. Identical results were obtained when the eIF4GII mutants were treated with purified recombinant CVB4 2Apro (data not shown), demonstrating that this enzyme also recognizes the same site.
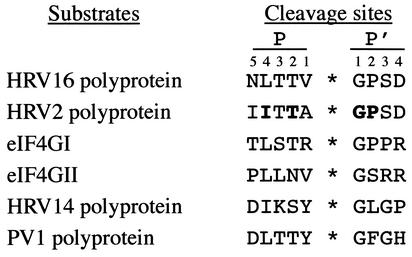
These results identify the cleavage site of HRV2 and CVB4 2Apro in eIF4GII as PLLNV699*700GS, lying 7 aa downstream of the cleavage site identified in eIF4GI as TLSTR641*642GP (13). As the two isoforms are cleaved equally well during infection by HRV2 (26) and HRV16 (data not shown), both eIF4G isoforms must exist in a cleavable form. Moreover, the binding to eIF4E, which is required for efficient cleavage, is not limiting for eIF4G cleavage. In contrast, during HRV14 and PV1 infection, eIF4GI is cleaved more rapidly than eIF4GII, indicating that the respective 2Apro poorly recognizes the eIF4GII cleavage site, compared to recognition of the eIF4GI site. The reason for this discrimination remains, however, unclear. An analysis of the cleavage sites recognized on the viral polyprotein and eIF4G isoforms by HRV2, -14, and -16 2Apro as well as that of PV1 2Apro (Fig. 3) does not indicate any particular pattern which could explain the discrimination. This suggests that the overall conformations of the two isoforms may be important.
FIG. 3.
Comparison of 2Apro cleavage sites on the respective polyproteins and eIF4G homologues. The viral polyprotein sequences represent the C terminus of VP1 and the N terminus of 2Apro, respectively, and are taken from the public database http://www.iah.bbsrc.ac.uk/virus/picornaviridae/sequencedatabase/index.html. The residues shown to be important for HRV2 2Apro substrate recognition by peptide cleavage, mutational analysis, or crystallography are in boldface. P and P′ designate the N-terminal and the C-terminal cleavage products, respectively. Numbers indicate the positions of amino acid residues relative to the cleavage site.
Indeed, the experiments presented here provide evidence that a significant conformational component in cleavage site recognition is present. Efficient cleavage of the recombinant GST-eIF4GII fragments was achieved in the absence of eIF4E (Fig. 1C). In contrast, the cleavage of eIF4GII, as well as eIF4GI (6), in HeLa extracts could be prevented by the addition of the GST-4E-BP1 fusion protein, which can sequester the eIF4E in the extract and prevent its binding to both eIF4G isoforms (data not shown). Thus, although the cleavage site is present in eIF4GII, it is no longer in a conformation which can be recognized by the enzyme. Interestingly, this is not the case when fragments of eIF4GII are used as in Fig. 2C, as efficient cleavage of recombinant GST-eIF4GII by purified 2Apro was observed in the absence of eIF4E. Thus, the remainder of eIF4GII, as well as other factors which may be bound to it, affects the conformation of the cleavage site. Further experiments will be required to examine the nature of these conformational changes and to elucidate in which way they are responsible for the differential cleavage of the eIF4G isoforms during HRV14 and PV infection.
Acknowledgments
We thank Graham Belsham for critical reading of the manuscript and for support and insights during the course of this work. We are grateful to Colin Lister for exceptional technical assistance. We are indebted to F. Hayden and R. Rueckert for HeLa-I cells and HRV16, respectively.
This work was supported by a grant from the Canadian Institute of Health Research (CIHR) to N.S., who is the recipient of a CIHR Distinguished Scientist Award and a Howard Hughes Medical Institute International Scholarship. A.G. was supported by a fellowship of Istituto Superiore di Sanitá, Italy.
REFERENCES
- 1.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 20:4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 22:4499-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haghighat, A., Y. Svitkin, I. Novoa, E. Kuechler, T. Skern, and N. Sonenberg. 1996. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70:8444-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hambidge, S. J., and P. Sarnow. 1992. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc. Natl. Acad. Sci. USA 89:10272-10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 9.Hewlett, M. J., J. K. Rose, and D. Baltimore. 1976. 5′-terminal structure of poliovirus polyribosomal RNA is pUp. Proc. Natl. Acad. Sci. USA 73:327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid stretch of human eIF4G binds poly(A) binding protein and functions in poly(A) dependent translation. EMBO J. 17:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang, S. K., H.-G. Kräusslich, M. J. H. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 13.Lamphear, B. J., R. Yan, F. Yang, D. Waters, H. D. Liebig, H. Klump, E. Kuechler, T. Skern, and R. E. Rhoads. 1993. Mapping the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 268:19200-19203. [PubMed] [Google Scholar]
- 14.Liebig, H. D., E. Ziegler, R. Yan, K. Hartmuth, H. Klump, H. Kowalski, D. Blaas, W. Sommergruber, L. Frasel, B. Lamphear, R. E. Rhoads, E. Kuechler, and T. Skern. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4γ and influence on in vitro translation. Biochemistry 32:7581-7588. [DOI] [PubMed] [Google Scholar]
- 15.Lomakin, I. B., C. U. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez de Quinto, S., E. Lafuente, and E. Martinez-Salas. 2001. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7:1213-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez de Quinto, S., and E. Martinez-Salas. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6:1380-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcotrigiano, J., A.-C. Gingras, N. Sonenberg, and S. K. Burley. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89:951-961. [DOI] [PubMed] [Google Scholar]
- 20.Marcotrigiano, J., I. B. Lomakin, N. Sonenberg, T. V. Pestova, C. U. Hellen, and S. K. Burley. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7:193-203. [DOI] [PubMed] [Google Scholar]
- 21.Matsuo, H., H. Li, A. M. McGuire, C. M. Fletcher, A.-C. Gingras, N. Sonenberg, and G. Wagner. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4:717-724. [DOI] [PubMed] [Google Scholar]
- 22.Nomoto, A., Y. F. Lee, and E. Wimmer. 1976. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc. Natl. Acad. Sci. USA 73:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 24.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 25.Rozen, F., I. Edery, K. Meerovitch, T. E. Dever, W. C. Merrick, and N. Sonenberg. 1990. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10:1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seipelt, J., H. D. Liebig, W. Sommergruber, C. Gerner, and E. Kuechler. 2000. 2A proteinase of human rhinovirus cleaves cytokeratin 8 in infected HeLa cells. J. Biol. Chem. 275:20084-20089. [DOI] [PubMed] [Google Scholar]
- 27.Shatkin, A. J. 1976. Capping of eucaryotic mRNAs. Cell 9:645-653. [DOI] [PubMed] [Google Scholar]
- 28.Skern, T., B. Hampölz, A. Guarné, I. Fita, E. Bergmann, J. Petersen, and M. N. G. James. 2002. Structure and function of picornavirus proteinases, p. 199-212. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 29.Sonenberg, N., K. M. Rupprecht, S. M. Hecht, and A. J. Shatkin. 1979. Eukaryotic mRNA cap binding protein: purification by affinity chromatography on Sepharose-coupled m7GDP. Proc. Natl. Acad. Sci. USA 76:4345-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan, R., W. Rychlik, D. Etchison, and R. E. Rhoads. 1992. Amino acid sequence of the human protein synthesis initiation factor eIF-4γ. J. Biol. Chem. 267:23226-23231. [PubMed] [Google Scholar]
- 32.Ziegler, E., A. M. Borman, R. Kirchweger, T. Skern, and K. M. Kean. 1995. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol. 69:3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]