Abstract
The vesicular stomatitis virus (VSV) matrix (M) protein plays a major role in the virus-induced inhibition of host gene expression. It has been proposed that the inhibition of host gene expression by M protein is responsible for suppressing activation of host interferon gene expression. Most wild-type (wt) strains of VSV induce little if any interferon gene expression. Interferon-inducing mutants of VSV have been isolated previously, many of which contain mutations in their M proteins. However, it was not known whether these M protein mutations were responsible for the interferon-inducing phenotype of these viruses. Alternatively, mutations in other genes besides the M gene may enhance the ability of VSV to induce interferons. These hypotheses were tested by transfecting cells with mRNA expressing wt and mutant M proteins in the absence of other viral components and determining their ability to inhibit interferon gene expression. The M protein mutations were the M51R mutation originally found in the tsO82 and T1026R1 mutant viruses, the double substitution V221F and S226R found in the TP3 mutant virus, and the triple substitution E213A, V221F, and S226R found in the TP2 mutant virus. wt M proteins suppressed expression of luciferase from the simian virus 40 promoter and from the beta interferon (IFN-β) promoter, while M proteins of interferon-inducing viruses were unable to inhibit luciferase expression from either promoter. The M genes of the interferon-inducing mutants of VSV were incorporated into the wt background of a recombinant VSV infectious cDNA clone. The resulting recombinant viruses were tested for their ability to activate interferon gene expression and for their ability to inhibit host RNA and protein synthesis. Each of the recombinant viruses containing M protein mutations induced expression of a luciferase reporter gene driven by the IFN-β promoter and induced production of interferon bioactivity more effectively than viruses containing wt M proteins. Furthermore, the M protein mutant viruses were defective in their ability to inhibit both host RNA synthesis and host protein synthesis. These data support the idea that wt M protein suppresses interferon gene expression through the general inhibition of host RNA and protein synthesis.
Virus infections usually trigger an antiviral response in host cells that functions to inhibit virus replication. As a result, most viruses have evolved mechanisms to suppress the antiviral response of the host. The balance between the ability of host cells to mount an antiviral response and the ability of the virus to suppress that response is a major determinant of the evolution of infection and viral tissue tropism in intact animal hosts (reviewed in reference 31). For many viruses, a major factor in the host antiviral response is the production of alpha and beta interferon (IFN-α and -β). Once IFNs are secreted by infected cells, signal transduction events are stimulated, both in the infected cells and in neighboring uninfected cells, which lead to the activation of genes whose products interfere with various steps in the viral life cycle (reviewed in reference 21). However, many viruses have evolved diverse mechanisms to combat the host defense mounted by IFNs. In general, these mechanisms can be divided into two types: those that inhibit the production of IFNs and those that inhibit the response to IFNs. Vesicular stomatitis virus (VSV), the prototype rhabdovirus, is a classic example of a virus that inhibits the production of IFNs (33). The goal of the experiments presented here was to determine whether the activity of the viral matrix (M) protein aids in the suppression of IFN gene expression during VSV infection.
The M protein of VSV plays a major role in virus assembly by binding the viral nucleocapsid to the cytoplasmic surface of the host plasma membrane during the budding process (17, 18, 28) and by inducing budding of virus envelopes (22, 23, 25). However, M protein is also responsible for many of the cytopathic effects associated with VSV infection. These include the characteristic rounding of cells, as well as the shutoff of host-directed gene expression (reviewed in reference 31). The ability of M protein to repress host gene expression is genetically separable from its viral assembly function (6, 11, 26). Furthermore, M protein is capable of inhibiting host gene expression independently of other viral components (5, 16, 36). This inhibition occurs at the level of host transcription, as well as nuclear-cytoplasmic transport of host RNAs and proteins (1, 5, 24, 37, 39). M protein also plays a major role in the inhibition of host translation (19, 25, 27). However, M protein is not able to inhibit translation in transfected cells in the absence of other viral components (4). Therefore, it is likely that the inhibition of host translation in VSV-infected cells is due to the combined effects of M protein and additional viral factors.
It has been proposed that the ability of M protein to inhibit host gene expression is responsible for the ability of VSV to suppress activation of host IFN gene expression (reviewed in reference 31). According to this model, there must be other products of virus infection, such as viral double-stranded RNA (dsRNA), that activate IFN gene expression, which is then suppressed by the activity of M protein. In support of this idea, transfection experiments have shown that M protein inhibits expression of a reporter gene from a plasmid containing the IFN-β promoter as effectively as it inhibits expression from other promoters (16). Despite the extensive evidence that M protein can inhibit host gene expression in the absence of other viral components, the role of M protein versus other viral components in the shutoff of host gene expression in the context of a viral infection has been questioned (40). In addition, it has been proposed that M protein plays little, if any, role in the suppression of IFN production by VSV (34).
Earlier studies have demonstrated the feasibility of isolating VSV mutants with strong IFN-inducing phenotypes (19, 34). Many of these IFN-inducing mutants contain point mutations in their M proteins (13, 34). However, it was not known whether the M protein mutations were responsible for the ability of these viruses to induce IFN. Alternatively, it has been proposed that mutations in other genes besides the M gene may account for their IFN-inducing phenotype (34). In this paper, mutant M proteins from these IFN-inducing viruses were used to resolve the question of whether M protein is the VSV factor that suppresses IFN induction during the virus infection. Furthermore, we tested whether the ability of M protein to inhibit IFN induction is due to its potent ability to shut off host gene expression. These hypotheses were tested in the experiments presented here by transfecting cells with M mRNA expressing wild-type (wt) M proteins or mutant M proteins from the IFN-inducing viruses and determining their ability to inhibit IFN gene expression. Results indicated that wt M proteins effectively suppressed luciferase expression from plasmids containing either the simian virus 40 (SV40) or IFN-β promoters, while M proteins of IFN-inducing viruses were unable to inhibit luciferase expression from either promoter. The M genes of several of these IFN-inducing mutants were incorporated into the wt background of an infectious VSV cDNA clone. Each of the recombinant viruses containing mutant M proteins induced expression of a luciferase reporter gene driven by the IFN-β promoter and induced the production of IFN bioactivity more effectively than viruses containing wt M proteins. These results indicate that M protein plays a major role in the inhibition of host IFN gene expression in VSV-infected cells. Furthermore, the M protein mutant viruses were defective in their ability to inhibit both host RNA synthesis and host protein synthesis. Thus, the IFN-inducing phenotype of the viruses containing M protein mutations was genetically correlated with a defect in their ability to inhibit host gene expression, suggesting that the suppression of IFN activity in VSV-infected cells is due in part to a global inhibition of host gene expression by M protein.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells and PC-3 prostate cells were from the American Type Culture Collection. wt VSV (Indiana serotype, Orsay strain) and the M protein mutant tsO82 (11) were grown in BHK cells as described previously (32). The recombinant viruses, rwt and rM51R, contain the San Juan strain of M protein and were isolated from infectious VSV cDNA clones as described elsewhere (27). Plasmids containing cDNA copies of the M genes of the wt HR strain and the T1026R1, TP2, and TP3 mutant viruses have been described previously (13). The M genes were modified by PCR with Pwo DNA polymerase (Boerhinger-Mannheim, Inc.) using the primers 5′GGGCTTAAGGAAGATTCTCGGTCTG3′ and 5′TTTGGCGCGCCAATTAGGAGAGAC3′. The PCR products were digested with AflII and BssHII and were inserted into the infectious VSV cDNA clone as described previously (27). The recombinant viruses isolated from these cDNA clones were designated rHR-M, r1026-M, rTP2-M, and rTP3-M viruses. All viruses were plaque isolated twice in BHK cells, and the sequences of the M genes were confirmed by reverse transcription-PCR and automated DNA sequencing as described elsewhere (27).
Plasmids and in vitro transcription of mRNA.
The pβlux plasmid contains the firefly luciferase gene under control of the IFN-β promoter (35), and the pGL3 control vector expresses luciferase constitutively from the SV40 promoter (Promega). The plasmid pSD.OM, used for in vitro transcription of mRNA encoding wt M protein (Orsay strain) together with a 3′ poly(A) sequence, has been described previously (4). The M gene cDNAs from wt HR, T1026R1, TP2, and TP3 viruses were modified by PCR with Pwo DNA polymerase by using the primers described previously (6). The PCR products were digested with HindIII and were cloned into the pSD4.2 vector for in vitro transcription of wt HR and mutant M mRNAs. In the in vitro transcription reactions, M mRNAs containing 5′ caps and 3′ poly(A) were synthesized in the presence of the cap analog 7mG(5′)ppp(5′)G from linearized plasmid DNA by the bacteriophage SP6 RNA polymerase (Message Machine; Ambion, Inc.).
Transfections and luciferase assays.
HeLa cells in 35-mm-diameter dishes (or six-well plates) were transfected using Lipofectin reagent (GIBCO-BRL) according to the manufacturer's instructions. To determine the effect of M protein on expression of luciferase from the SV40 promoter, cells were transfected with various amounts of in vitro-transcribed M mRNA together with 250 ng of pGL3 plasmid DNA and various amounts of yeast RNA to normalize RNA levels to 750 ng. At 16 h posttransfection, cells were washed with phosphate-buffered saline (PBS) and harvested. Luciferase activity was determined using the Promega luciferase assay system.
To determine the effect of wt and mutant M proteins on expression of luciferase from the IFN-β promoter, cells were transfected with either 100 ng of wtHR, TP2, and TP3 M mRNAs or 300 ng of T1026 M mRNA together with 1 μg of pβlux. At 16 h posttransfection, cells were washed with PBS and harvested, and luciferase activity was measured. Transfections were carried out both in the presence and absence of poly(I)-poly(C). However, in most cases, the transfection protocol itself partially induced the IFN promoter, and the addition of dsRNA had little if any effect in activation of the IFN promoter.
To determine the effect of wt and M protein mutant viruses on activation of the IFN-β promoter, HeLa cells in 35-mm dishes were transfected with 1 μg of pβlux plasmid DNA. At 24 h posttransfection, cells were infected with viruses containing wt or mutant M proteins at a multiplicity of 20 PFU/cell. Cells were mock infected as negative controls or were treated with poly(I)-poly(C) (200 μg/ml; Sigma Chemical Co.) at 24 h posttransfection as positive controls. Cells were harvested at 3, 6, and 9 h postinfection, and luciferase activity was measured.
IFN bioassay.
To determine the IFN activity produced by cells infected with wt and mutant viruses, supernatants (100 μl) were collected from HeLa and PC-3 cells infected with wt and mutant viruses at the times indicated below in Fig. 5. Infectious virus was inactivated by acid treatment, the acid was neutralized, and serial dilutions were incubated with HeLa cells in 96-well plates overnight at 37°C. As a standard, cells were incubated with serial fivefold dilutions of IFN (Universal type I IFN; PBL Biomedical Laboratories, New Brunswick, N.J.). The samples were aspirated, and cells were challenged with wt VSV at 2.24 × 104 PFU/ml in 100 μl of medium. Controls included cells infected with VSV alone and cells that were not challenged with VSV. Cells were incubated overnight at 37°C, medium was aspirated, and cells were fixed with 95% ethanol. Cells were then stained with a 0.1% crystal violet solution in methanol. Absorbance was read at 550 nm on an ELISA reader.
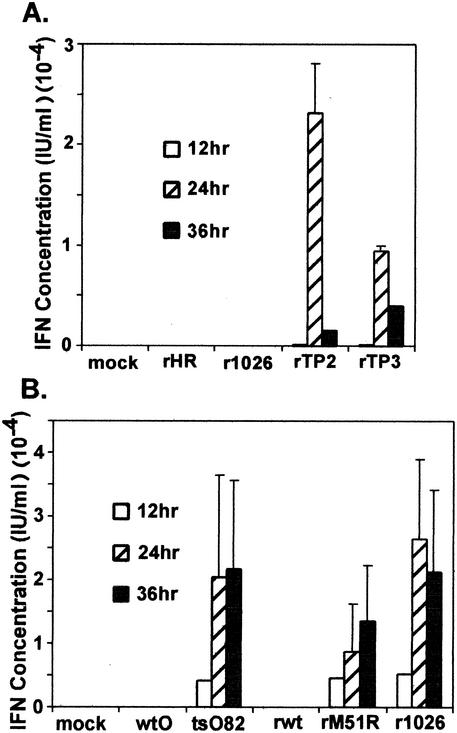
FIG. 5.
IFN bioactivity produced by cells infected with wt and mutant M protein viruses. HeLa cells were incubated overnight at 37°C with serial dilutions of supernatants (100 μl) collected from HeLa (A) and PC-3 (B) cells infected with wt and mutant viruses. The samples were aspirated, and cells were challenged with wt VSV at 2.24 × 104 PFU/ml in 100 μl of medium. Cells were incubated overnight at 37°C, medium was aspirated, and cells were fixed and stained with crystal violet. Absorbance was read at 550 nm on an ELISA reader. The IFN concentration (in international units per milliliter) was quantitated by comparing results to those in cells incubated with serial fivefold dilutions of an IFN standard. Data shown are means ± standard errors of the means for three independent experiments.
Radiolabeling of infected and transfected cells.
To analyze host and viral protein synthesis during virus infections, HeLa cells in 35-mm dishes were infected with viruses containing wt or mutant M proteins at a multiplicity of 20 PFU/cell in Dulbecco's modified essential medium (DMEM) with 2% fetal calf serum (FCS). At 4, 8, and 12 h postinfection, cells were labeled with a 15-min pulse of [35S]methionine (100 μCi/ml) in a total of 0.3 ml of methionine-free medium. Cells were washed with PBS and harvested in radioimmunoprecipitation assay (RIPA) buffer (0.15 M NaCl, 1% deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 10 mM Tris [pH 7.4]). Cell extracts were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and phosphorimaging as described elsewhere (27).
To determine the amount of M protein expressed by mRNAs encoding wt or mutant M proteins, cells were transfected with various amounts of M mRNA or with 250 ng of pGL3 vector alone. At 5 h posttransfection, cells were washed with PBS and harvested in RIPA buffer. Cell extracts were immunoprecipitated with the anti-M protein monoclonal antibody, 23H12, and processed for electrophoresis as described previously (6). Data were quantitated by phosphorimaging.
RNA synthesis in infected cells.
HeLa cells were infected with viruses containing wt or mutant M proteins at a multiplicity of 20 PFU/cell in DMEM plus 2% FCS at 37°C or were mock infected as a control. The virus was allowed to adsorb for 1 h, and cells were fed with medium containing 2% FCS. Parallel samples were incubated in the presence of actinomycin D (total concentration of 5 μg/ml). At 2, 4, and 6 h postinfection, cells were labeled with [3H]uridine (20 μCi/ml) for 30 min, washed in PBS, and harvested. Cells were resuspended in SDS-lysis buffer containing RNase-proteinase degrader (Invitrogen), and DNA was sheared with a 20-gauge needle. Samples were then precipitated with 7% trichloroacetic acid on ice and washed twice with 7% trichloroacetic acid. Acid-precipitable radioactivity was measured by scintillation counting.
Growth curve assay.
HeLa cells in 35-mm dishes were infected with viruses containing wt or mutant M proteins (multiplicity of infection = 10 PFU/cell) in DMEM containing 2% FCS. At 1 h postinfection, the medium was removed and cells were washed twice with PBS and then fed with 2 ml of DMEM containing 10% FCS. At the indicated times postinfection, 100 μl of medium was removed from the dishes and stored at −70°C. The yield of virus was determined by plaque assays on BHK cells and was expressed as PFU per milliliter.
RESULTS
Effect of mutant M proteins expressed from transfected mRNA on expression from the SV40 promoter.
To determine whether the ability of M protein to inhibit host gene expression is responsible for the ability of the virus to suppress activation of IFN gene expression, we asked if the mutant M proteins from the previously isolated IFN-inducing VSV mutants (19) were defective in their ability to inhibit host gene expression. Therefore, we tested the effect of wt and mutant M proteins expressed from transfected M mRNA, in the absence of other viral components, on expression of luciferase from a plasmid containing the SV40 promoter. These IFN-inducing viruses are derived from the HR strain of VSV, and their M protein mutations are depicted in the diagram in Fig. 1A. The M protein of the IFN-inducing mutant T1026R1 virus (M-T1026) has a substitution of arginine for methionine at position 51 (M51R mutation) of the 229-amino-acid M protein (13, 16). This mutation has been shown previously to render the M protein defective in its ability to inhibit host gene expression (1, 6, 16, 37, 45). M-TP2 and M-TP3 are the M proteins from the TP2 and TP3 mutant viruses, respectively, which were independently isolated from the HR strain based on their IFN-inducing phenotypes (19). These M proteins contain the double substitution V221F and S226R (TP3) and the triple substitution E213A, V221F, and S226R (TP2), which are near the carboxy terminus of the 229-amino-acid M protein (13). The ability of these mutant M proteins to inhibit host gene expression in the absence of other viral components had not been tested previously.
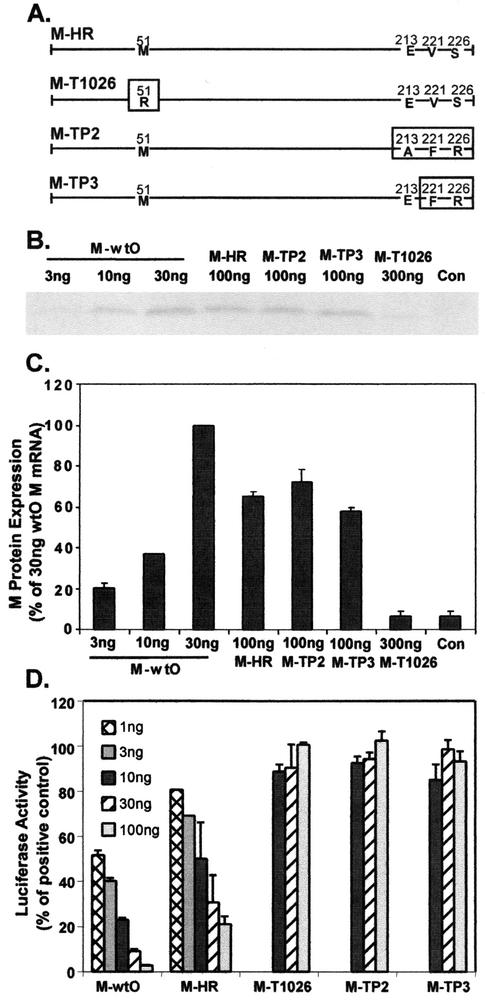
FIG. 1.
Effect of wt and mutant M proteins on expression of luciferase from the SV40 promoter. (A) Diagram representing sequences of M proteins from IFN-inducing mutant viruses of the HR strain of VSV. M-HR is the wt M protein of the HR strain. Mutations in the M-T1026, M-TP2, and M-TP3 proteins are indicated by boxes. (B) Representative image of M proteins expressed from cells transfected with wt and mutant M mRNAs. L cells were transfected with the indicated amounts of wt and mutant M mRNAs for 5 h. Cells transfected with pGL3 plasmid DNA alone were used as a negative control. Cells were labeled with [35S]methionine (200 μCi/ml) for 1 h, and cell extracts were prepared. Extracts were immunoprecipitated with the anti-M protein monoclonal antibody 23H12 and processed for SDS-PAGE and phosphorimaging. (C) Quantitation of labeled M proteins. Results are expressed as the percentage of M protein expressed in cells transfected with 30 ng of wtO mRNA. Data are the means ± standard errors of the means for four experiments. (D) Effect of wt and mutant M proteins on expression of luciferase from the SV40 promoter. HeLa cells were transfected with the indicated amounts of in vitro-transcribed M mRNA together with 250 ng of pGL3 plasmid DNA containing a luciferase gene driven by the SV40 promoter. At 24 h posttransfection, cells were harvested and luciferase activity was measured. Data are presented as the percentage of the activity of controls transfected with pGL3 plasmid DNA in the absence of M mRNA and are the means ± standard errors of the means for eight independent experiments.
M protein inhibits its own transcription when expressed from DNA vectors that depend on host cell transcription, making it difficult to express from recombinant plasmid DNA vectors (5, 7, 29). To circumvent this problem, M protein can be more effectively expressed by transfecting cells with in vitro-transcribed M mRNA instead of plasmid DNA (4). This is because M protein does not inhibit translation of transfected mRNAs in the absence of other viral components. In fact, wt M protein actually stimulates translation of transfected mRNAs, including its own mRNA (4). This leads to higher levels of expression of wt M protein compared to mutant M proteins when cells are transfected with equivalent amounts of M mRNAs (as shown below and in reference 32).
The relative levels of expression from transfected mRNAs encoding M proteins derived from the HR strain (M-HR, M-TP2, M-TP3, and M-T1026) were compared to those of the M protein of the Orsay strain of VSV (M-wtO), which we had studied previously (1, 5, 6). Cells were transfected with 100 ng of wtHR, TP2, or TP3 M mRNAs or 300 ng of T1026 M mRNA and compared to cells transfected with 3, 10, or 30 ng of wtO M mRNA. At 5 h posttransfection, cells were radiolabeled with [35S]methionine for 1 h and lysed. Lysates were immunoprecipitated with the anti-M monoclonal antibody, 23H12, and processed for electrophoresis and phosphorescence imaging. Similar results were obtained in L cells and HeLa cells. However, only the data from L cells were quantitated due to the fact that immunoprecipitates from HeLa cell lysates contained high levels of background proteins, making it difficult to quantitate the amount of M protein expressed from M mRNA. An image of the labeled M protein bands is shown in Fig. 1B. The wt and mutant M proteins synthesized by each of the M mRNAs were quantitated and are shown as a percentage of the M protein expressed in cells transfected with 30 ng of wtO M mRNA (Fig. 1C). The M proteins derived from the HR strain were less effectively expressed than the wtO M protein, so that at least three times more wtHR, TP2, or TP3 M mRNA was needed to achieve levels of M protein expression comparable to that of wtO M protein. The amount of M protein obtained by transfecting 300 ng of T1026 M mRNA was close to background levels. Therefore, it appears that the T1026 M protein was not expressed efficiently from transfected mRNA. In contrast, the TP2 and TP3 M proteins were expressed as efficiently as wtHR M protein. The low level of detection of the T1026 M protein is not likely to be due to lack of antibody reactivity with the mutant M protein. Even though the M51R mutation in this M protein is near the epitope recognized by this antibody against M protein (37), the M51R mutant M protein of the Orsay strain is immunoprecipitated as efficiently as wt M protein (37; H. Yuan and D. S. Lyles, unpublished data).
To determine the ability of wt and mutant M proteins to inhibit host-directed gene expression, HeLa cells were transfected with 250 ng of plasmid DNA encoding luciferase expressed from the SV40 promoter, together with varying amounts of wt or mutant M mRNA. At 24 h posttransfection, cell extracts were prepared, and luciferase activity was measured. Data are expressed as a percentage of the luciferase activity obtained from cells transfected with the luciferase plasmid in the absence of M mRNA (Fig. 1D). The wtO M protein inhibited luciferase expression from the plasmid containing the SV40 promoter with approximately 50% inhibition when cells were transfected with 1 to 3 ng of M mRNA. We have previously shown by nuclear runoff experiments that M protein inhibits expression from the SV40 promoter at the transcriptional level (1, 5). It is also possible that inhibition of nuclear-cytoplasmic mRNA transport contributes to the inhibition of luciferase expression (5, 24, 37, 39). Similar to wtO M protein, the wt HR M protein also inhibited luciferase expression, although 10 ng of HR M mRNA was required to achieve 50% inhibition. Since more than threefold more HR M mRNA was required to give levels of expression equivalent to those of wtO M mRNA (Fig. 1C), these data indicate that the potency of the wtHR M protein is similar to that of the wtO M protein, when the relative expression levels are considered. All of the M protein mutants of the HR strain (TP2, TP3, and T1026) were defective in their ability to inhibit luciferase expression. In fact, luciferase activity remained constant in cells transfected with concentrations of each of the mutant M mRNAs that were 10-fold higher than those used with the wt M mRNAs (Fig. 1D). In the case of the TP2 and TP3 M proteins, the inability to inhibit luciferase expression cannot be accounted for by low levels of expression, since they were expressed as efficiently as the wtHR M protein (Fig. 1C). In the case of the T1026 M protein, the low level of expression could be responsible, in part, for its failure to inhibit luciferase expression. Taken together these results indicate that the mutant M proteins are defective in their ability to inhibit host-directed gene expression when expressed in transfected cells in the absence of other viral gene products.
Mutant M proteins induce expression of luciferase from the IFN-β promoter.
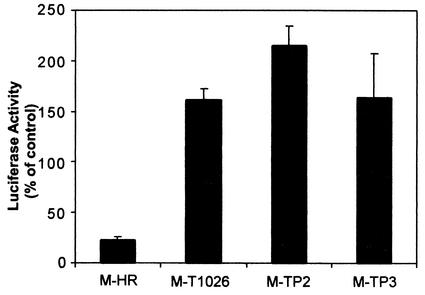
Data in Fig. 1 indicate that mutant M proteins from IFN-inducing VSV mutants are defective in their ability to shut off luciferase gene expression driven from the SV40 promoter. In a similar assay, we also tested the effect of wt and mutant M proteins on expression of luciferase from a plasmid containing the IFN-β promoter. HeLa cells were transfected with 1 μg of plasmid DNA encoding luciferase expressed from the IFN-β promoter (pβlux), together with 100 ng of wtHR, TP2, and TP3 M mRNAs or 300 ng of T1026 M mRNA. At 16 h posttransfection, cell extracts were prepared, and luciferase activity was measured. In order to compare data between different experiments, luciferase activities are expressed as a percentage of the activity in cells transfected with pβlux alone. Results in Fig. 2 show that the wt M protein of the HR strain inhibited expression of luciferase from the plasmid expressing the IFN-β promoter. This result is similar to previous results obtained using a different reporter gene driven by the IFN-β promoter (16). However, each of the mutant M proteins failed to inhibit luciferase expression from the plasmid containing the IFN-β promoter. In fact, luciferase was expressed at somewhat higher levels in cells expressing mutant M proteins than in control cells. Such stimulation of gene expression by mutant M proteins has been observed previously (2, 6). However, the basis for this effect has not been explored. The important conclusion from Fig. 2 is that the M proteins of these IFN-inducing VSV mutants are defective in their ability to inhibit luciferase activity driven by the IFN-β promoter, similar to their inability to inhibit the activity from the SV40 promoter (Fig. 1).
FIG. 2.
Effect of wt and mutant M proteins on expression of luciferase from the IFN-β promoter. HeLa cells were transfected with 1 μg of pβlux encoding luciferase expressed from the IFN-β promoter, together with 100 ng of wt, TP2, and TP3 M mRNA or 300 ng of T1026 M mRNA. At 16 h posttransfection, cell extracts were prepared, and luciferase activity was measured. Luciferase activities are expressed as a percentage of the activity in cells transfected with pβlux alone and are the means ± standard errors of the means for four independent experiments.
M protein mutations contribute to defects in IFN suppression in the context of a virus infection.
The M genes from the IFN-inducing mutants of VSV were incorporated into the wt background of a recombinant VSV infectious cDNA clone to determine whether the M protein mutations contribute to their IFN-inducing phenotype in the context of the virus infection. In addition to the wt and mutant M proteins derived from the HR strain of VSV, we also tested the effects of mutations in M genes of additional virus strains (Orsay and San Juan). The viruses used in our study are diagramed in Fig. 3. Viruses 1 and 2 are naturally occurring viruses derived from the Orsay strain of VSV, and the remaining viruses are recombinant viruses isolated from VSV infectious cDNA clones. Virus 1 is the wt Orsay strain (wtO), and virus 2 is the tsO82 mutant derived from wtO virus, which was shown previously to induce higher levels of IFN production than wtO virus (34). tsO82 virus contains the methionine-to-arginine substitution in position 51 (M51R substitution) in the M protein sequence (11). The M51R mutation was introduced into the wt background of our recombinant wt virus (rwt virus; number 3 in Fig. 3) to generate rM51R-M virus (number 4), using infectious cDNA clones modified slightly (27) from the one described by Whelan et al. (42). The M proteins of these viruses are derived from the San Juan strain of VSV. The San Juan M protein differs from the HR M protein by seven amino acid substitutions, as indicated in Fig. 3.
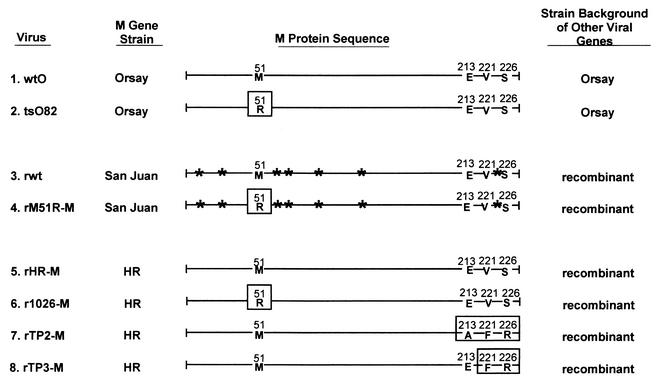
FIG. 3.
Viruses used in this study. The diagram represents the sequences of the M proteins of the viruses used in this study. Mutations in the M proteins are indicated by boxes. The tsO82 virus (number 2) is a naturally occurring mutant of the Orsay strain of VSV (number 1) containing the M51R mutation (11). The remaining viruses are recombinants isolated from VSV infectious cDNA clones, which differ only in their M genes. The M genes of the original recombinant wt (rwt) virus (3) and rM51R-M mutant (4) are derived from the San Juan strain (27, 42). The viruses containing the wt M protein from the HR strain (rHR-M virus) (5), the M51R mutation in the HR M protein (r1026-M virus) (6), and the TP2 and TP3 mutations in the HR M protein (7 and 8) were generated for this study. Sites of amino acid differences between the San Juan and HR strains are indicated by asterisks.
Viruses 5 to 8 are recombinant viruses containing M proteins derived from the HR strain of VSV (as shown in Fig. 1A). Virus 5 (rHR-M virus) contains the wtHR M protein, and virus 6 (r1026-M virus) contains the M protein of the IFN-inducing mutant virus T1026R1. As mentioned previously, the M protein of r1026-M contains the same M51R mutation found in the tsO82 and rM51R-M viruses. Viruses 7 and 8 contain the M proteins of the TP2 and TP3 mutant viruses, which contain mutations near the carboxy terminus of the M protein (13).
The effect of M protein mutations on the ability of these viruses to stimulate IFN gene expression was determined by transfecting HeLa cells with the plasmid encoding luciferase under control of the IFN-β promoter (pβlux). At 24 h posttransfection, cells were infected with viruses containing wt or mutant M proteins. Cells were harvested at 3, 6, or 9 h postinfection, and lysates were tested for luciferase activity. Data are expressed as a percentage of the luciferase activity in uninfected cells transfected with a control plasmid in which luciferase was expressed from the SV40 promoter (Fig. 4). Other controls included cells transfected with pβlux alone to determine unstimulated luciferase levels and cells treated with the dsRNA analog poly(I)-poly(C) to stimulate IFN gene expression.
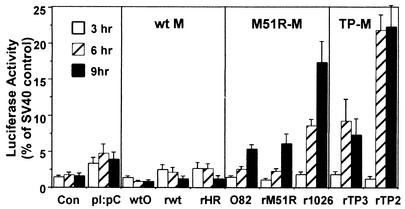
FIG. 4.
Effect of viruses containing wt or mutant M proteins on the activity of the IFN-β promoter. HeLa cells were transfected with 1 μg of pβlux plasmid DNA encoding luciferase under control of the IFN-β promoter. At 24 h posttransfection, cells were infected with viruses containing wt or mutant M proteins at a multiplicity of 20 PFU/cell. Cells were harvested at 3, 6, and 9 h postinfection, and luciferase activity was determined. Cells were transfected with pβlux DNA and mock infected as negative controls (Con), and cells were transfected with pβlux DNA and then treated with poly(I)-poly(C) (pI:pC) as positive controls. Data are expressed as a percentage of the luciferase activity expressed by uninfected cells transfected with 250 ng of pGL3 plasmid DNA to detect constitutive luciferase activity from the SV40 promoter. Data shown are means ± standard errors of the means for four independent experiments.
All of the viruses containing wt M proteins (wtO, rwt, and rHR-M viruses) were unable to activate IFN gene expression, as demonstrated by little if any increase in luciferase levels over those of the negative control. In contrast, all of the viruses containing the M51R M protein mutation (tsO82, rM51R-M, and r1026-M viruses) induced luciferase activity to levels as high as or higher than that of the positive control treated with poly(I)-poly(C). Likewise, the rTP2-M and rTP3-M viruses, containing the carboxy-terminal substitutions in M protein, induced high levels of luciferase expression. Quantitatively, luciferase activity in cells infected with M protein mutant viruses was stimulated 2- to 10-fold over that seen with viruses containing wt M proteins. Pairwise comparison of recombinant viruses containing wt versus mutant M proteins derived from the same virus strain indicated that mutations in M protein are responsible for the IFN-inducing phenotypes of the recombinant viruses. For example, rM51R-M virus induced higher levels of luciferase than its wt control, rwt virus, and r1026-M virus induced higher levels of luciferase than its wt control, rHR-M virus.
The principal conclusion to be drawn from Fig. 4 is that viruses with wt M proteins inhibit expression of luciferase from the IFN promoter, while viruses with mutant M proteins activate luciferase expression. However, the data also show that differences in the virus strains from which the M proteins were derived play a role in dictating the amount of stimulation of IFN gene expression by M protein mutant viruses. For example, r1026-M virus, which contains the M51R mutation in the M gene from the HR strain, induced higher levels of luciferase than the rM51R-M virus, which contains the same M51R mutation in the M gene from the San Juan strain. The M proteins of these two recombinant viruses differ by seven amino acid substitutions, but the other viral genes besides the M gene are identical. Thus, the strain differences in the M proteins of these two viruses are responsible for the difference in induction of luciferase expression.
To confirm the results of the luciferase assay, we analyzed the IFN activity produced by cells infected with wt and mutant M protein viruses by an IFN bioassay. This assay is based on the reduction of VSV cytopathic effect by supernatants collected from infected cells. HeLa cells were infected with wt and mutant M protein viruses, and aliquots of the supernatant media at 12, 24, and 36 h postinfection were tested for IFN activity (Fig. 5A). The rTP2-M and rTP3-M viruses, containing carboxy-terminal mutations in the M gene, induced IFN activity in HeLa cells (Fig. 5A). However, the r1026-M virus, containing the M51R mutation, and the rHR-M virus, containing wt M protein, did not induce detectable IFN activity in HeLa cells. We noted that the rTP2-M and rTP3-M viruses required approximately 24 h to induce detectable levels of IFN activity. By this time, HeLa cells infected with the r1026-M virus were already dead due to virus-induced cellular apoptosis, similar to previous data (27). Therefore, we tested the ability of wt and M51R mutant viruses to induce IFN activity in PC-3 cells, a human prostate tumor cell line that is more resistant to VSV-induced killing (unpublished data). Results in Fig. 5B indicate that both viruses containing wt M proteins (wtO and rwt) were unable to induce IFN activity in PC-3 cells. However, the tsO82, rM51R-M, and r1026-M viruses containing the M51R mutation in M proteins from different virus strains induced IFN activity to varying levels. Therefore, the overall conclusion from these data is that viruses containing wt M proteins are effective suppressors of IFN activity, while viruses with M protein mutations induce IFN activity. Once again, M proteins derived from different virus strains appear to play a role in dictating the degree of IFN activity induced by viruses with M protein mutations. Not only did the r1026-M virus induce higher levels of luciferase expression from the IFN-β promoter than the rM51R-M virus (Fig. 4), it also induced higher levels of IFN activity in the bioassay (Fig. 5B).
M protein mutant viruses are defective at inhibiting host RNA and protein synthesis.
The data in Fig. 4 and 5 indicate that M protein mutations are responsible for the ability of the recombinant M protein mutant viruses to activate IFN gene expression. Therefore, we can conclude that M protein mutations contribute to the IFN-inducing phenotypes of the original mutant viruses from which these M proteins were derived. The data in Fig. 1 and 2 suggest that the IFN-inducing phenotype of these viruses is due to defects in the inhibition of host gene expression. To test this hypothesis, synthesis of host RNA and proteins in cells infected with wt and M protein mutant viruses was determined by pulse-labeling experiments. Cells were infected with wt and mutant viruses and labeled with [3H]uridine at 2, 4, and 6 h postinfection to determine the ratio of viral and host RNA synthesis. The time of the pulse (30 min) was short compared to the RNA turnover rate, so that labeling primarily reflected the rates of synthesis rather than turnover. Cells were lysed, and trichloroacetic acid-insoluble radioactivity was measured to determine the total cellular RNA synthesis (host plus viral). Parallel samples were treated with actinomycin D to inhibit host RNA synthesis. In these samples only viral RNA would be labeled, since actinomycin D does not affect viral RNA synthesis. Host RNA synthesis was calculated by subtracting radioactivity in samples treated with actinomycin D from the total radioactivity in the absence of actinomycin D, and it is expressed as a percentage of the mock-infected controls (Fig. 6). Viruses with wt M proteins effectively inhibited host RNA synthesis, so that by 6 h postinfection levels of host RNA synthesis were 10 to 20% of controls. However, all of the M protein mutant viruses were defective in their ability to inhibit host RNA synthesis. Comparison of recombinant M protein mutant viruses with their isogenic counterparts with wt M proteins (rM51R-M versus rwt viruses, and r1026-M, rTP2-M, or rTP3-M versus rHR-M viruses) shows that M protein plays a major role in the inhibition of host RNA synthesis at early times postinfection.
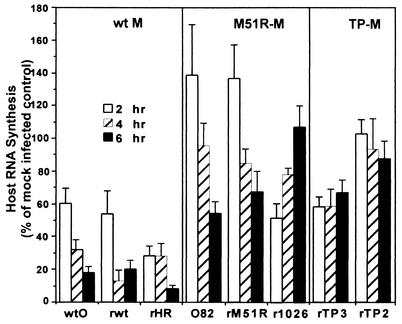
FIG. 6.
Inhibition of host RNA synthesis by viruses containing wt or mutant M proteins. HeLa cells were infected with viruses containing wt or mutant M proteins at a multiplicity of 20 PFU/cell. At 2, 4, and 6 h postinfection, cells were labeled with [3H]uridine (20 μCi/ml) for 30 min. Cells were lysed in SDS-lysis buffer, and aliquots were precipitated with trichloroacetic acid to measure acid-insoluble radioactivity. Parallel samples were incubated in the presence of actinomycin D, so that only viral RNA would be labeled. The rate of host RNA synthesis was calculated by subtracting the radioactivity in viral RNA from the total radioactivity. Data are expressed as a percentage of the uninfected cell control and are means ± standard errors of the means for five experiments.
The data in Fig. 6 also show that M protein mutants from different virus strains have slightly different effects on host RNA synthesis, similar to their differences in IFN gene induction. The tsO82 and rM51R-M viruses actually stimulated host RNA synthesis at early times postinfection (2 h), which then declined to about 60 to 70% of control by 6 h. In contrast, the r1026-M virus had an unusual effect in that host RNA synthesis decreased to 60% of control at 2 h postinfection, but host RNA synthesis increased over the time course of the experiment to 100% of control by 6 h postinfection. Cells infected with the rTP3-M and rTP2-M viruses maintained a constant level of RNA synthesis over the time course at around 60 and 90% of controls, respectively.
The inhibition of host gene expression in VSV-infected cells also involves an inhibition of host protein synthesis. The inhibition of host protein synthesis is not due to depletion of cellular mRNAs as a result of the inhibition of host transcription or transport. In fact, the cytoplasm of infected cells contains normal amounts of cellular mRNAs that can be effectively translated in vitro (30). Instead, the inhibition is due to inactivation of host translation factors (10, 14). To determine the ability of wt and mutant viruses to inhibit host translation, HeLa cells were infected and then were pulse-labeled with [35S]methionine for 10 min at 4, 8, or 12 h postinfection. As in the case of RNA synthesis, the time of the pulse-label was short (10 min) compared to the turnover rates of viral and host proteins, so that labeling reflected primarily rates of synthesis. Proteins were solubilized and analyzed by SDS-PAGE and phosphorescence imaging. A representative image from analysis at 8 h postinfection is shown in Fig. 7A. All of the viruses containing wt M proteins (wtO, rwt, and rHR-M) effectively inhibited host protein synthesis compared to the mock-infected control. This can be clearly seen in regions of the gel that are devoid of viral proteins, such as the region between the L and G proteins. In contrast, the viruses containing the M51R M protein mutation (tsO82, rM51R-M, and r1026-M) were much less effective in their ability to inhibit host protein synthesis. It is also apparent from Fig. 7A that the viruses containing wt M protein synthesized viral proteins at a very high level, despite the inhibition of host protein synthesis. The M protein mutants synthesized viral proteins at levels at least as high as their corresponding wt controls.
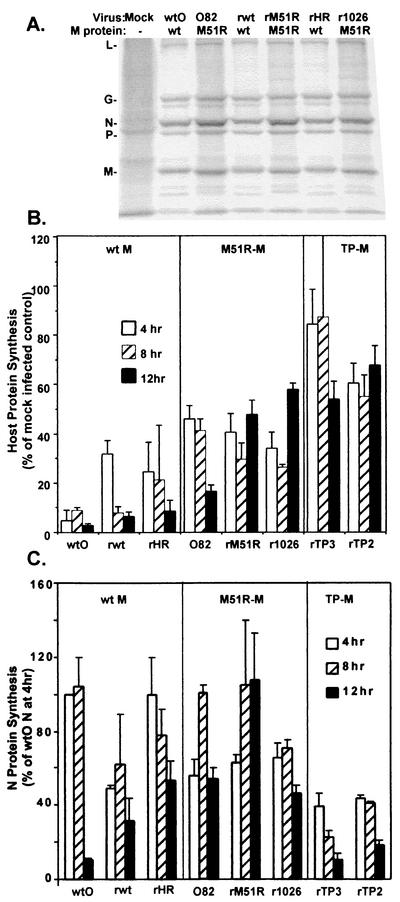
FIG. 7.
Inhibition of host protein synthesis by viruses containing wt or mutant M proteins. HeLa cells were infected with viruses containing wt or mutant M proteins at a multiplicity of 20 PFU/cell or were mock infected as a control. Cells were labeled with a 15-min pulse of [35S]methionine (100 μCi/ml) at 4, 8, and 12 h postinfection. Lysates were subjected to SDS-PAGE, and labeled proteins were quantitated by phosphorimaging. (A) Representative image from analysis of viruses containing wt or M51R mutant M proteins at 8 h postinfection. Positions of viral proteins are indicated on the left. (B) Host protein synthesis was determined from images similar to that in panel A in regions of the gel devoid of viral proteins between the L and G proteins and between the P and M proteins. Results are shown as a percentage of the mock-infected control and are the mean ± standard error of the mean of four independent experiments. (C) Effect of M protein mutations on viral protein synthesis. HeLa cells infected with viruses containing wt or mutant M proteins were labeled with [35S]methionine, and the labeled proteins were analyzed by SDS-PAGE and phosphorimaging as described in the legend for Fig. 4. The labeled M proteins in images similar to those shown in Fig. 4A were quantitated and are expressed as a percentage of the wtO M protein labeled at 4 h postinfection. Data are the mean ± standard error of the mean of four experiments.
Host protein synthesis in infected cells at 4, 8, and 12 h postinfection was determined from images similar to Fig. 7A by quantitation of the radioactivity in two regions of the gel that were devoid of viral proteins (between L and G and between P and M) and is shown in Fig. 7B as the percentage of a mock-infected control. Each of the viruses containing wt M proteins (wtO, rwt, and rHR-M) effectively inhibited host protein synthesis, so that by 12 h postinfection host protein synthesis levels were 5 to 10% of the mock-infected control. In contrast, the M51R M protein mutant viruses (tsO82, rM51R-M, and r1026-M) were less effective than their wt controls at repressing host translation, which was maintained at a level of 40 to 50% of control throughout the 12-h time course of the experiments. The viruses containing the TP2 and TP3 M proteins were even more defective in their ability to inhibit host protein synthesis, which was maintained at a level of 60 to 80% of control. These data together with the data in Fig. 6 demonstrate that the ability of the mutant M protein viruses to induce IFN gene expression is correlated with a reduction in their capacity to shut off both host transcription and host translation.
Ability of recombinant viruses to synthesize viral proteins and produce infectious progeny.
The rates of synthesis of viral proteins in cells infected with VSVs containing wt or mutant M proteins were determined from images similar to those in Fig. 7A. Radioactivity in the N protein band is shown in Fig. 7C and is expressed as a percentage of the N protein synthesis at 4 h postinfection with the wtO virus, which was near the maximum amount. Cells infected with recombinant viruses containing wt M protein (rwt and rHR-M viruses) synthesized N protein at different rates throughout the 12-h time course. Synthesis of N protein by rwt virus was around 55% of the level of the wtO virus control at 4 h postinfection, while that of rHR-M virus was around 80% of control. Since these viruses are isogenic except for the strain differences in their M proteins, these results reflect the influence of M protein on viral gene expression. Similar results were obtained with analysis of M protein synthesis (data not shown). The N protein/M protein ratios in cells infected with all of the viruses were the same as the ratio for wtO virus, with the exception of cells infected with rM51R-M virus, in which the ratio of N protein to M protein was approximately 40% higher. Since this difference was not observed with other viruses containing the M51R M protein mutation (tsO82 and r1026-M viruses), this provides further evidence that the virus strain from which the M protein was derived influenced viral gene expression.
We have previously shown that the M51R-M proteins display no differences in their turnover rates compared to wt-M proteins (6). Results shown in this paper also indicate that, as a general trend, the viruses containing the M51R M protein mutation synthesized viral proteins at levels similar to or greater than that of their wt counterparts (Fig. 7C). The tsO82 virus synthesized viral proteins at levels similar to that of the wtO virus through 8 h postinfection. However, by 12 h, the tsO82 virus expressed greater amounts of N protein than wtO did. Similarly, the rM51R-M virus expressed viral proteins at levels comparable to those of rwt at early times postinfection. However, by 8 h postinfection, rM51R-M actually expressed greater amounts of N protein than any of the other viruses. Interestingly, the r1026-M virus expressed lower amounts of viral proteins at early times postinfection, but by 8 h postinfection it expressed levels of N protein comparable to its wt counterpart (rHR-M). In contrast, the recombinant viruses containing the TP2 and TP3 M proteins synthesized less viral protein than their rHR-M control at all time points (Fig. 7C). This appeared to be due to a defect in the ability of these viruses to synthesize viral RNA (data not shown). Thus, the inability of M protein mutant viruses to inhibit host RNA and protein synthesis was accompanied by defects in virus replication in the case of the rTP2-M and rTP3-M viruses, but not in the case of the viruses containing the M51R M protein mutation. The defect in viral RNA and protein synthesis exhibited by the rTP2-M and rTP3-M viruses is not dependent on IFN production, since similar results were obtained in BHK cells, which are unresponsive to IFN (data not shown). However, it is possible that a more rapid turnover of these mutant M proteins could be partly responsible for their replication defects.
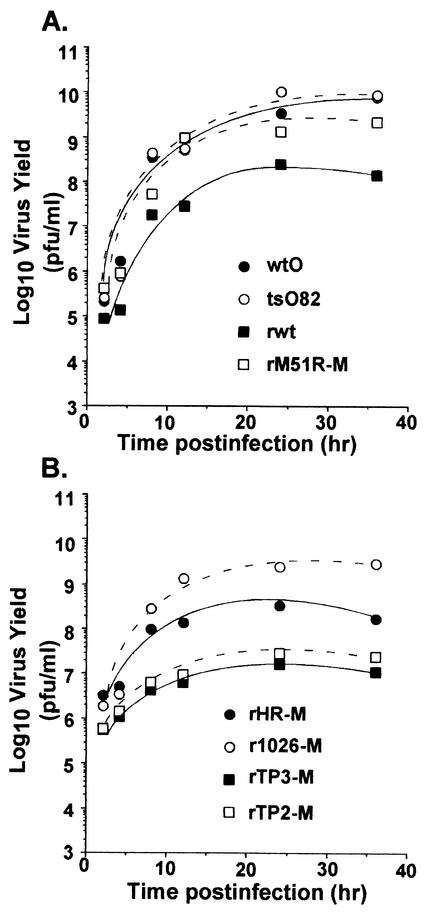
Single-cycle growth experiments were done to determine the ability of wt and mutant M protein viruses to produce infectious progeny (Fig. 8). HeLa cells were infected with wt and mutant viruses at a high multiplicity of infection (10 PFU/cell). At the indicated times postinfection, supernatants were collected and viral titers were determined by plaque assay on BHK cells. The tsO82 virus (Fig. 8A) grew to titers as high as wtO virus. These data are similar to previous results showing that tsO82 virus is not temperature sensitive for virus growth in HeLa cells (22). The rM51R-M (Fig. 8A) and r1026-M (Fig. 8B) mutant M protein viruses actually produced higher levels of infectious progeny than their wt counterparts rwt and rHR-M, respectively. These higher yields of infectious progeny also correlated with higher levels of viral protein synthesis at late times postinfection in the case of cells infected with the rM51R-M virus, as shown in Fig. 7C. Therefore, the data in Fig. 7 and 8 indicate that viruses containing the M51R M protein mutations are not defective in their ability to produce viral proteins or infectious viral progeny. This result suggests that the IFN induced by these viruses has little if any ability to inhibit virus replication in a single-cycle growth experiment. This is the expected result, since most of the viral replicative cycle occurs before the IFN can be produced and then induce the antiviral state.
FIG. 8.
Single-cycle growth analysis. HeLa cells were infected with viruses containing wt or mutant M proteins at a multiplicity of 20 PFU/cell. At 1 h postinfection, the medium was removed, and cells were washed twice. Fresh medium was added to the infected cells, and a small aliquot of the supernatant was removed at the indicated times postinfection to determine the amount of progeny virus by plaque assay. Data are the average of two independent experiments.
In contrast to results obtained by the M51R M protein mutants, recombinant viruses containing the TP2 and TP3 mutations grew to lower titers in single-cycle growth experiments (Fig. 8B) than did rHR-M (Fig. 8B). The lower yield of infectious progeny produced in cells infected with rTP2-M and rTP3-M viruses also correlated with reduced rates of viral RNA and protein synthesis. Therefore, the inability of these mutant M protein viruses to shut off host RNA and protein synthesis may be due in part to defects in their ability to replicate (Fig. 7) and produce infectious progeny (Fig. 8B) in infected cells.
DISCUSSION
The data presented here show that M protein plays a major role in the inhibition of IFN gene expression in VSV-infected cells. Furthermore, the ability of M protein to inhibit IFN production is genetically correlated with the overall inhibition of host RNA and protein synthesis. Previous data had shown that four IFN-inducing mutants of VSV under consideration here, tsO82, T1026R1, TP2, and TP3 viruses, have mutations in their M proteins (11, 13, 16). However, it was not known whether these viruses have additional mutations in genes other than their M genes that account for their IFN-inducing phenotype. It had been shown previously that the M51R mutation in the M proteins of tsO82 and T1026R1 viruses render these M proteins defective in their ability to inhibit host gene expression in the absence of other viral components (1, 6, 16, 37). These results were extended here to show that the mutant M proteins of TP2 and TP3 viruses are also defective in the inhibition of host gene expression (Fig. 1), including expression from the IFN-β promoter (Fig. 2). Thus, all of the M proteins of the IFN-inducing mutant viruses tested in this study are defective in their ability to inhibit host gene expression.
The fact that M protein inhibits expression of luciferase from a plasmid containing the IFN-β promoter in transfected cells (15) (Fig. 2) still left open the possibility that M protein is not responsible for suppression of IFN induction in virus-infected cells. For example, if the inhibition of host gene expression by M protein were to occur only at late times postinfection, as has been proposed (40), this inhibition may be too late to prevent IFN synthesis. Indeed, the argument has been made that the IFN-inducing activity of these viruses is due entirely to mutations in genes other than the M gene (34). This issue was addressed here by incorporating the M protein mutations onto the wt background of a VSV infectious cDNA clone. The resulting recombinant mutant M protein viruses induced expression of a luciferase reporter gene driven by the IFN-β promoter (Fig. 4) and induced IFN activity as measured by an IFN bioassay (Fig. 5), while the recombinant viruses containing wt M proteins did not. Furthermore, the recombinant mutant M protein viruses were defective in the inhibition of host RNA and protein synthesis (Fig. 6 and 7). These data indicate that M protein plays a main role in the inhibition of host gene expression in VSV-infected cells and is a major suppressor of IFN gene expression.
The data presented in Fig. 4 and 5 serve to establish that the M protein mutations in the original IFN-inducing viruses account at least in part for their IFN-inducing phenotypes. However, these results do not rule out the possibility that these viruses contain additional mutations that contribute to their ability to induce IFNs. Indeed, there are several examples of IFN-inducing mutants of VSV that do not contain M protein mutations (13, 19, 34), indicating that other viral genes also play a role in determining the extent of IFN gene activation in virus-infected cells. We propose that these mutations enhance the activity of viral inducers of IFN to the extent that they overcome the inhibitory effects of M protein. Alternatively, these mutations may affect other inhibitors of IFN gene expression besides M protein.
Viruses containing the same M51R substitution in the context of M proteins derived from different virus strains (rM51R-M and r1026-M viruses) differ in their ability to induce IFN gene expression (Fig. 4 and 5). This suggests that the relative contribution of M protein versus other viral proteins in regulating IFN gene expression may be dependent on strain differences. For example, the effect of the M51R mutation may be modulated by variable surrounding amino acids in the M protein. We have also found that in different cell lines the extent of IFN induction by each of the M51R mutants also varies (unpublished data). Therefore, it is possible that the role of M protein in the activation of IFN gene expression is also dependent on the presence of specific host factors in different cell types.
The ability of mutant M protein viruses to induce IFN gene expression was correlated with defects in the ability of the mutant M proteins to inhibit host RNA and protein synthesis (Fig. 4, 5, 6, and 7). These results support our model in which wt M protein functions as a suppressor of IFN gene expression as a result of its general ability to inhibit host RNA and protein synthesis (31). According to this model, there must be other products of virus infection, such as viral dsRNA, that activate IFN gene expression, which is then suppressed by the activity of wt M protein. In the case of the mutant M protein viruses, the enhanced activation of IFN gene expression compared to that of viruses with wt M proteins would be a result of the absence of this inhibitory activity. In support of this model, the IFN-suppressing activity of wt VSV is dominant over the IFN-inducing activity of the M protein mutant T1026R1 virus in mixed infections (33). Likewise, coinfection with wt VSV and heterologous IFN-inducing viruses suppresses IFN production (33). It is also possible that the induction of IFN by mutant M protein viruses may stimulate antiviral genes, including nucleoporins, that contribute to their inability to further shut off host gene expression at the level of RNA transport (15).
How does M protein suppress IFN gene expression? VSV inhibits host gene expression at multiple levels, including inhibition of host transcription, inhibition of nuclear-cytoplasmic transport of host RNA, and inhibition of host translation (31). M protein inhibits host transcription and nuclear-cytoplasmic RNA transport both in virus-infected cells and when expressed in transfected cells in the absence of other viral components (1, 5, 24, 37, 39). Inhibition at the transcriptional level has been demonstrated by nuclear runoff assays (1, 5). The inhibition of host RNA polymerase II-dependent transcription is due, at least in part, to inactivation of the general transcription initiation factor TFIID, which is the transcription factor that binds to the TATA box upstream of most RNA polymerase II-dependent promoters (44, 45).
The M protein-induced inhibition of host nuclear-cytoplasmic RNA transport has been attributed to the interaction of M protein with a nuclear pore component, which has been identified as the nucleoporin Nup98 (37, 39). The M protein-induced inhibition of nuclear-cytoplasmic RNA transport has been demonstrated convincingly in Xenopus laevis oocytes, in which there is little if any inhibition of transcription (24, 37, 39). However, it has been difficult to quantitate the contribution of the inhibition of transport in transfected mammalian cells, due to the concomitant inhibition of transcription (15, 39). The block in nuclear-cytoplasmic transport in VSV-infected cells is evident from its effects on the processing of small nuclear RNAs (20) and rRNAs (43). However, a careful series of biochemical experiments, including pulse-chase and subcellular fractionation experiments, suggested that changes in transport or turnover of host RNA in VSV-infected cells were minor compared to the profound inhibition of host transcription (41).
In addition to the inhibition of host RNA synthesis, the inhibition of host gene expression in VSV-infected cells involves a dramatic inhibition of host protein synthesis. This inhibition is not due to depletion of cellular mRNAs resulting from the inhibition of transcription (30). Instead, the inhibition is due to inactivation of host translation factors (10, 14). In contrast to the M protein-induced inhibition of host transcription and nuclear-cytoplasmic transport, M protein cannot inhibit host translation when expressed in transfected cells in the absence of other viral components (4). However, mutant M protein viruses fail to inhibit host protein synthesis as effectively as viruses containing wt M proteins (Fig. 4) (25, 27). This suggests that M protein does play a role in inhibition of host translation, but that one or more additional viral components are required to inhibit host translation. There are multiple translation initiation factors whose activity is reduced in VSV-infected cells, including eIF2, eIF4F, and eIF4B (10, 14), although it has not been determined which of these factors is inhibited in response to M protein versus other viral components.
The ability of M protein to inhibit host gene expression at multiple levels is analogous to the activity of other viral suppressors of the host IFN response, which also function at multiple levels (31). This appears to reflect the fact that no single inhibitory mechanism is completely effective at suppressing IFN production. As an example, the influenza A virus NS1 protein contains an RNA-binding domain which suppresses IFN gene activation by sequestering viral dsRNA (reviewed in reference 21). In addition, the NS1 protein contains an activation domain that enables the protein to suppress the processing and nuclear-cytoplasmic transport of host mRNAs, which appears to also play a role in suppressing the host antiviral response (reviewed in reference 31). Similarly, the vaccinia virus E3L protein suppresses activation of IFN gene expression by sequestering viral dsRNA (38). In addition, vaccinia virus encodes the K3L protein, which functions as an inhibitor of the IFN-inducible protein kinase R (12), and the B18R protein, which inhibits cellular responses to IFN by acting as a decoy receptor (3). Thus, the idea that viral proteins, such as M protein, suppress IFN gene expression by multiple mechanisms is a common theme in virus-host interactions.
All of the mutant M proteins analyzed here were defective in their ability to inhibit host gene expression (Fig. 8). However, the viruses containing these mutations were not phenotypically identical in terms of the levels of viral protein expression or the levels of progeny virus produced. The viruses containing the M51R M protein mutation expressed viral proteins at levels at least as high as their wt controls. In fact, at late times postinfection, the tsO82 and rM51R-M viruses expressed even higher levels of viral proteins than their wt counterparts (Fig. 7), leading to correspondingly higher levels of progeny virus production as seen clearly in the case of the rM51R-M virus (Fig. 8). This result indicates that the IFN induced by these viruses has little if any ability to inhibit virus replication in a single-cycle growth experiment. However, the effects of these mutations on the multiple cycles of virus infection that occur in intact animals may be quite profound, since the IFN response is a major determinant of viral pathogenesis and tissue tropism in vivo (reviewed in references 21 and 31). This issue will be addressed in our future experiments.
In contrast to viruses containing the M51R mutation, the viruses containing the TP2 and TP3 M protein mutations expressed lower levels of viral proteins than their control virus containing wt M protein (rHR-M virus), and they produced correspondingly lower levels of viral progeny. These data indicate that M protein mutations affect the level of viral gene expression as well as the level of host gene expression. This idea has been put forth previously, based on the observation that temperature-sensitive (ts) mutations in M protein increase the level of viral mRNA synthesis at the nonpermissive temperature (9). However, in contrast to ts M protein mutations, the TP2 and TP3 M protein mutations appear to decrease the level of viral RNA synthesis (unpublished data).
A role for M protein as a regulator of viral gene expression is also supported by the effects of M proteins from different virus strains. The rM51R and r1026-M viruses are identical except for differences in the virus strains from which their M proteins were derived. Nevertheless, viral protein expression in cells infected with the rM51R virus is approximately 40% greater at 8 to 12 h postinfection than in cells infected with the r1026-M virus (Fig. 7), providing further evidence that M protein can also regulate viral gene expression. The strain differences in their M proteins also account for the fact that r1026-M virus induced higher levels of IFN gene expression than rM51R-M virus (Fig. 4 and 5).
The results presented here support the idea that M protein inhibits IFN gene expression in parallel with the general inhibition of host RNA and protein synthesis. However, it is also possible that M protein inhibits additional steps in the production of IFNs upstream of the general transcription factor TFIID. For example, the activation of NF-κB, one of the factors required for IFN gene transcription, is delayed in cells infected with wt VSV compared to cells infected with the T1026R1 mutant (8). This suggests that the wt M protein delays activation of NF-κB, which could play a role in the suppression of IFN gene expression. Future experiments will determine the effect of M protein on upstream activators of IFN gene expression.
Acknowledgments
We thank Griffith Parks, David Ornelles, and members of the Lyles laboratory for helpful advice and comments on the manuscript. We also thank Margie McKenzie for isolating virus from the infectious clones.
This work was supported by Public Health Service grants AI 32983 and AI 15892 from the National Institute of Allergy and Infectious Diseases, by grant PC010058 from the U.S. Army Medical Research and Materiel Command to D. S. Lyles, and by funds from the Natural Sciences and Engineering Council of Canada and Université du Québec à Montréal to L. Poliquin.
REFERENCES
- 1.Ahmed, M., and D. S. Lyles. 1998. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J. Virol. 72:8413-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, M., and D. S. Lyles. 1997. Identification of a consensus mutation in M protein of vesicular stomatitis virus from persistently infected cells that affects inhibition of host-directed gene expression. Virology 237:378-383. [DOI] [PubMed] [Google Scholar]
- 3.Alcami, A., J. A. Symons, and G. L. Smith. 2000. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J. Virol. 74:11230-11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black, B. L., G. Brewer, and D. S. Lyles. 1994. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J. Virol. 68:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, B. L., and D. S. Lyles. 1992. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J. Virol. 66:4058-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, B. L., R. B. Rhodes, M. McKenzie, and D. S. Lyles. 1993. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J. Virol. 67:4814-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blondel, D., G. G. Harmison, and M. Schubert. 1990. Role of matrix protein in cytopathogenesis of vesicular stomatitis virus. J. Virol. 64:1716-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulares, A. H., M. C. Ferran, and J. Lucas-Lenard. 1996. NF-κB activation is delayed in mouse L929 cells infected with interferon suppressing, but not inducing, vesicular stomatitis virus strains. Virology 218:71-80. [DOI] [PubMed] [Google Scholar]
- 9.Clinton, G. M., S. P. Little, F. S. Hagen, and A. S. Huang. 1978. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell 15:1455-1462. [DOI] [PubMed] [Google Scholar]
- 10.Connor, J. H., and D. S. Lyles. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein, 4E-BP1. J. Virol. 76:10177-10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulon, P., V. Deutsch, F. Lafay, C. Martinet-Edelist, F. Wyers, R. C. Herman, and A. Flamand. 1990. Genetic evidence for multiple functions of the matrix protein of vesicular stomatitis virus. J. Gen. Virol. 71:991-996. [DOI] [PubMed] [Google Scholar]
- 12.Davies, M. V., O. Elroy-Stein, R. Jagus, B. Moss, and R. J. Kaufman. 1992. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J. Virol. 66:1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desforges, M., J. Charron, S. Berard, S. Beausoleil, D. F. Stojdl, G. Despars, B. Laverdiere, J. C. Bell, P. J. Talbot, C. P. Stanners, and L. Poliquin. 2001. Different host-cell shutoff strategies related to the matrix protein lead to persistence of vesicular stomatitis virus mutants on fibroblast cells. Virus Res. 76:87-102. [DOI] [PubMed] [Google Scholar]
- 14.Dratewka-Kos, E., I. Kiss, J. Lucas-Lenard, H. B. Mehta, C. L. Woodley, and A. J. Wahba. 1984. Catalytic utilization of eIF-2 and mRNA binding proteins are limiting in lysates from vesicular stomatitis virus infected L cells. Biochemistry 23:6184-6190. [DOI] [PubMed] [Google Scholar]
- 15.Enninga, J., D. E. Levy, G. Blobel, and B. M. A. Fontoura. 2002. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 295:1523-1525. [DOI] [PubMed] [Google Scholar]
- 16.Ferran, M. C., and J. M. Lucas-Lenard. 1997. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J. Virol. 71:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flood, E. A., and D. S. Lyles. 1999. Assembly of nucleocapsids with cytosolic and membrane-derived matrix proteins of vesicular stomatitis virus. Virology 261:295-308. [DOI] [PubMed] [Google Scholar]
- 18.Flood, E. A., M. O. McKenzie, and D. S. Lyles. 2000. Role of M protein aggregation in defective assembly of temperature-sensitive M protein mutants of vesicular stomatitis virus. Virology 278:520-533. [DOI] [PubMed] [Google Scholar]
- 19.Francoeur, A. M., L. Poliquin, and C. P. Stanners. 1987. The isolation of interferon-inducing mutants of vesicular stomatitis virus with altered viral P function for the inhibition of total protein synthesis. Virology 160:236-245. [DOI] [PubMed] [Google Scholar]
- 20.Fresco, L. D., M. G. Kurilla, and J. D. Keene. 1987. Rapid inhibition of processing and assembly of small nuclear ribonucleoproteins after infection with vesicular stomatitis virus. Mol. Cell. Biol. 7:1148-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 22.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Her, L. S., E. Lund, and J. E. Dahlberg. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science 276:1845-1848. [DOI] [PubMed] [Google Scholar]
- 25.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations on the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaptur, P. E., M. O. McKenzie, G. W. Wertz, and D. S. Lyles. 1995. Assembly functions of vesicular stomatitis virus matrix protein are not disrupted by mutations at major sites of phosphorylation. Virology 206:894-903. [DOI] [PubMed] [Google Scholar]
- 27.Kopecky, S. A., M. C. Willingham, and D. S. Lyles. 2001. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. J. Virol. 75:12169-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenard, J. 1996. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology 216:289-298. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., L. Z. Luo, R. M. Snyder, and R. R. Wagner. 1988. Expression of the M gene of vesicular stomatitis virus cloned in various vaccinia virus vectors. J. Virol. 62:776-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lodish, H. F., and M. Porter. 1980. Translational control of protein synthesis after infection by vesicular stomatitis virus. J. Virol. 36:719-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyles, D. S. 2000. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 64:709-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyles, D. S., and M. O. McKenzie. 1997. Activity of vesicular stomatitis virus M protein mutants in cell rounding is correlated with the ability to inhibit host gene expression and is not correlated with virus assembly function. Virology 229:77-89. [DOI] [PubMed] [Google Scholar]
- 33.Marcus, P. I., and M. J. Sekellick. 1985. Interferon induction by viruses. XIII. Detection and assay of interferon induction-suppressing particles. Virology 142:411-415. [DOI] [PubMed] [Google Scholar]
- 34.Marcus, P. I., M. J. Sekellick, C. F. Spiropoulou, and S. T. Nichol. 1993. Interferon induction by viruses. XXII. Vesicular stomatitis virus Indiana: M-protein and leader RNA do not regulate interferon induction in chicken embryo cells. J. Interferon Res. 13:413-418. [DOI] [PubMed] [Google Scholar]
- 35.Noah, D. L., M. A. Blum, and B. Sherry. 1999. Interferon regulatory factor 3 is required for viral induction of beta interferon in primary cardiac myocyte cultures. J. Virol. 73:10208-10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paik, S. Y., A. C. Banerjea, G. G. Harmison, C. J. Chen, and M. Schubert. 1995. Inducible and conditional inhibition of human immunodeficiency virus proviral expression by vesicular stomatitis virus matrix protein. J. Virol. 69:3529-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen, J. M., L. S. Her, V. Varvel, E. Lund, and J. E. Dahlberg. 2000. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol. Cell. Biol. 20:8590-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 39.von Kobbe, C., J. M. A. van Deursen, J. P. Rodrigues, D. Sitterlin, A. Bachi, X. Wu, M. Wilm, M. Carmo-Fonseca, and E. Izaurralde. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell 6:1243-1252. [DOI] [PubMed] [Google Scholar]
- 40.Wagner, R. R., and J. K. Rose. 1996. Rhabdoviridae: the viruses and their replication, p. 561-604. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed)., Fundamental virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 41.Weck, P. K., and R. R. Wagner. 1978. Inhibition of RNA synthesis in mouse myeloma cells infected with vesicular stomatitis virus. J. Virol. 25:770-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whelan, S. P., L. A. Ball, J. N. Barr, and G. T. Wertz. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. USA 92:8388-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, F. S., and J. M. Lucas-Lenard. 1980. Inhibition of ribonucleic acid accumulation in mouse L cells infected with vesicular stomatitis virus requires viral ribonucleic acid transcription. Biochemistry 19:804-810. [DOI] [PubMed] [Google Scholar]
- 44.Yuan, H., S. Puckett, and D. S. Lyles. 2001. Inhibition of host transcription by vesicular stomatitis virus involves a novel mechanism that is independent of phosphorylation of TATA-binding protein (TBP) or association of TBP with TAF subunits. J. Virol. 75:4453-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan, H., B. K. Yoza, and D. S. Lyles. 1998. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results from inactivation of TFIID. Virology 251:383-392. [DOI] [PubMed] [Google Scholar]