Abstract
Circular concatemerization of the recombinant adeno-associated virus (rAAV) genome has been suggested as the predominant process facilitating long-term rAAV transduction in muscle. A recent study (S. Song, P. J. Laipis, K. I. Berns, and T. R. Flotte, Proc. Natl. Acad. Sci. USA 98:4084-4088, 2001) with SCID mice, which are defective in the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), has suggested that DNA-PKcs regulates the removal of free rAAV vector ends in muscle tissue. In the present study, we have sought to evaluate whether a lack of DNA-PKcs activity reduces circularization of rAAV genomes in SCID muscle and whether such a reduction alters the directivity of heterodimerization. Consistent with the previous report, linear rAAV genomes and free vector ends were detected only in DNA-PKcs-deficient muscle by Southern blotting. Appreciable amounts of circular rAAV genomes were detected in both DNA-PKcs-deficient and wild-type muscle samples by Southern blotting and bacterial trapping experiments. The existence of double-D inverted terminal repeat circular intermediates in SCID and wild-type muscles was also supported by their sensitivity to T7 endonuclease I digestion. However, DNA-PKcs-deficient muscle did demonstrate a ∼50% reduction in the abundance of rescued circular genomes, despite equivalent levels of single rAAV transduction seen in wild-type animals. Dual trans-splicing lacZ vectors were used to functionally evaluate directional head-to-tail intermolecular viral genome concatamerization in vivo. Although AAV genomes are processed differently in SCID and wild-type muscles, a comparable level of trans-splicing-mediated β-galactosidase expression was observed in both strains, suggesting that both circular and linear AAV concatemers may have contributed to the trans-splicing-mediated transgene expression. In summary, we have shown that SCID skeletal muscle retains a fairly high capacity to form circular genomes, despite a significant increase in linear vector genomes. Furthermore, the alteration in equilibrium between circular and linear concatemer genomes caused by the lack of DNA-PKcs activity does not appear to significantly affect the efficiency of dual-vector gene expression from head-to-tail linear and/or circular heterodimers.
The development of adeno-associated virus (AAV)-based gene transfer techniques has proven to be an extremely powerful tool for treating many genetic diseases (1, 12). Due to the relatively small packaging size of this single-stranded virus, AAV-based gene therapies have been restricted to diseases that involve small genes, such as that for factor IX in hemophilia B. The development of novel recombinant AAV (rAAV) dual-vector approaches, in which two portions of a transgene cassette are reconstituted in vivo from two independent viral vectors, has offered tremendous potential for expanding AAV gene therapy to larger disease genes (8, 9, 11, 19, 26, 30). Using these approaches, many diseases, such as Duchenne muscular dystrophy and cystic fibrosis, which have large disease genes are now more feasible targets for AAV-mediated treatment.
The most critical molecular process for dual-vector approaches is intermolecular concatamerization and recombination. The process of intermolecular rAAV genome concatamerization and recombination was first identified in muscle tissue. This finding demonstrated the feasibility of bringing two independent vector genomes into a stable covalently linked intermediate as the foundation for dual-vector approaches (31). Despite the great potential of this finding, the mechanisms of vector genome recombination in vivo are only poorly understood. Two distinct classes of intermolecular intermediates have been identified, including linear and circular concatemer genomes. Our group and others have previously shown that circular AAV episomes are responsible for long-term AAV transduction in normal skeletal muscle (6, 27). Similarly, circular AAV genomes have also been observed in other tissues such as liver, although it appears that episomal linear genomes are responsible for most of the transgene expression in this tissue (18, 20, 21). It remains unknown whether tissue-specific processing of rAAV genomes exists and whether this accounts for the reported differences between muscle and liver.
DNA-dependent protein kinase (DNA-PK) consists of a DNA binding subunit and a catalytic subunit (DNA-PKcs). DNA-PK has been shown to catalyze the nonhomologous end-to-end ligation in V(D)J recombination (14, 16). Severe combined immunodeficient (SCID) mice (3) carry a nonsense mutation in the DNA-PKcs gene and are defective in rejoining double-strand DNA breaks (2). The involvement of DNA-PKcs in rAAV vector genome concatamerization and circularization was recently observed in a series of elegant experiments that evaluated the molecular structure of rAAV genomes in SCID and normal mice (24). That report demonstrated that rAAV infection in SCID mouse skeletal muscle gives rise to enhanced levels of linear viral genomes (24).
Our laboratory has previously reported that head-to-tail circular AAV concatemers are the predominant AAV genome conversion products in muscle tissue (6, 7). Furthermore, we have shown that these head-to-tail circular concatemers contribute to dual-vector trans-spliced transgene expression (8, 30). However, the extent to which linear AAV concatemers contribute to dual-vector reconstitution of a transgene product is presently unclear. In this study, we have utilized molecular assays and dual lacZ trans-splicing reporter vectors to functionally evaluate the influence of DNA-PKcs deficiency on AAV genome processing and concatamerization in skeletal muscle. Findings from these studies demonstrated a substantial increase in linear vector genomes and free vector ends in SCID compared to wild-type animals. Furthermore, in contrast to the preferential head-to-tail orientation in wild-type muscle, the directivity of linear vector concatamerization is affected in SCID muscle, favoring a more random orientation of vector genomes. Marginal reduction in both circular viral genomes and functional head-to-tail vector genomes were seen in SCID mice by using a shuttle rescue vector and dual trans-splicing vectors, respectively. In summary, our study has confirmed and extended the report by Song et al. (24) and suggests that DNA-PKcs deficiency in SCID mice does alter the equilibrium between circular and linear AAV concatamerization in skeletal muscle. This alteration appears to only marginally affect the functional expression of a dual-vector-encoded gene product from head-to-tail circular and/or linear intermolecular concatemers.
MATERIALS AND METHODS
rAAV production.
In an effort to be consistent with the previous studies evaluating episomal AAV genomes in SCID and wild-type mice (6, 24), each of the viruses used in the present study contained type 2 AAV inverted terminal repeats (ITR) and was packaged with type 2 capsid. Methods used for generating rAAV were as previously described (8). In brief, pcisEGFPori3 was used to generate the shuttle AAV vector for rescuing circular AAV genomes in bacteria (5, 6). pcisLacZDonor and pcisLacZAcceptor, each carrying half of the lacZ gene, were used to produce β-galactosidase trans-splicing viruses (8). The recombinant viral stocks were generated with an adenovirus-free transient-transfection system as previously described (28). The viral fractions were pooled and dialyzed in HEPES-buffered saline (20 mM HEPES, 150 mM NaCl [pH 7.8]). Viral aliquots were stored at −80°C in 5% glycerol until use. The viral titers were determined by quantitative slot blots with cis plasmid standards. The average yield was 5 × 1012 viral particles/ml. The contamination of wild-type type 2 AAV was less than 1 functional particle per 1010 rAAV particles (4).
Animal procedures.
All animal experiments were carried out in accordance with National Institutes of Health (NIH) and institutional guidelines. Eight-week-old C57BL/6J (BL6) and C57BL/6J-SCID (SCID) mice were purchased from The Jackson Laboratory (Bar Harbor, Maine). To analyze the trans-splicing efficiency, 8 × 1010 particles of each vector, AV.LacZDonor and AV.LacZAcceptor, were mixed at an equal particle ratio and injected into the anterior tibialis muscles of mice (total of 30 μl; 1.6 × 1011 rAAV particles per muscle) (6). Mock infection with HEPES-buffered saline was used as a negative control. Additional negative controls included injection with 8 × 1010 particles of AV.LacZDonor alone or AV.LacZAcceptor alone. To avoid interanimal variability, coinfection and single infections were performed with the same animal, using one of the two anterior tibialis muscles for each experimental group. In total, six to nine animals of each genotype were evaluated by X-Gal (isopropyl-β-d-thiogalactopyranoside) staining, and three animals were evaluated by β-galactosidase enzyme assays. To ensure similar injection volumes, singly infected virus was diluted with an equal volume of HEPES-buffered saline prior to injection. trans-splicing AAV vector-infected muscles were harvested at 10 months postinfection to examine β-galactosidase expression. To analyze the molecular fate of rAAV genomes in muscle tissue, 1011 particles of AV.EGFPori3 were injected into the anterior tibialis muscles (n = 16 animals for each mouse strain). Mice were sacrificed 6 weeks later, and the DNA was extracted for Southern blot and transformation assays.
DNA extraction.
The low-molecular-weight Hirt DNA preparations were used as previously described to identify circular AAV transduction intermediates in murine skeletal muscles (6). However, Song et al. previously used a total DNA extraction method for analyzing episomal viral genome structure in rAAV-infected mouse skeletal muscles (24). To exclude possible experimental variations caused by different DNA extraction methods, Hirt DNA was prepared from half of each AV.EGFPori3-infected muscle sample and total DNA was prepared from the remaining half. Briefly, the entire anterior tibialis muscle was frozen in liquid nitrogen and pulverized in an ice-cold porcelain mortar. To extract Hirt DNA, muscle samples were incubated in Hirt buffer (10 mM Tris [pH 8.0], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], 10 μg of DNase-free RNase) at 37°C for 90 min. Pronase (Sigma) and proteinase K (Roche) were then added to final concentrations of 0.5 and 1 mg/ml, respectively. Following a 5-hour incubation at 37°C, NaCl was added to the digestion mixture at a final concentration of 1.1 M. After overnight incubation at 4°C, samples were centrifuged at 16,100 × g for 20 min and Hirt DNA was purified from the supernatants by standard phenol-chloroform extraction and ethanol precipitation. To isolate total DNA, muscle samples were digested at 55°C overnight in a buffer containing 50 mM Tris (pH 8.0), 100 mM EDTA, 1% SDS, 100 mM NaCl, and 500 μg of proteinase K (Roche) per ml. NaCl was added at a final concentration of 1.6 M at the end of digestion, and samples were spun at 16,100 × g for 10 min. The total DNA was then purified from the supernatant by standard phenol-chloroform extraction and ethanol precipitation.
Physical and functional analysis of episomal AAV genome in muscle DNA.
To physically analyze the AAV genome structure from AAV-infected BL6 and SCID muscles, half of the DNA from one piece of anterior tibialis muscle was digested with 40 U of AgeI (New England Biolabs) in a total volume of 200 μl for 15 h. DNA was then purified from the digestion mixture by standard phenol-chloroform extraction and ethanol precipitation. The other half of DNA was treated similarly except for the omission of AgeI restriction enzyme. The purified DNA was electrophoresed in a 0.8% agarose gel and then transferred to N+ Nylone membrane. Southern hybridization was performed with 5 × 106 cpm of radiolabeled probe per ml and with washing at a stringency of 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at 60°C for 20 min twice. To rescue the circular AAV genomes, one-fourth of the DNA from one piece of anterior tibialis muscle was transformed in 50 μl of SURE strain Escherichia coli (Stratagene). Sensitive T7 endonuclease I digestion was performed to confirm the presence of circular AAV genomes. In this case, one-third of the DNA from one piece of anterior tibialis muscle was digested with either 20 U of T7 endonuclease I or 20 U of lambda exonuclease, or without DNA-modifying enzymes, at 37°C for 5 h. Following digestion, DNA was purified by standard phenol-chloroform extraction and ethanol precipitation. One-half of the purified DNA was used to transform 50 μl of SURE strain E. coli. To increase the detection sensitivity for the circular AAV genome, additional Southern blot analyses were performed with pooled muscle DNA samples. In this case, DNAs extracted from two independent pieces of muscle were coprecipitated as one experimental sample. These concentrated samples were digested with 100 U of either T7 endonuclease I or lambda exonuclease in a total volume of 250 μl for 5 h at 37°C. At the end of digestion, the DNA was purified by standard phenol-chloroform extraction and ethanol precipitation and loaded as one lane for Southern analysis. The untreated controls were incubated with 1× lambda exonuclease buffer and examined simultaneously with the DNA-modifying-enzyme-treated samples.
Evaluation of reporter gene expression.
The β-galactosidase expression from trans-splicing vector-infected muscles was evaluated by both in situ X-Gal staining and total lysate enzymatic assays. Mice were sacrificed at 10 months postinfection. Central cross-sectional pieces of freshly harvested muscles (1 mm thick) were fixed en bloc in 0.5% glutaraldehyde at 37°C for 30 min. Muscle samples were then incubated in 1 mM MgCl2-phosphate-buffered saline for an additional 30 min. Finally, samples were stained in X-Gal solution for 12 h at room temperature. Staining was terminated by transferring muscle samples to 10% neutral-buffered formalin after washing with phosphate-buffered saline. The percentage of X-Gal-positive area was evaluated from two en face photomicrographs (each side of the stained tissue slice) of each experimental specimen (n = 6 SCID mice and 8 BL6 mice) by using NIH image software. The β-galactosidase enzyme activity assays (n = 3 SCID mice and 3 BL6 mice) were performed with a Galacto-Light Plus system (Applied Biosystems, Bedford, Mass.) essentially as we reported before (8). To ensure equal comparison, the β-galactosidase activity in each muscle sample was normalized to the protein concentration. The enhanced green fluorescent protein (EGFP) expression in AV.EGFPori3-infected muscles was analyzed by direct immunofluorescence microscopy of freshly excised tissues (n = 8 SCID mice and 8 BL6 mice).
RESULTS
Dual-vector trans-splicing-mediated β-galactosidase expression is observed in both SCID and BL6 skeletal muscles.
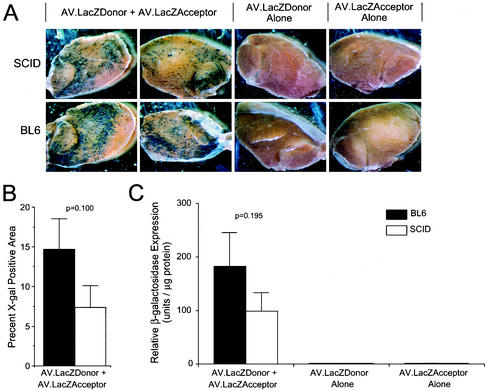
To test whether the reported difference in AAV genome structure in SCID muscle alters dual-vector reconstitution of a transgene product, the anterior tibialis muscles of both BL6 and SCID mice were infected with an equal amount of AV.LacZDonor and AV.LacZAcceptor viruses. Muscle tissues were harvested at 10 months postinfection, and LacZ expression was evaluated. As shown in Fig. 1A, en bloc histochemical staining revealed a significant amount of LacZ-positive myofibers in both BL6 and SCID muscles infected with both vectors. Quantification of the X-Gal-positive area revealed a twofold reduction in gene expression that did not reach significance (P = 0.100, Student t test) (Fig. 1B). As expected, no transgene expression was detected following single-vector infection. These observations were further confirmed by the quantitative β-galactosidase enzyme activity assay (Fig. 1C). The 46% reduction in rAAV trans-spliced β-galactosidase expression seen in SCID muscle was also not statistically significant (P = 0.195, Student t test).
FIG. 1.
Quantitative analysis of AAV dual-vector trans-splicing in SCID and BL6 muscles. The β-galactosidase expression from a pair of rAAV trans-splicing vectors (AV.LacZDonor and AV.LacZAcceptor) was evaluated at 10 months after infection of muscle tissue. (A) Photomicrographs of en face X-Gal-stained muscle samples infected with the various trans-splicing vectors. (B) Morphometric quantification of X-Gal staining SCID (n = 6) and BL6 (N = 9) mouse muscles coinfected with AV.LacZDonor and AV.LacZAcceptor. The mean (± standard error of the mean) X-Gal-positive area of the entire muscle for each sample set is shown. (C) Enzymatic quantification of β-galactosidase activity. The mean (± standard error of the mean) relative light units per microgram (n = 3 samples for each group) is shown. No significant difference was seen between coinfected SCID and BL6 samples in panels B and C (Student t test; P values are given above each comparison).
Structural analysis of episomal AAV genomes in SCID and BL6 skeletal muscles.
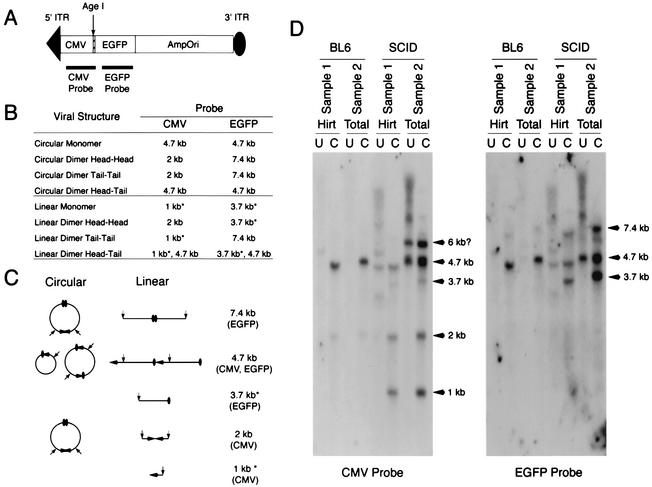
Our previous studies with BL6 mice have correlated the formation of head-to-tail circular concatemers with the efficiency of gene expression by dual-vector cis -activation and trans-splicing (9, 30). This conclusion is based on the direct bacterial rescue of circularized heterodimers and multimers from coinfected muscles and the observation that the abundance of these intermolecular intermediates increased with dual-vector-mediated gene expression. Previous Southern blot analysis of SCID mouse muscles has suggested that circular AAV genomes are substantially reduced as a result of a loss of DNA-PKcs activity (24). To correlate our trans-splicing result with the previous report, we performed a Southern blot analysis similar to that conducted by Song et al. To facilitate subsequent bacterial recovery of circular AAV genomes, we have opted to use a previously described AV.EGFPori3 shuttle vector in this analysis. Both Hirt DNA and total muscle DNA were extracted from virus-infected SCID and BL6 muscles at 6 weeks postinfection. DNA samples were either digested with the single-cutter restriction enzyme AgeI (Fig. 2A) or left undigested. The expected diagnostic bands for circular and linear AAV monomers and dimers are shown in Fig. 2B and C. In the case of circular AAV genomes, a 4.7-kb band will be expected following AgeI digestion from monomers or head-to-tail concatemers. In contrast, the existence of free-end DNA fragments following AgeI digestion serves as evidence for the presence of linear AAV genomes.
FIG. 2.
Southern blot analysis of episomal AAV genomes in SCID and BL6 muscles. (A) Schematic representation of the AV.EGFPori3 virus used in this study (not drawn to the scale). (B and C) Predicated restriction patterns and circular and linear structures of episomal AAV monomers and dimers. The 1-kb free head-end fragment (detected by the CMV probe) and the 3.7-kb free tail-end fragment (detected by the EGFP probe) are the diagnostic bands for the existence of linear AAV genomes and are indicated by asterisks. (D) Southern blots of Hirt and total DNAs harvested at 6 weeks postinfection with AV.EGFPori3. The membrane was initially hybridized with a CMV probe and then stripped and reprobed with an EGFP probe. Hirt, low-molecular-weight Hirt DNA; Total, total DNA; U, undigested; C, AgeI digested.
Consistent with our previous reports, a 4.7-kb AgeI-digested dominant band was detected in BL6 muscles. None of the diagnostic bands for linear AAV genomes were observed with either a cytomegalovirus (CMV) probe or an EGFP probe in BL6 samples (Fig. 2D). These results indicate that head-to-tail recombination is the primary pathway for rAAV genome processing in normal skeletal muscle. Interestingly, two additional faint bands, representing head-to-head recombination (a 2-kb band detected by the CMV probe) and tail-to-tail recombination (a 7.4-kb band detected by the EGFP probe), were also observed in BL6 samples. This substantiates previous findings from our laboratory demonstrating a lower abundance of rescued head-to-head and tail-to-tail circular AAV genomes following dual-vector infection (6, 31).
Consistent with the finding by Song et al., free ITR ends were detected only in SCID muscles and were not seen in BL6 animals. These bands are observed as 1-kb free head-end bands (detected with a CMV probe) and 3.7-kb free tail-end bands (detected with an EGFP probe) following AgeI digestion (Fig. 2D). The presence of the 4.7-kb band in undigested DNA (only in SCID muscles) also supported the selective presence of linear monomer genomes in DNA-PKcs-deficient muscles. The intensity of this 4.7-kb band increased following AgeI digestion. This result was not mutually exclusive with the existence of circular AAV genomes in SCID muscles, since the increase could result from either circular or linear head-to-tail AAV concatemers. Interestingly, the levels of head-to-head recombination (a 2-kb band detected with the CMV probe) and tail-to-tail recombination (a 7.4-kb band detected with the EGFP) were substantially enhanced in SCID samples (Fig. 2). Nearly identical Southern hybridization patterns were observed with Hirt and total DNA samples, with the exception of a slower-migrating band (6 kb) seen only in total SCID DNA (CMV probe). The nature of this 6-kb band, which hybridized only to the CMV probe, was unclear (Fig. 2D).
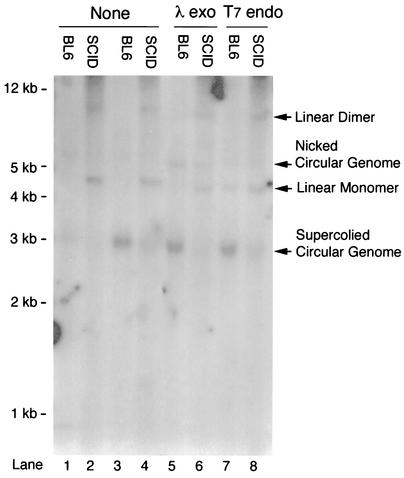
To visualize circular AAV genomes, DNA samples from two independently infected muscles were pooled for Southern blot analysis. Consistent with the restriction analysis data in Fig. 2, circularized AAV genomes were detected in both BL6 and SCID samples, although they were less abundant in SCID muscle (Fig. 3, lanes 1 to 4). The 4.7-kb double-stranded linear AAV monomers were observed only in SCID samples (Fig. 3, lanes 2 and 4).
FIG. 3.
Southern blot characterization of pooled DNA samples from SCID and BL6 muscles. Lanes 1 to 4, pooled DNA samples from two independently infected muscles were directly examined by Southern blotting without additional enzymatic treatment. Supercoiled circular AAV genomes were detected in both BL6 and SCID samples. A 4.7-kb double-stranded linear monomer band was seen only in SCID samples (lanes 2 and 4). Lanes 5 to 8, pooled DNA samples were digested with either lambda exonuclease (lanes 5 and 6) or T7 endonuclease (lanes 7 and 8) before Southern analysis. The double-stranded linear AAV genome in SCID samples was resistant to lambda exonuclease digestion (lane 6). The T7 endonuclease I treatment released some supercoiled circular AAV genomes into 4.7-kb double-stranded linear fragments (lane 7). None, samples were not treated with DNA-modifying enzymes; λ exo, samples were predigested with lambda exonuclease before being loaded on the gel; T7 endo, samples were predigested with T7 endonuclease I before being loaded on the gel.
Shuttle vector rescue of circular AAV genomes from both SCID and BL6 muscles.
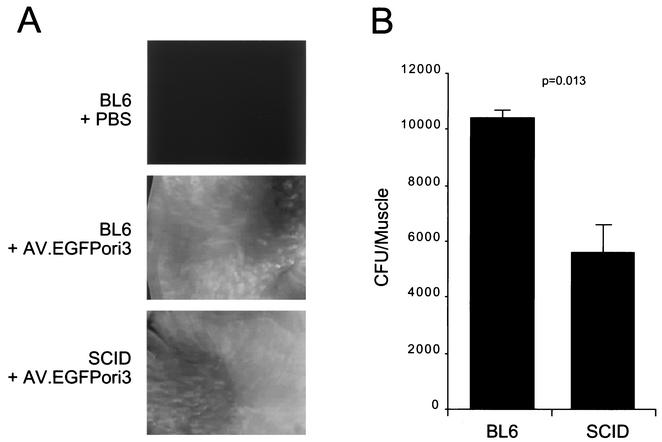
As an alternative method for evaluating the existence of circular AAV transduction intermediates, we have previously utilized a shuttle vector trapping approach to isolate and expand circularized viral genomes in bacteria. This method is based on the theory that circular AAV genomes should behave identically to plasmids if an antibiotic selective marker and the E. coli replication origin are included in the viral genome. Using this bacterial trapping strategy, we have successfully isolated circular AAV genomes from normal mouse skeletal muscle (6). This approach was also used to indirectly quantify total circular viral genomes in BL6 and SCID muscles following AV.EGFPori3 infection. In these studies, Hirt DNA and total DNA from muscle tissues were extracted at 6 weeks postinfection and used to transform E. coli. Consistent with previous reports (24, 25), BL6 and SCID muscles displayed comparable levels of EGFP expression (Fig. 4A). Interestingly, circular AAV genomes were obtained from both BL6 and SCID DNAs following E. coli transformation. A lack of DNA-PKcs activity in SCID muscle resulted in a moderate but significant reduction in the total number of retrieved clones (Fig. 4B) (∼50%; P = 0.013, Student t test). This decrease closely paralleled the Southern blot finding (Fig. 3). To determine whether the clones retrieved from SCID muscles were different from those from BL6 muscles, we randomly prepared 100 plasmids from each population. Detailed restriction analyses and Southern hybridization revealed consistent and indistinguishable double-D circular structures from both groups (data not shown).
FIG. 4.
Transgene expression and circular AAV genome recovery from AV.EGFPori3-infected muscles. (A) Representative en face photomicrographs of EGFP expression in freshly isolated muscles at 6 weeks postinfection. (B) CFU quantification of circular episomal AAV genomes from BL6 and SCID muscles (mean ± standard error of the mean; n = 4). The statistical evaluation between SCID and BL6 samples was performed with the Student t test.
To conclusively address the presence of circular AAV genomes in SCID muscle, it is essential to exclude the artificial CFU produced by linear AAV genome circularization in bacteria. Abundant double-stranded linear AAV genomes, which we observed in SCID muscle Southern blots (Fig. 2 and 3), might serve as substrates for bacterially mediated religation and circularization. We have previously shown that the in vitro-reconstituted double-stranded linear AAV genomes were incapable of transforming E. coli (5, 6). However, linear AAV genomes in SCID muscle might be structurally different from the ones generated by in vitro reconstitution. To confirm circular AAV genome formation in SCID muscles, a series of functional transformation assays were performed with T7 endonuclease I-predigested Hirt DNA. T7 endonuclease I specifically binds to four-way junctions (such as Holiday structures) in DNA and resolves cruciform DNA junctions into linear molecules (22). Since the double-D structure in the ITR junction mimics the Holiday configuration (7, 29), we predicted that circular AAV genomes should be recognized and cleaved by T7 endonuclease I. As a result, this enzymatic cleavage should reduce the number of clones retrieved from transformation. On the other hand, if AAV genome circularization occurred in bacteria, rather than in muscle, treatment of muscle DNA before transformation should have no effect on CFU. Another DNA-modifying enzyme, lambda exonuclease, was also included in the experiment as a control. Lambda exonuclease is a highly processive exonuclease; it rapidly degrades double-stranded linear DNA, especially linear DNA with a 5′ phosphate (15, 17). Importantly, lambda exonuclease has no endonuclease activity and will not hydrolyze DNA at nicked sites. Therefore, we predicted that lambda exonuclease treatment should not reduce CFU derived from circular AAV genomes that already exist in AAV-infected muscle.
To test this hypothesis, a rescued clone (pDD212) from AV.EGFPori3-infected muscle was digested with either T7 endonuclease I or lambda exonuclease. The restriction enzyme XhoI digests the AV.EGFPori3 genome only once. The XhoI-predigested pDD212 was used as linear DNA template. As shown in Fig. 5A, T7 endonuclease I treatment indeed released the 4.7-kb linear AAV band from the circular pDD212. Interestingly, linearized pDD212 was resistant to T7 endonuclease I digestion. We suspected that the supercoiling of the circular genome might be required for cruciform DNA junction formation at the double-D ITR. As expected, the 4.7-kb XhoI-predigested linear molecules were efficiently digested by lambda exonuclease (Fig. 5A). To further extend these in vitro observations, we digested the muscle DNA with either T7 endonuclease I or lambda exonuclease. As shown in Fig. 3, T7 endonuclease I digestion was indeed able to resolve supercoiled circular AAV genomes to linear fragment (Fig. 3, lane 7). Interestingly, lambda exonuclease digestion had no apparent effect on double-stranded linear AAV genomes presented in SCID muscle (Fig. 3, lane 6). Similar resistance has also been observed in replicative forms of linear AAV genomes (data not shown). It is possible that the unusual structure of the terminal ITR cannot be recognized by lambda exonuclease. Alternatively, terminal AAV ITR could have been protected by certain cellular proteins such as D-sequence binding protein (23) or DNA binding subunit (Ku70/Ku80) of DNA-PK.
FIG. 5.
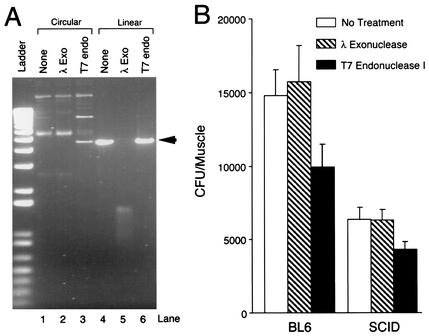
Functional confirmation of circular AAV genomes in SCID and BL6 muscles by use of DNA-modifying enzymes. (A) Effect of lambda exonuclease and T7 endonuclease I on episomal AAV genomes. Circular and linear forms (the linear form was obtained by XhoI digestion of a representative circular AAV monomer, pDD212) were treated with lambda exonuclease and T7 endonuclease I, respectively. Approximately one-third of circular genomes were converted to the 4.7-kb linear form (arrow) by T7 endonuclease I digestion (lane 3). Lambda exonuclease treatment did not affect circular genomes, but it completely digested the 4.7-kb linear DNA into small residual fragments. (B) CFU quantification of circularized AAV genomes following lambda exonuclease or T7 endonuclease I treatment (mean ± standard error of the mean; n = 4). The Student t test revealed a significant difference between untreated samples and T7 endonuclease I-treated samples (P = 0.02 for the BL6 group; P = 0.018 for the SCID group). No statistical difference was obtained between untreated samples and lambda exonuclease-treated samples from the same mouse strain.
Having confirmed the sensitivity of the supercoiled circular AAV genome to T7 endonuclease I treatment, we next examined its effect on CFU recovery from muscle DNA transformation (Fig. 5B). Consistent with the data in Fig. 4, transformation of undigested SCID muscle Hirt DNA demonstrated a ∼50% reduction in total CFU. As expected, lambda exonuclease treatment did not affect CFU recovery in either BL6 or SCID samples. Importantly, T7 endonuclease I pretreatment of Hirt DNA led to a 30% decrease in recovered CFU in both BL6 and SCID samples (Fig. 5B).
DISCUSSION
Single-stranded rAAV genomes require a conversion process to double-stranded DNA in order to express encoded transgenes. Many conversion products have been identified in the context of rAAV infection, including episomal linear and circular double-stranded monomer and concatemer genomes. Furthermore, these concatemer genomes have the potential to be oriented in a head-to-tail, head-to-head, or tail-to-tail fashion. It is presently unclear whether all of these genome conversion products contribute to long-term integrated or episomal transgene expression in the setting of latent rAAV infections. In the present study, we have attempted to utilize molecular assays and dual-vector gene delivery approaches to evaluate the involvement of DNA-PKcs (a key enzyme in the DNA repair pathway involved in nonhomologous DNA end joining) in the circularization and heterodimerization of rAAV genomes in mouse skeletal muscle.
Dual-vector delivery based on trans-splicing heterodimers requires directional head-to-tail covalent joining of two independent genomes. We have previously demonstrated the functional involvement of circular rAAV genome concatemers in dual-vector trans-splicing (9, 30). However, the extent to which linear head-to-tail concatemers contribute to dual-vector gene expression with this approach remains unclear. Since DNA-PKcs-deficient SCID muscle has been previously demonstrated to have a substantial increase in free-end vector genomes, we hypothesized that rAAV vector genome circularization may be reduced in this model. Southern blot analysis supported previous findings, demonstrating a substantial increase in free vector ends and linear concatemer genomes in SCID muscle compared to the BL6 control (Fig. 2). Furthermore, these data suggested that end-to-end ligation seen in SCID mice proceeds through a random process, leading to nearly equal abundances of head-to-head, tail-to-tail, head-to-tail, and tail-to-head linear rAAV concatemer genomes. In contrast, BL6 concatemers demonstrated a preference for head-to-tail concatamerization (Fig. 2).
Since head-to-tail linear and circular concatemer genomes give similar restriction patterns on Southern blots, an alternative approach was required to evaluate the abundance of circular concatemers. Using a bacterial shuttle vector capable of trapping rAAV circular genomes, we determined that BL6 mice had significantly (twofold; P = 0.013) more abundant circular genomes than SCID mice (Fig. 4 and 5). This difference was seen despite equivalent levels of transgene expression from the GFP-expressing shuttle vector. This decrease of CFU in SCID muscle also agreed with Southern data from pooled samples (Fig. 3). To further determine the circular AAV genome in SCID muscle, DNA samples were digested with sensitive T7 endonuclease I prior to transformation. The unique activity of T7 endonuclease I in linearizing the circular AAV genome provided conclusive evidence for the presence of circular AAV genomes in both BL6 and SCID muscles.
Dual trans-splicing vectors provide the ideal framework for evaluating the directivity of vector genome concatamerization events in vivo, since only head-to-tail concatemers will effectively express the dually encoded gene product. Given the observed DNA-PKcs-dependent alterations in the equilibrium between circular and linear concatemer genomes, we hypothesized several potential outcomes from dual-vector expression studies. First, if circular genomes are the sole contributors of functional gene expression, we would predict a ∼50% decrease in β-galactosidase production from LacZ trans-splicing vectors in DNA-PKcs-deficient SCID muscles. Second, if both linear and circular concatemer genomes equally contribute to gene expression, a marginal reduction in dual-vector β-galactosidase gene expression would be expected, since more random end-end joining is seen in the absence of DNA-PKcs. Although results from these studies demonstrated a twofold trend toward reduced β-galactosidase activity in the absence of DNA-PKcs, this trend did not demonstrate statistical significance (Fig. 1). These findings support the notion that both linear and circular head-to-tail concatemers are capable of expressing a trans-spliced transgene product.
Several interpretations might explain the marginal, but not significant, reduction in dual-vector trans splicing in SCID mice. If this reduction has a biologic basis other than assay variability, it could be due to a shift in equilibrium to a more random end-end joining of viral genomes in SCID mice that decreases the pool of head-to-tail concatemers. If the total number of genomes is equivalent in SCID and BL6 mice, then one would predict a reduction in the total abundance of head-to-tail genomes and subsequent trans-spliced gene expression in SCID muscle. However, our results suggest that SCID muscles might contain two to three times more episomal viral genomes than BL6 controls. Since fewer circular genomes were retrieved from SCID tissues (Fig. 3 to 5), the majority of episomal AAV in SCID muscle must be linear genomes. Even in the setting of more random end-end joining in SCID muscle, the higher level of total vector genomes would still lead to significant head-to-tail concatemers. Assuming that transcription and splicing efficiency are not different for linear and circular genomes, we would anticipate comparable levels of transgene expression from dual-vector infection in SCID and BL6 muscles. On the other hand, an overall increase in the total amount of double-stranded AAV genomes in SCID did not seem to dramatically enhance single AAV vector transduction efficiency (Fig. 4A). Although visual EGFP evaluation is a less quantitative way to assay transgene expression, studies by Song et al. have shown a consistently comparable level of transgene expression over a 1-year period in SCID and BL6 muscles (24).
Only the catalytic subunit of DNA-PK is lost in SCID mice, while the DNA binding subunit (Ku70/Ku80) is not affected by the SCID mutation (2). The Ku70/Ku80 heterodimer is the most abundant DNA end binding protein in mammalian cells. This heterodimer directly binds to free DNA ends and aligns them for ligation (10). However, end-joining enzyme activity is dependent on the recruitment of DNA-PKcs. Since self-ligation is a favored reaction stoichiometrically, it is possible that in DNA-PKcs-positive BL6 myofibers, the dominant early product will be self-ligated circular monomer. In SCID muscle cells, however, the loss of DNA-PKcs may have compromised formation of a functional enzymatic complex and subsequent AAV circularization. An additional explanation for the reduced amount of circular genomes found in SCID mice is that it is due to an enhanced linear concatamerization as opposed to a reduced circularization. It is also important to appreciate that single gene defects may cause compensatory alterations in other DNA repair enzymes. Such alterations may account for either the reduced degradation of linear forms in SCID muscle or the enhanced end-to-end linear concatamerization.
It is presently uncertain how the lack of DNA-PKcs augments linear monomer or concatemer formation. Nakai et al. (20) have previously reported that plus- and minus-strand annealing is responsible for the initial-stage double-stranded AAV genome formation in hepatocytes. Although this process has not been proven to occur in muscle, such double-stranded linear annealed genomes could be the substrate for linear concatamerization in the absence of DNA-PKcs. The loss of DNA-PKcs in SCID mice excludes the possibility of DNA-PK-catalyzed rejoining of free-end AAV genomes. However, an alternative DNA-PK-independent pathway for repairing double-stranded DNA breaks (13) may have contributed to the linear AAV concatemer formation in SCID muscles. Similarly, the DNA-PK-independent pathway might also be responsible for the remaining circular AAV genome formation in SCID mice, although this pathway might be kinetically less efficient than the DNA-PK pathway.
Our data suggest that DNA-PKcs activity has a minimal but measurable effect on rAAV genome circularization. We favor an interpretation that rAAV genome circularization and concatamerization are likely catalyzed by multiple DNA repair pathways and that the equilibrium of these pathways is altered by the absence of DNA-PKcs. In support of our present findings, M. A. Kay and colleagues have found similar alterations in the equilibrium of linear- and circular-form rAAV genomes in SCID liver (personal communication). Linear DNA molecules introduced into the liver by transfection were efficiently removed by both circularization and concatamerization in normal mice but were removed exclusively by concatamerization in SCID mice. The exact molecular pathway(s) for circular AAV genome formation remains elusive. In contrast to the direct end-end joining model (31), a potential rolling-cycle-like strand replacement model has also been proposed on the basis of the electron microscopic visualization of the isolated AAV genome (K. I. Berns and T. J. Kelly, Jr., Letter, J. Mol. Biol. 82:267-271, 1974) and the sequence analysis of the head-to-tail ITR junction (7). Further illustration of these pathways may eventually clarify the molecular mechanism(s) underlying circular AAV genome formation in mammalian cells.
Acknowledgments
This work was supported by a research grant from the Muscular Dystrophy Association (to D.D.) and NIH grant HL58340 (to J.F.E.).
We gratefully acknowledge K. Wyne and W. Ding for editorial and/or morphometric quantification assistance.
REFERENCES
- 1.Acland, G. M., G. D. Aguirre, J. Ray, Q. Zhang, T. S. Aleman, A. V. Cideciyan, S. E. Pearce-Kelling, V. Anand, Y. Zeng, A. M. Maguire, S. G. Jacobson, W. W. Hauswirth, and J. Bennett. 2001. Gene therapy restores vision in a canine model of childhood blindness. Nat. Genet. 28:92-95. [DOI] [PubMed] [Google Scholar]
- 2.Blunt, T., D. Gell, M. Fox, G. E. Taccioli, A. R. Lehmann, S. P. Jackson, and P. A. Jeggo. 1996. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl. Acad. Sci. USA 93:10285-10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosma, G. C., R. P. Custer, and M. J. Bosma. 1983. A severe combined immunodeficiency mutation in the mouse. Nature 301:527-530. [DOI] [PubMed] [Google Scholar]
- 4.Duan, D., K. J. Fisher, J. F. Burda, and J. F. Engelhardt. 1997. Structural and functional heterogeneity of integrated recombinant AAV genomes. Virus Res. 48:41-56. [DOI] [PubMed] [Google Scholar]
- 5.Duan, D., P. Sharma, L. Dudus, Y. Zhang, S. Sanlioglu, Z. Yan, Y. Yue, Y. Ye, R. Lester, J. Yang, K. J. Fisher, and J. F. Engelhardt. 1999. Formation of adeno-associated virus circular genomes is differentially regulated by adenovirus E4 ORF6 and E2a gene expression. J. Virol. 73:161-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle. J. Virol. 72:8568-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan, D., Z. Yan, Y. Yue, and J. F. Engelhardt. 1999. Structural analysis of adeno-associated virus transduction intermediates. Virology 261:8-14. [DOI] [PubMed] [Google Scholar]
- 8.Duan, D., Y. Yue, and J. F. Engelhardt. 2001. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 4:383-391. [DOI] [PubMed] [Google Scholar]
- 9.Duan, D., Y. Yue, Z. Yan, and J. F. Engelhardt. 2000. A new dual-vector approach to enhance recombinant adeno-associated virus-mediated gene expression through intermolecular cis activation. Nat. Med. 6:595-598. [DOI] [PubMed] [Google Scholar]
- 10.Dynan, W. S., and S. Yoo. 1998. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 26:1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halbert, C. L., J. M. Allen, and A. D. Miller. 2002. Efficient mouse airway transduction following recombination between AAV vectors carrying parts of a larger gene. Nat Biotechnol. 20:697-701. [DOI] [PubMed] [Google Scholar]
- 12.Kay, M. A., C. S. Manno, M. V. Ragni, P. J. Larson, L. B. Couto, A. McClelland, B. Glader, A. J. Chew, S. J. Tai, R. W. Herzog, V. Arruda, F. Johnson, C. Scallan, E. Skarsgard, A. W. Flake, and K. A. High. 2000. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat Genet. 24:257-261. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. E., R. A. Mitchell, A. Cheng, and E. A. Hendrickson. 1997. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell. Biol. 17:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieber, M. R., J. E. Hesse, S. Lewis, G. C. Bosma, N. Rosenberg, K. Mizuuchi, M. J. Bosma, and M. Gellert. 1988. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 55:7-16. [DOI] [PubMed] [Google Scholar]
- 15.Little, J. W. 1981. Lambda exonuclease. Gene Amplif. Anal. 2:135-145. [PubMed] [Google Scholar]
- 16.Malynn, B. A., T. K. Blackwell, G. M. Fulop, G. A. Rathbun, A. J. Furley, P. Ferrier, L. B. Heinke, R. A. Phillips, G. D. Yancopoulos, and F. W. Alt. 1988. The scid defect affects the final step of the immunoglobulin VDJ recombinase mechanism. Cell 54:453-460. [DOI] [PubMed] [Google Scholar]
- 17.Mitsis, P. G., and J. G. Kwagh. 1999. Characterization of the interaction of lambda exonuclease with the ends of DNA. Nucleic Acids Res. 27:3057-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakai, H., Y. Iwaki, M. A. Kay, and L. B. Couto. 1999. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J. Virol. 73:5438-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakai, H., T. A. Storm, and M. A. Kay. 2000. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nat. Biotechnol. 18:527-532. [DOI] [PubMed] [Google Scholar]
- 20.Nakai, H., T. A. Storm, and M. A. Kay. 2000. Recruitment of single-stranded recombinant adeno-associated virus vector genomes and intermolecular recombination are responsible for stable transduction of liver in vivo. J. Virol. 74:9451-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakai, H., S. R. Yant, T. A. Storm, S. Fuess, L. Meuse, and M. A. Kay. 2001. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 75:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons, C. A., and S. C. West. 1990. Specificity of binding to four-way junctions in DNA by bacteriophage T7 endonuclease I. Nucleic Acids Res. 18:4377-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qing, K., B. Khuntirat, C. Mah, D. M. Kube, X. S. Wang, S. Ponnazhagan, S. Zhou, V. J. Dwarki, M. C. Yoder, and A. Srivastava. 1998. Adeno-associated virus type 2-mediated gene transfer: correlation of tyrosine phosphorylation of the cellular single-stranded D sequence-binding protein with transgene expression in human cells in vitro and murine tissues in vivo. J. Virol. 72:1593-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song, S., P. J. Laipis, K. I. Berns, and T. R. Flotte. 2001. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc. Natl. Acad. Sci. USA 98:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song, S., M. Morgan, T. Ellis, A. Poirier, K. Chesnut, J. Wang, M. Brantly, N. Muzyczka, B. J. Byrne, M. Atkinson, and T. R. Flotte. 1998. Sustained secretion of human alpha-1-antitrypsin from murine muscle transduced with adeno-associated virus vectors. Proc. Natl. Acad. Sci. USA 95:14384-14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun, L., J. Li, and X. Xiao. 2000. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nat. Med. 6:599-602. [DOI] [PubMed] [Google Scholar]
- 27.Vincent-Lacaze, N., R. O. Snyder, R. Gluzman, D. Bohl, C. Lagarde, and O. Danos. 1999. Structure of adeno-associated virus vector DNA following transduction of the skeletal muscle. J. Virol. 73:1949-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao, X., W. Xiao, J. Li, and R. J. Samulski. 1997. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J. Virol. 71:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan, Z., Y. Zhang, D. Duan, and J. F. Engelhardt. 2000. From the cover: trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. USA 97:6716-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, J., W. Zhou, Y. Zhang, T. Zidon, T. Ritchie, and J. F. Engelhardt. 1999. Concatamerization of adeno-associated viral circular genomes occurs through intermolecular recombination. J. Virol. 73:9468-9477. [DOI] [PMC free article] [PubMed] [Google Scholar]