Abstract
The genetic basis for susceptibility or nonsusceptibility of plants to viruses is understood poorly. Two selectable tobacco etch virus (TEV) strains were developed for identification of Arabidopsis thaliana mutants with either gain-of-susceptibility or loss-of-susceptibility phenotypes. These strains conferred a conditional-survival phenotype to Arabidopsis based on systemic expression of herbicide resistance or proherbicide sensitivity genes, thereby facilitating mass selections and screens for Arabidopsis mutants that enhance or suppress TEV replication, cell-to-cell movement, or long-distance movement. A multicomponent mechanism that restricts systemic invasion of TEV was identified through isolation of gain-of-susceptibility mutants with alterations at two loci.
Keywords: tobacco etch virus, host, mutant selections
Virus–host interactions are governed by complex sets of viral and host genes that specify compatibility or incompatibility (1). Compatibility functions control virus genome replication, cell-to-cell movement, and long-distance movement through the plant vascular system. Incompatible interactions can involve virus recognition and activation of host defense responses, such as the hypersensitive response, systemic acquired resistance, and homology-dependent gene silencing (2). However, incompatibility in virus–host interactions may also occur by passive means (3).
The genetic bases of virus–host interactions controlling susceptibility and resistance is poorly understood. Although many examples of natural variation in susceptibility to plant viruses exist and are important agronomic traits, very few host mutants have been identified from systematic screens for plants with altered susceptibility to viruses. Mutations in the N gene, which encodes resistance to tobacco mosaic virus, were identified in screens for loss of the hypersensitive response after tobacco mosaic virus (TMV) infection (4). Characterization of one unstable mutant line that could revert to TMV resistance led to the transposon tagging and cloning of N. The tom1, tom2, and cum1 mutations were identified by immunological screens for Arabidopsis mutants that accumulated TMV or cucumber mosaic virus coat protein to reduced levels (5–7). The wild-type TOM1 and TOM2 loci are necessary for efficient replication of TMV in protoplasts and systemic infection in plants (5, 6), whereas CUM1 is required by cucumber mosaic virus for efficient cell-to-cell movement in inoculated leaves (7). The vsm1 mutation, which restricts TMV to the inoculated leaves, was identified from a screen for mutants that were either asymptomatic or delayed in the onset of symptoms after TMV infection (8).
Identification and analysis of host-cell factors with compatibility and incompatibility functions have been limited, in part because versatile genetic systems to isolate a range of susceptibility mutants have yet to be established (1). To this end, the Arabidopsis/tobacco etch virus (TEV) pathosystem was established. Long-distance movement of TEV is inhibited in certain Arabidopsis ecotypes (e.g., Col-0), depending on the allele at the RTM1 locus (9). Arabidopsis ecotypes such as Col-0, which possess RTM1, restrict TEV to inoculated leaves, whereas rtm1-bearing ecotypes, such as C24 and La-er, allow long-distance movement by TEV. The RTM1-mediated restriction of TEV does not appear to function by classic mechanisms involving the hypersensitive response or induction of systemic acquired resistance (2). The identification of ecotypes that are differentially infected by TEV provides the opportunity to study the genetics of both resistance and susceptibility to this virus.
Here, we describe the development of systems that overcome many of the limitations to recovery of large numbers of altered virus-susceptibility mutants. Engineered TEV strains that confer positive selection for either gain-of-susceptibility or loss-of-susceptibility mutants of Arabidopsis and high-throughput inoculation methods were devised and tested. The selectable TEV-bar strain was used to isolate gain-of-susceptibility mutants with RTM1-suppressed phenotypes. Genetic characterization of these mutants revealed that at least two loci, RTM1 and RTM2, cooperate to condition a restricted TEV movement phenotype. The counter-selectable TEV-P450 strain was also developed for the isolation of Arabidopsis mutants with loss-of-susceptibility phenotypes.
MATERIALS AND METHODS
Construction of Selectable Viruses.
To construct TEV-bar, the bar gene was amplified from pBIO122 (a gift from B. Baker) by using Pfu polymerase (Stratagene) and the oligonucleotide primers BAR1 (5′-gcaatcgatatgagcccagaacga-3′) and BAR2 (5′-accacgcgtgatctcggtgacggg-3′), which added ClaI and MluI sites to the 5′ and 3′ ends of the bar sequence, respectively. The PCR product was digested with ClaI and MluI, gel purified, and ligated into the intermediate cloning vector, p7SN.0823-CMK, to yield p7SN.0823-bar. The insert was cleaved from p7SN.0823-bar, gel purified, and ligated into pTEV7DA-CMK that was digested with ClaI and MluI, generating pTEV-bar. To construct TEV-P450, the RbcS transit peptide fusion with the 5′ end of P450SU1 was amplified with Pfu polymerase from pSSU-SU12 (10) by using the primers SU12NCO (5′-gacaccatggcttcctctgtga-3′) and P450NCO* (5′-gcagatcaccatagagggcaccggca-3′). These primers added an NcoI site to the 5′ end of the gene and destroyed the NcoI site at nucleotide 466 of the P450SU1 coding region. The PCR product was used as a primer with SU12R (5′-tcgacgcgtccaggtgaccgggagttc-3′) in a second PCR reaction to amplify the full-length RbcS leader/P450SU1 fusion. This PCR product was digested with NcoI and MluI, gel purified, and ligated into p7SN.0823-CMK, creating p7SN.0823-P450. p7SN.0823-P450 was digested with XbaI, the insert was gel purified and ligated into pTEV7DA-CMK, generating pTEV-P450.
Propagation of TEV Strains.
Infectious transcripts corresponding to TEV-bar, TEV-P450, and TEV-GUS were generated from BglII-linearized pTEV-bar, pTEV-P450 and pTEV7DA-G↓H (11) DNA, respectively, by using SP6 polymerase. Nicotiana tabacum cv. Xanthi-nc plants were inoculated with TEV transcripts to produce inoculum used for experiments with Arabidopsis. Large-scale inoculum was prepared by grinding TEV-infected tissue from 500 Xanthi-nc plants in a Waring blender for 1.5 min with 2 vol/wt of 20 mM Hepes (pH 7.5) buffer containing 18% vol/vol 1-butanol and 0.1% wt/vol sodium sulfite. Ground tissue was expressed through cheesecloth and insoluble material was removed by centrifugation in a JA-10 rotor (Beckman Coulter) at 4,500 rpm. Virus was precipitated on ice for 2 h with 4% polyethylene glycol (8,000 molecular wt), 1.0% Triton X-100, and 0.1 M NaCl and collected by centrifugation at 8,000 rpm in a JA-14 rotor (Beckman Coulter) for 10 min. The pellet was resuspended in one-fourth the original volume of 20 mM Hepes buffer, and virus was further purified by a second precipitation on ice for 1 h with 8% polyethylene glycol and 0.1 M NaCl. Virus was collected by centrifugation at 8,000 rpm, and pellets were resuspended in 1 ml of 20 mM Tris⋅HCl (pH 8.0) per 10 plants used in the preparation. The inoculum preparations were stored in 1-ml aliquots at −80°C.
Arabidopsis Inoculation and Herbicide Application.
The TEV-bar, TEV-P450, or TEV-GUS inoculum was diluted in 20 mM Tris⋅HCl (pH 8.0) containing 10 g/liter of carborundum. Rosette leaves of 4-week-old Col-3, C24, and ethylmethane sulfonate- or fast neutron bombardment-mutagenized Col-0 M2 (Lehle Seed, Round Rock, TX) plants were inoculated by using an artist’s airbrush (Paasche, Harwood Heights, IL, model VL80) (4) with an air pressure of 75 psi (1 psi = 6.89 kPa). For small-scale glufosinate-ammonium (GA) (AgrEvo, Montvale, NJ; 0.01% solution in deionized water) applications, inflorescence tissue was removed with a razor blade, and a spray bottle was used to soak the plants. For large-scale mutant selections, inflorescence tissue was removed with a hedge trimmer (Black & Decker, Towson, MD), and GA (0.067% in deionized water) was applied by using a spray chamber with herbicide delivered at a rate of 480 liters per hectare. The proherbicide R7402 (DuPont) was diluted to 100 μg/liter (wt/vol) in deionized water containing 100 μl/liter Silwet-L77 (Vac-In-Stuff, Lehle Seed) and was applied manually with a spray bottle.
Immunoblot and GUS Activity Assays.
Inflorescence tissue (approximately 50–100 mg) was ground in 100 μl of protein dissociation buffer. Samples were boiled for 3 min, spun in a microcentrifuge for 3 min at 14,000 rpm, and 10 μl was run on SDS/PAGE gels (5% stacking gel/12.5% resolving gel). Proteins were electrophoretically transferred to Protran nitrocellulose membranes (Schleicher and Schuell) at 100 V for 1 h. Membranes were blocked for 1 h with nonfat milk (5%), incubated 1 h with capsid antibody, incubated 30 min with anti-rabbit-Ig horseradish peroxidase conjugate (Amersham Pharmacia), and developed by using an enhanced chemiluminescence system (Amersham Pharmacia).
Assays for GUS activity were performed with approximately 100 mg of Arabidopsis inflorescence tissue, as previously described (9).
Genomic DNA Extraction and Mapping.
Genomic DNA was isolated from inflorescence tissue as described (12). PCR reactions by using simple sequence-length polymorphism markers (13) were performed as described for nga6, nga8, nga76, nga111, nga112, nga158, nga168, nga172, and nga249 [http://cbil.humgen.upenn.edu/≈atgc/genetic-mapping/gen_maps.html]. PCR reactions with the CTR1 cleaved amplified polymorphic sequence (CAPS) marker were performed as described [http://genome-www.stanford.edu/Arabidopsis/maps/CAPS Chr5.html]. A CAPS marker was developed for the right end of yeast artificial chromosome clone CIC3A2 [http://kazusa.or.jp/arabi/endseq/]. Additional PCR-based markers were generated from the sequences of P1 and TAC clones (14) MUK11 (GenBank accession AB008271), MUG13 (GenBank accession AB005245), K18I23 (GenBank accession AB010692), and MOP10 (GenBank accession AB005241). Primer sequences are available on request. CAPS (15) and dCAPS (16, 17) polymorphisms were identified by comparison of the published Col-0 sequence with the sequence of the Ws-2 allele. All sequences were analyzed for restriction-site polymorphisms by using the DNA strider program (18) and sequence alignments were performed with the clustal w program (19).
Bacterial Artificial Chromosome (BAC) Library Screen.
A filter containing the Texas A&M University BAC library [Arabidopsis Biological Resource Center; (20)] was screened by using the MUK11–3′ PCR product. MUK11–3′ was amplified from Col-3 genomic DNA, gel purified, and radiolabeled by using random hexamer primers and the Klenow fragment of DNA polymerase I (21). The MUK11–3′ probe was hybridized at 42°C in aqueous hybridization solution (22) and washed twice for 20 min in 0.1× SSC, 1% SDS at 65°C. Positive clones were identified by autoradiography.
RESULTS
Selectable TEV Strains and Mass Inoculation.
The identification of plant genes involved in virus–host interactions has been limited because of the lack of development of general genetic approaches to identify mutants with a range of altered susceptibility phenotypes. Several barriers to isolation of informative mutants exist. First, many host genes required to support virus replication and movement may encode essential cellular proteins (23); isolation of viable mutants with defects at these loci may require selections and screens of extremely large populations. Second, traditional methods of inoculation and screening for susceptibility have relied on appearance of symptoms or immunological assays (5–8). Symptom phenotypes are often misleading or noninformative with regard to systemic infection, and immunological and biochemical assays limit the overall throughput in mutant screens. Third, versatile model genetic systems with which virus–host interactions can be dissected have been limited.
To overcome these limitations, we have devised a new system to identify TEV susceptibility mutants in Arabidopsis. Strains of TEV were engineered to provide herbicide-based positive selections for mutants with either gain-of-susceptibility or loss-of-susceptibility to TEV (Fig. 1). Positive selection for host mutants by using virus-dependent herbicide survival should provide an effective approach for identification of susceptibility variants from large populations. The TEV-bar strain contains the bacterial bar gene, which confers resistance to the herbicide GA (24). Fully susceptible Arabidopsis plants (C24 ecotype) infected by TEV-bar were resistant to GA and regenerated inflorescence tissue efficiently after two applications of herbicide (Fig. 2A). The regenerated tissue contained TEV capsid protein, indicating that virus was present in upper noninoculated tissue (Fig. 2B, lane 3). Restrictive Arabidopsis plants (Col-3 ecotype), in which TEV is unable to move out of inoculated leaves infected by TEV-bar were killed by GA application (Fig. 2A). Capsid protein was not detected in upper noninoculated tissue from these plants (Fig. 2B, lane 4). A low percentage of Col-3 plants slowly regenerated inflorescence tissue after one herbicide treatment, but these “escapes” were killed effectively by a second GA application. Noninoculated C24 and Col-3 plants were highly sensitive to GA (data not shown). TEV-bar, therefore, should provide a potent selection for gain-of-susceptibility mutants of Col-3 or another normally restrictive ecotype.
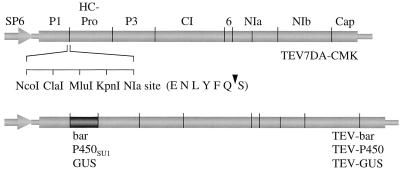
Figure 1.
Schematic representation of recombinant TEV genomes. The bar, P450SU1, and uidA (GUS) genes were fused in-frame within the TEV open-reading frame (thick bar) between the coding sequences for P1 and HC-Pro proteins in the pTEV7DA-CMK cloning vector. The bar, P450SU1, and GUS polypeptides are cleaved from the nascent TEV polyprotein by two viral encoded proteinases, P1 and NIa. The pTEV7DA-CMK cloning vector contains a polylinker cloning site adjacent to an artificial NIa proteinase cleavage site (amino acid sequence given in single-letter code, with arrow indicating the scissile bond). TEV-GUS was described previously, as were the logic and principles used for vector design. The names of TEV proteins are indicated above the TEV7DA-CMK genome map. SP6, SP6 polymerase promoter for producing infectious RNA transcripts in vitro.
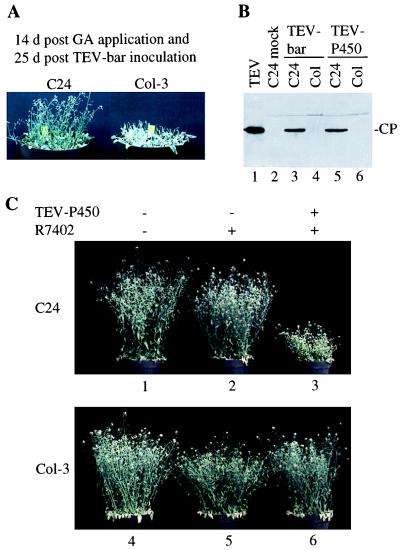
Figure 2.
TEV-bar and TEV-P450 selection strategies. (A) Ten A. thaliana C24 or Col-3 plants were inoculated with TEV-bar, and inflorescences were removed at 11 days p.i., then sprayed with 0.01% GA at 11 and 18 days p.i. At 14 days after the initial GA application, C24 plants had produced new inflorescences, whereas Col-3 plants were dead. (B) Capsid protein (CP) immunoblot analysis of purified TEV capsid protein (lane 1), inflorescence tissue from a mock-inoculated C24 plant (lane 2), or inflorescence tissue from C24 and Col-3 plants inoculated with TEV-bar (lanes 3, 4) or TEV-P450 (lanes 5, 6). The TEV-bar- and TEV-P450-inoculated plants were treated with GA and R7402, respectively. (C) Sets of 10 C24 and Col-3 plants were either inoculated with TEV-P450 (3, 6) or mock-inoculated (1, 2, 4, 5). At 16 days p.i., inflorescence tissue was removed from each plant and two pots of each ecotype were sprayed with 100 μg/l R7402. At 16 days after R7402 application, C24 plants that were inoculated with TEV-P450 were severely stunted and chlorotic, whereas all other plants grew similar to nontreated control plants.
The counter-selectable TEV-P450 virus encodes a chloroplast-targeted cytochrome P450SU1, which conditions sensitivity to the sulfonylurea proherbicide, R7402 (10). In contrast to TEV-bar, TEV-P450 should confer proherbicide-dependent lethality only if plants are systemically infected, which should be useful as a strong selection for loss-of-susceptibility mutants of an ecotype that is normally fully susceptible. TEV-P450-infected C24 plants contained TEV capsid protein in upper noninoculated tissues (Fig. 2B, lane 5) and were highly sensitive to R7402 application (Fig. 2C). These plants were stunted, turned yellow within 16 days of proherbicide application, and were nearly infertile. In contrast, neither TEV-P450-infected Col-3 plants, which lacked capsid protein in noninoculated tissue (Fig. 2B, lane 6), nor noninfected Col-3 or C24 plants were affected by the proherbicide (Fig. 2C). These data also demonstrate that TEV is an effective vector for targeting foreign genes to chloroplasts, because P450SU1 is nonfunctional in planta unless it is localized to chloroplasts (10).
To overcome the limitations associated with manual inoculation of individual plants, a mass inoculation procedure by using an artist’s airbrush was developed. In preliminary experiments, TEV-GUS was used to optimize this high-pressure aerial delivery method. Air pressure was identified as a key variable, with 75 psi providing the most efficient inoculation (data not shown). In four independent experiments (44 plants total), all plants were successfully inoculated, yielding 13.6 ± 9.5 to 33.2 ± 18.2 infection foci per plant by using standard inoculum. This efficiency was judged to be satisfactory for application in large-scale mutant searches.
Isolation of Gain-of-Susceptibility Mutants by Using TEV-bar.
The TEV-bar selectable virus was applied in large-scale selections and screens for Arabidopsis Col-0 mutants with gain-of-susceptibility phenotypes. The Col-0 ecotype restricts TEV to inoculated leaves because of the presence of a dominant RTM1 locus, which maps to position ≈16 cM on chromosome 1 (9). Gain-of-susceptibility, or increased capacity to support long-distance movement of TEV, should result from mutations at the RTM1 locus or at loci required for the RTM1-mediated restriction. Approximately 81,150 M2 plants from 31 populations of ethyl methanesulfonate- or fast neutron bombardment-mutagenized Col-0 were inoculated by the high-pressure aerial delivery method (Table 1). At 14 days postinoculation (p.i.), inflorescences were removed and the plants were sprayed with GA herbicide. Sixty-four plants survived the herbicide treatment (Table 1). These individuals, which are referred to as GA survivors, were detected easily as the only plants to regenerate inflorescences and set seed after herbicide application (Fig. 3A). Of these plants, 15 were confirmed to be systemically infected by TEV-bar because they contained TEV capsid protein in upper noninoculated tissue (Fig. 3B, lanes 5–7; data not shown) and were confirmed to be the Col-0 ecotype based on simple sequence-length polymorphism analysis (13) (Table 1). The progeny from self-fertilization (M3 generation) of 12 of the capsid-protein-containing plants were tested for susceptibility to another engineered TEV strain, TEV-GUS, which encodes the reporter protein β-glucuronidase (Fig. 1). Each of 10 M3 plants from 10 lines (A130, A133, A138, A154, A155, A156, A157, A158, A159, and A161) were susceptible to systemic infection by TEV-GUS as measured by GUS activity assay in noninoculated tissue (Table 1, Fig. 3C). These susceptible lines, therefore, exhibited heritable gain-of-susceptibility mutant phenotypes. Mutant lines A155, A156, A157, A158, and A159 were identified from the same M2 population and were assumed to be siblings; only the A158 and A159 lines from this group were subjected to further analysis. Of the remaining two lines, A152 yielded M3 plants of which approximately 50% exhibited susceptibility to systemic infection by TEV-GUS, and A153 yielded M3 plants, of which none were susceptible to TEV-GUS. These two lines were not characterized further.
Table 1.
Summary of mutant screens using TEV-bar
| Screen | Mutagen* | Number screened | GA survivors† | Capsid positive‡ | Col-0 ecotype§ | M3 progeny susceptible¶ |
|---|---|---|---|---|---|---|
| 1 | EMS | 7,400 | 7 | 4 | 3 | 3 |
| 2 | FNB | 16,900 | 12 | 10 | 0 | ND‖ |
| 3 | EMS | 13,600 | 3 | 1 | 1 | 1 |
| 4 | EMS | 12,000 | 5 | 1 | 1 | 0 |
| 5 | EMS | 13,250 | 7 | 6 | 6 | 6 |
| 6 | EMS | 13,000 | 19 | 3 | 1 | 1 |
| 7 | EMS | 5,000 | 11 | 3** | 3 | ND‖ |
| Total | 81,150 | 64 | 28 | 15 | 11 |
EMS, ethyl methanesulfonate; FNB, fast-neutron bombarded.
Number of plants that regenerated inflorescence tissue after GA application.
Number of GA survivors that possessed capsid protein in newly regenerated inflorescence tissue.
Number of TEV capsid protein-positive GA survivors that were verified to be of the Col-0 ecotype based on the genotype for four different SSLP markers (nga6, nga76, nga111, and nga172).
Number of capsid protein-positive Col-0 M2 GA survivors whose M3 progeny were systemically infected by TEV-GUS. These plants were considered to have heritable mutations resulting in a gain-of-susceptibility phenotype after TEV infection.
ND, not determined.
In screen 7, only the three GA survivors that had the Col-0 genotype were tested for TEV capsid protein in systemic tissue. Therefore, a total of 56 GA survivors were tested for systemic movement by TEV.
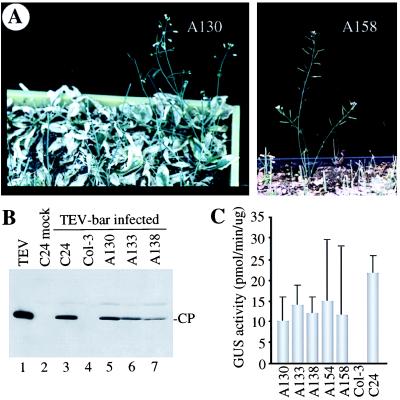
Figure 3.
Isolation of gain-of-susceptibility mutants of A. thaliana Col-0 by using the TEV-bar selectable virus. (A) Identification of mutants A130 and A158 as survivors after GA application. The photographs were taken 13 days after GA application. (B) Capsid protein (CP) immunoblot analysis of purified TEV capsid protein (lane 1), inflorescence tissue from a mock-inoculated C24 plant (lane 2), or inflorescence tissue from C24, Col-3, or GA survivors A130, A133, and A138 inoculated with TEV-bar (lanes 3–7). (C) β-Glucuronidase (GUS) activity assays of inflorescence tissue from M3 progeny of GA survivors (A130, A133, A138, A154, and A158) or restrictive (Col-3) and susceptible (C24) control plants inoculated with TEV-GUS. GUS assays were done at 15 days p.i. or 16 days p.i. for A154 and A158.
Genetic Characterization of Gain-of-Susceptibility Mutants.
A series of crosses and progeny analyses were done to genetically characterize the gain-of-susceptibility mutants. The A130, A133, A138, A154, A158, and A159 M3 plants were backcrossed with Col-3 plants. The F1 progeny plants from each cross restricted systemic infection by TEV-GUS, as indicated by a lack of GUS activity in upper noninoculated tissue at 15 and 22 days p.i. (Table 2), although each plant contained infection foci in inoculated leaves. The F1 plants from the A130, A133, and A138 crosses were allowed to self-fertilize to generate the F2 generation. The ratio of TEV-GUS restrictive:susceptible plants in each F2 population was statistically indistinguishable from a 3:1 ratio, indicating that the gain-of-susceptibility phenotype of these mutants was likely monogenic and recessive in each case (Table 2).
Table 2.
Genetic characterization of gain-of-susceptibility mutants
| Cross | TEV-GUS infection phenotype*
|
Ratio | |
|---|---|---|---|
| Restrictive | Susceptible | ||
| Test of dominance (F1) | |||
| Col-0 × A130 | 5 | 0 | |
| Col-0 × A133 | 4 | 0 | |
| Col-0 × A138 | 5 | 0 | |
| A154 × Col-0 | 9 | 0 | |
| A158 × Col-0 | 23† | 0 | |
| A159 × Col-0 | 8 | 0 | |
| Segregation analyses (F2) | |||
| Col-0 × A130 | 41 | 12 | 3:1 (χ2 = 0.16)‡ |
| Col-0 × A133 | 71 | 23 | 3:1 (χ2 = 0.015)‡ |
| Col-0 × A138 | 105 | 32 | 3:1 (χ2 = 0.2)‡ |
| Complementation (F1) | |||
| La-er × A130 | 0 | 10 | |
| La-er × A133 | 0 | 10 | |
| La-er × A138 | 10 | 0 | |
| A154 × La-er | 0 | 12 | |
| A158 × La-er | 0 | 12 | |
| A159 × La-er | 0 | 12 | |
| A130 × A133 | 0 | 23† | |
| A130 × A138 | 17† | 0 | |
| A138 × A133 | 6 | 1§ | |
Plants were inoculated with TEV-GUS and inflorescence tissue was assayed at 15 and 21 days p.i. for GUS activity. All plants regardless of whether they contained GUS activity in inflorescence tissue, were confirmed to be successfully inoculated by detection of infection foci in inoculated leaves using the GUS colorimetric assay (9).
Data shown are combined from two reciprocal crosses.
P > 0.5.
This plant was likely the result of accidental self-pollination of A138 at the time of crossing. All other data indicated that A138 carried a mutation that could complement rtm1 plants and that segregated as a recessive monogenic locus.
Genetic complementation tests were done by crossing M3 progeny plants of the A130, A133, A138, A154, A158, and A159 mutants to one another and to the La-er ecotype [systemically susceptible to TEV because of a recessive rtm1 allele (9)]. The F1 progeny of these crosses were inoculated with TEV-GUS and GUS assays were performed on upper noninoculated tissue at 15 and 22 days p.i. The F1 progeny plants of each cross between A130, A133, A154, A158, A159, and La-er were susceptible to systemic TEV-GUS infection, suggesting that these mutants contained defects at the RTM1 locus (Table 2). The mutant alleles in A130, A133, A154, and A158/A159 were designated rtm1-1, rtm1-2, rtm1-3, and rtm1-4, respectively. In contrast, the F1 progeny of A138 crossed with A130, A133, or La-er were not systemically susceptible to TEV-GUS (Table 2), suggesting that A138 contains a gain-of-susceptibility mutation at a locus other than RTM1.
Genetic Mapping of a Second Locus Required for Restriction of TEV Movement in Arabidopsis.
To determine the chromosomal position of the mutation in the A138 mutant, a mapping population was generated by crossing A138 to the Ws-2 ecotype (TEV restrictive because of RTM1 locus). The four F1 plants tested were resistant to systemic infection by TEV-GUS (data not shown). A total of 196 susceptible individuals from the F2 generation were analyzed by using simple sequence-length polymorphism and CAPS (15) markers distributed throughout each of the Arabidopsis chromosomes. The TEV susceptibility phenotype segregated closely with markers near the top of chromosome 5, mapping centromeric to CTR1 (five recombination events in 392 chromosomes) and CIC3A2R (two recombinants), and telomeric to nga158 (six recombinants) (Fig. 4). The wild-type locus at this site was designated Restricted TEV Movement-2 (RTM2); the A138 mutant allele was designated rtm2–1. The restricted infection phenotype in Col-0 plants, therefore, requires at least two interdependent loci—RTM1 and RTM2.
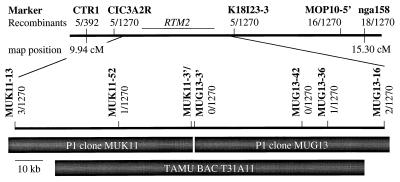
Figure 4.
Map position of RTM2 between CTR1 and nga158 on chromosome 5. PCR-based markers are indicated in bold type. The number of recombination events scored among 1,270 meiotic events in 635 TEV-GUS-susceptible individuals is given below each marker. Only 392 meiotic events were scored for recombination between CTR1 and RTM2. The RI map positions for CTR1 and nga158 were published in the August 1998 Lister & Dean RI map [http://genome-www.stanford.edu/Arabidopsis/ww/home.html].
To facilitate the construction of a high-resolution map and positional cloning of RTM2, the mapping population was increased to 635 TEV-susceptible F2 and F3 individuals, and nine new PCR-based markers were developed throughout the interval between CIC3A2R and nga158 (Fig. 4). RTM2 was bracketed by single recombination events detected at the MUK11–52 locus telomeric to RTM2 and the MUG13–36 locus centromeric to RTM2. The MUK11 and MUG13 P1 clones do not overlap (Fig. 4; ref. 14). The extent of the gap between these two clones was determined to be approximately 1 kb by PCR analysis by using the MUK11–3′ forward and MUG13–3′ forward primers. Therefore, the physical distance between MUK11–52 and MUG13–36 is approximately 78 kb. To identify clones spanning the interval between MUK11–52 and MUG13–36, a filter containing the TAMU BAC library (20) was probed with the MUK11–3′ PCR product. Of the three BAC clones identified, the T31A11 BAC clone was determined to span the 78-kb interval by comparison of T31A11 end sequences (GenBank accession numbers B78540 and B96655) with MUK11 and MUG13 by using the blast program (25).
DISCUSSION
The selectable virus strategy enables efficient recovery of mutants with altered susceptibility phenotypes. By using the TEV-bar or TEV-P450 strains, positive selection for either gain-of-susceptibility of loss-of-susceptibility mutants is provided. Combined with the aerial inoculation method, this system overcomes many of the limitations that have plagued attempts to recover large numbers of mutants with virus susceptibility defects. Most importantly, the strategy enables high-throughput selections among plants by using herbicide or proherbicide survival steps, eliminating reliance on visual symptoms or biochemical and immunological assays as an initial screen for infection. The high-throughput nature of this system will be essential for recovery of rare mutants, as well as for identification of large numbers of mutants with defects at a single locus. We envision that the gain-of-susceptibility selections and screens by using TEV-bar will be particularly useful for recovery of mutants with defects in defense pathways. The loss-of-susceptibility selections by using TEV-P450 should be amenable to isolation of three classes of mutants. TEV-P450 selections should aid in identification of mutants with defects in genes required for virus replication, cell-to-cell movement, and long-distance movement. The concept of isolating mutants with host compatibility function defects was illustrated nicely (5) by using brome mosaic virus replicons expressing a counter-selectable marker in yeast. The TEV-P450 virus should also be useful in identification of enhanced disease-resistant mutants. An enhanced disease-resistant mutant would not carry a mutation in a gene controlling a compatibility function, but would result in the induction of a host defense pathway that may specifically or nonspecifically restrict TEV infection. In addition, the TEV-P450 virus should prove valuable in isolation of intra- and extragenic suppressors of the rtm1 or rtm2 alleles carried in susceptible plants.
The gain-of-susceptibility selections and screens with TEV-bar in this study demonstrate the feasibility and efficacy of the selectable virus strategy. As the Col-0 ecotype carries a TEV-restricting RTM1 allele, the GA survivors were expected to include mutants with defects at the RTM1 locus or at loci encoding factors associated with an RTM1-dependent restriction pathway. The selection by using GA herbicide was very efficient for identification of systemically infected plants (50% of GA survivors tested) (Table 1). The selection also revealed the presence of a significant number of fully susceptible ecotype plants as contaminants in the mutagenized seed lots, although these were easily distinguished from Col-0 plants by using PCR-based markers. The vast majority of the susceptible Col-0 plants identified in the screen contained heritable gain-of-susceptibility mutations. An initial concern of using this selection strategy was that TEV-bar mutants with the ability to overcome the RTM1-mediated restriction would be recovered with a high frequency, thus limiting the value of this strategy as a means to recover host mutants. However, this potential problem proved not to be a major impediment to use of the screen because no gain-of-virulence mutants of TEV-bar were identified.
The identification of two loci, RTM1 and RTM2, which function cooperatively in Arabidopsis to block long-distance movement of TEV in a nonhypersensitive response-dependent manner reveals the existence of a multicomponent pathway or complex with virus-suppressing activity that clearly differs from the signaling pathway activated by typical dominant viral resistance genes [such as N; (4)]. The interdependence of RTM1 and RTM2 is suggested by the requirement for both loci in TEV-restrictive plants. As Col-0 is susceptible to systemic infection by a broad range of other viruses (26), the RTM1/RTM2-mediated restriction is virus-specific. The long-distance movement block could be caused by interference with the vasculature-dependent movement functions of TEV. Four TEV proteins — HC-Pro, CI, NIa, and capsid protein — are required either directly or indirectly for long-distance movement (27), providing multiple targets against which a movement-restricting response could act. Alternatively, the restriction could be caused by a local or systemic induced response that affects vasculature-dependent transport or establishment of infection at sites distal to the inoculated leaves. In fact, there is evidence that nonhypersensitive response defense responses are induced on infection by viruses (reviewed by Carrington and Whitham, ref. 1). For example, virus-inhibiting processes with the characteristics of posttranscriptional gene silencing are induced on infection by a range of viruses (28, 29). It is interesting to note that one of the functions of TEV HC-Pro is to suppress posttranscriptional gene silencing (30–32), which may represent a counterdefensive strategy to circumvent this antiviral response. Mutant TEV strains with defects in HC-Pro exhibit the ability to infect inoculated leaves, but lack the ability to move long distances (33), a phenotype similar to wild-type TEV strains in RTM1/RTM2 plants (9). The hypothesis that RTM1 and/or RTM2 mediate specific recognition of TEV and elicitation of silencing is worth testing. Isolation of RTM1 and RTM2 will be crucial to understanding the biochemical functions of these two proteins.
Acknowledgments
We thank Jon Holt for assistance with plant maintenance and inoculations and Jenny Kleene for assistance with GUS assays. We are grateful for the contributions of plasmids from Barbara Baker and Joan Odell. This work was supported by grants from the National Institutes of Health (GM18529 to S.A.W. and AI43288 to J.C.C.), by a Washington State University Plant Biology Research Training Center fellowship to M.L.Y., and by the United States Department of Agriculture (95–37303-1867 to J.C.C.).
ABBREVIATIONS
- TEV
tobacco etch virus
- TMV
tobacco mosaic virus
- GA
glufosinate-ammonium
- CAPS
cleaved amplified polymorphic sequence
- BAC
bacterial artificial chromosome
- p.i.
postinoculation
- GUS
β-glucuronidase
References
- 1.Carrington J C, Whitham S A. Curr Opin Plant Biol. 1998;1:336–341. doi: 10.1016/1369-5266(88)80056-6. [DOI] [PubMed] [Google Scholar]
- 2.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 3.Dawson W O, Hilf M E. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:527–555. [Google Scholar]
- 4.Whitham S, Dinesh-Kumar S P, Choi D, Hehl R, Corr C, Baker B. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 5.Ishikawa M, Naito S, Ohno T. J Virol. 1993;67:5328–5338. doi: 10.1128/jvi.67.9.5328-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohshima K, Taniyama T, Yamanaka T, Ishikawa M, Naito S. Virology. 1998;243:472–481. doi: 10.1006/viro.1998.9078. [DOI] [PubMed] [Google Scholar]
- 7.Yoshii M, Yoshioka N, Ishikawa M, Naito S. Plant J. 1998;13:211–219. doi: 10.1046/j.1365-313x.1998.00024.x. [DOI] [PubMed] [Google Scholar]
- 8.Lartey R T, Ghoshroy S, Citovsky V. Mol Plant–Microbe Interact. 1998;11:706–709. doi: 10.1094/MPMI.1998.11.7.706. [DOI] [PubMed] [Google Scholar]
- 9.Mahajan S K, Chisholm S T, Whitham S, Carrington J C. Plant J. 1998;14:177–186. doi: 10.1046/j.1365-313x.1998.00105.x. [DOI] [PubMed] [Google Scholar]
- 10.O’Keefe D P, Tepperman J M, Dean C, Leto K J, Erbes D L, Odell J T. Plant Physiol. 1994;105:473–482. doi: 10.1104/pp.105.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrington J C, Haldeman R, Dolja V V, Restrepo-Hartwig M A. J Virol. 1993;67:6995–7000. doi: 10.1128/jvi.67.12.6995-7000.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dellaporta S L, Wood J, Hicks J B. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 13.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura Y, Sato S, Kaneko T, Kotani H, Asamizu E, Miyajima N, Tabata S. DNA Res. 1997;4:401–414. doi: 10.1093/dnares/4.6.401. [DOI] [PubMed] [Google Scholar]
- 15.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 16.Neff M M, Neff J D, Chory J, Pepper A E. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 17.Michaels S D, Amasino R M. Plant J. 1998;14:381–385. doi: 10.1046/j.1365-313x.1998.00123.x. [DOI] [PubMed] [Google Scholar]
- 18.Marck C. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi S, Creelman R A, Mullet J E, Wing R A. Weeds World. 1995;2:17–20. [Google Scholar]
- 21.Feinberg A P, Vogelstein B. Anal Biochem. 1983;257:8569–8572. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 22.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 23.Ishikawa M, Diez J, Restrepo-Hartwig M, Ahlquist P. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson C J, Movva N R, Tizard R, Crameri R, Davies J E, Lauwereys M, Botterman J. EMBO J. 1987;6:2519–2523. doi: 10.1002/j.1460-2075.1987.tb02538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Kunkel B N. Trends Genet. 1996;12:63–69. doi: 10.1016/0168-9525(96)81402-8. [DOI] [PubMed] [Google Scholar]
- 27.Carrington J C, Kasschau K D, Mahajan S K, Schaad M C. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratcliff F, Harrison B D, Baulcombe D C. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 29.Al-Kaff N S, Covey S N, Kreike M M, Page A M, Pinder R, Dale P J. Science. 1998;279:2113–2115. doi: 10.1126/science.279.5359.2113. [DOI] [PubMed] [Google Scholar]
- 30.Anandalakshmi R, Pruss G J, Ge X, Marathe R, Mallory A C, Smith T H, Vance V B. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasschau K D, Carrington J C. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 32.Brigneti G, Voinnet O, Li W X, Ji L H, Ding S W, Baulcombe D C. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Kasschau K D, Cronin S, Carrington J C. Virology. 1997;228:251–262. doi: 10.1006/viro.1996.8368. [DOI] [PubMed] [Google Scholar]