Abstract
The assembly and budding of human immunodeficiency virus type 1 (HIV-1) at the plasma membrane are directed by the viral core protein Pr55gag. We have analyzed whether Pr55gag has intrinsic affinity for sphingolipid- and cholesterol-enriched raft microdomains at the plasma membrane. Pr55gag has previously been reported to associate with Triton X-100-resistant rafts, since both intracellular membranes and virus-like Pr55gag particles (VLPs) yield buoyant Pr55gag complexes upon Triton X-100 extraction at cold temperatures, a phenotype that is usually considered to indicate association of a protein with rafts. However, we show here that the buoyant density of Triton X-100-treated Pr55gag complexes cannot be taken as a proof for raft association of Pr55gag, since lipid analyses of Triton X-100-treated VLPs demonstrated that the detergent readily solubilizes the bulk of membrane lipids from Pr55gag. However, Pr55gag might nevertheless be a raft-associated protein, since confocal fluorescence microscopy indicated that coalescence of GM1-positive rafts at the cell surface led to copatching of membrane-bound Pr55gag. Furthermore, extraction of intracellular membranes or VLPs with Brij98 yielded buoyant Pr55gag complexes of low density. Lipid analyses of Brij98-treated VLPs suggested that a large fraction of the envelope cholesterol and phospholipids was resistant to Brij98. Collectively, these results suggest that Pr55gag localizes to membrane microdomains that are largely resistant to Brij98 but sensitive to Triton X-100, and these membrane domains provide the platform for assembly and budding of Pr55gag VLPs.
The plasma membrane is partially composed of ordered domains, called “rafts,” which are enriched in sphingolipids and cholesterol and contain a specific set of proteins (5, 43). Rafts are resistant to extraction with nonionic detergents at low temperatures, and thus rafts and raft-associated proteins can be separated from detergent-solubilized material by fractionation of cell lysates on density gradients (6). The plasma membrane apparently contains different types of rafts, which exhibit differential sensitivities to different detergents (11, 41). The best-characterized rafts are Triton X-100-resistant rafts, which have been implicated in playing a critical role in numerous cellular processes (28, 43, 45). Triton X-100-resistant rafts have also been proposed to provide a platform for assembly and budding of several different enveloped viruses (38, 53, 55). One of these viruses is human immunodeficiency virus type 1 (HIV-1) (32, 35, 36, 42). Assembly and budding of HIV-1 occur at the plasma membrane and are directed by the viral core protein precursor Gag (Pr55gag), since Pr55gag expressed in the absence of other viral components induces formation of enveloped virus-like particles (VLPs) (18, 30). Pr55gag associates with the cytoplasmic leaflet of the plasma membrane via an amino-terminal dual motif that consists of a covalent myristic acid modification and a cluster of basic amino acid residues (7, 13, 18, 19, 26, 47, 48, 58). Through a mechanism that is poorly understood, the membrane-bound Pr55gag proteins oligomerize into core structures and concomitantly deform the membrane into a bud (14, 17). Five recent reports have concluded that Triton X-100-resistant rafts play an important role in this Pr55gag-mediated assembly and budding (32, 35, 36, 42, 57). Four of these reports assigned Pr55gag to the Triton X-100-resistant rafts, since density gradient analyses indicated that a significant fraction of intracellular Pr55gag displayed buoyant density in cold Triton X-100 cell lysates (32, 35, 36, 57). Results from Lindwasser and Resh (32), however, implied that Pr55gag does not localize to “classical” Triton X-100-resistant rafts, but instead localizes to distinct, dense Triton X-100-resistant rafts, which were given the name “barges.” It was speculated that higher density of barges was caused by the presence of extensive arrays of oligomeric Pr55gag assembly intermediates in a raft-like membrane. It was suggested that assembly of HIV-1 occurs at the raft-like barge-membranes, since Triton X-100-solubilized Pr55gag complexes from extracellular VLPs had a density similar to that of intracellular barges, and mutant Pr55gag proteins that exhibited increased affinity for barges were found to produce VLPs more efficiently than wild-type Pr55gag (32). In addition to localization of Pr55gag to rafts or barges, three other lines of evidence have been put forward in support of rafts playing a critical role in assembly and budding of HIV-1. (i) Cholesterol-depleting agents, which among other things cause alterations in raft structures, have been demonstrated to decrease the release of virus particles from HIV-1-infected cells (36), as well as to reduce the infectivity of released particles (36, 57). (ii) There is localization (partial) of HIV-1 Env proteins to Triton X-100-resistant rafts (42). (iii) The presence of raft-associated host proteins and lipids in HIV-1 particles has been interpreted to signify selective budding of HIV-1 through (Triton X-100-resistant) rafts (35).
In this study, we have analyzed raft association of HIV-1 Pr55gag by using Triton X-100 and Brij98 extractions as well as confocal fluorescence microscopy. Our results demonstrate that Triton X-100 extraction when cold readily solubilizes bulk of VLP envelope lipids. Thus, it is questionable whether the buoyancy of Triton X-100-treated Pr55gag complexes can be taken as evidence for localization of the protein to rafts. However, confocal fluorescence microscopy analyses indicated extensive colocalization of Pr55gag at the cell surface with patched GM1-positive rafts. Furthermore, Brij98 extractions indicated that a population of intracellular Pr55gag, as well as the VLP envelope, was largely resistant to this detergent. This raises the possibility that cell surface raft structures that are resistant to Brij98 but sensitive to Triton X-100 could provide a platform for Pr55gag-mediated assembly and budding of HIV-1.
MATERIALS AND METHODS
Cell culture.
Jurkat cells were maintained in RPMI 1640 medium containing 10% fetal calf serum, 10 mM HEPES (pH 7.4), 2 mM glutamine, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml (all tissue culture reagents were from Invitrogen Life Technologies). 293T cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 2 mM glutamine, 100 U of penicillin per ml, 0.1 mg of streptomycin per ml, nonessential amino acids, and 1 mM sodium pyruvate.
Preparation of infectious SFV-C/HIVgag.
Plasmid pSFV-C/HIVgag was used for the production of recombinant Semliki Forest virus (SFV) particles directing the synthesis of HIV-1 Pr55gag. In this plasmid, the Pr55gag coding sequence is fused to the C gene of SFV. The first step in the construction of pSFV-C/HIVgag was cloning of the Pr55gag open reading frame from pSP64-HIV111b-BH10 (20) as a 1,720-bp BamHI-StuI fragment into the BamHI-SmaI-linearized pSFV1 (31) to give pSFV1-HIVgag. The fusion between SFV-C and HIV-1 Pr55gag genes was engineered by PCR such that the second codon of Pr55gag was fused to the last codon of SFV-C by using Vent DNA polymerase (New England Biolabs, Beverly, Mass.) in accordance with the manufacturer's instructions (27, 54). The terminal primers used for the amplification of the fused DNA fragment were 5′-CAACGGAAAAACGCAGCAGC-3′ (the C 5′ primer) and 5′-CCCCCTGGCCTTAACCGAATTTTTCCC-3′ (the Pr55gag 3′ primer). The fusion primers were 5′-GGGGTCCGAAGAGTGGGGTGCGGAGAGCGTCA-3′ (C fragment primer) and 5′-TGACGCTCTCGCACCCCACTCTTCGGACCCC-3′ (Pr55gag fragment primer). After amplification, the fusion fragment was cut with AsuII and ClaI and cloned into AsuII-ClaI-digested pSFV1-HIVgag to give pSFV-C/HIVgag(1). Subsequently the pSFV-C/HIVgag (1) was modified by site-directed mutagenesis (9) with the primers 5′-CGTGCTCAAGAGTTCCAAGTTGGCC-3′ (SacI site eliminator) and 5′-GTACTGAGAGACAGGCTAACTTCCTGGGGAAGATCTGGCCTTCC-3′ to mutate the frameshift signal so that only Pr55gag sequences from the Gag gene insert could be translated. The mutations were silent mutations with respect to Pr55gag coding sequence. Another modification of pSFV-C/HIVgag(1) was to delete one of the two amino-terminal repeats (LQSRPEPTAPP) of the BH10 p6 domain by using the primer5′-GGAAGGCCAGGGAATTTTCTTCAGAGCAGACCAGGCCAACAGCCCCACCAGAAGAGAGCTTCAGGTCTGGG-3′. This gave the final version pSFV-C/HIVgag that was used in the study. Infectious SFV particles directing the synthesis of HIV-1 Pr55gag (SFV-C/HIVgag) were prepared by coelectroporating equal amounts of in vitro-transcribed SFV-C/HIVgag and helper 1 (31) RNAs into 107 BHK-21 cells as previously described (49, 50).
Analysis of raft association of Pr55gag. (i) Total cell lysates.
Jurkat cells (3.5 × 106) were infected with SFV-C/HIVgag in RPMI-BSA medium (RPMI 1640 containing 0.2% bovine serum albumin [BSA], 10 mM HEPES, pH 7.4, 2 mM glutamine, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml), and 4 h postinfection, cells were switched to methionine-free minimum essential medium (MEM; supplemented with 0.2% BSA, 10 mM HEPES, pH 7.4, 2 mM glutamine, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml) for 30 min. Cells were subsequently metabolically labeled in methionine-free modified Eagle's medium containing 100 μCi of [35S]methionine per ml for 15 min and chased for 0 to 60 min in RPMI-BSA medium containing 10-fold excess of cold methionine. After chase, cells were placed on ice and washed once with ice-cold phosphate-buffered saline (PBS; with MgCl2 and CaCl2), and then the cells were lysed in 200 μl of lysis buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 20 μg of phenylmethylsulfonyl fluoride per ml, 1 μg of CLAP [chymostatin, leupeptin, aprotinin, and pepstatin A] per ml). After 30 min of incubation at 0°C, lysate was mixed with 3 ml of 56% iodixanol (Optiprep; Nycomed Pharma A/S) and overlaid with 3 ml of 40% iodixanol, 5 ml of 30% iodixanol, and 0.5 ml of 5% iodixanol (all iodixanol solutions were in 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). Gradients were centrifuged in an SW41 rotor at 77,100 × g for 14 h at 4°C. One-milliliter fractions were collected from the top, aliquots of each fractions were mixed with sodium dodecyl sulfate (SDS) sample buffer, and equal amounts of fractions were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Radioactivity in Pr55gag bands was determined by using a Bas-III Image Plate and the Bio-Image analyzer system Bas 2000 (Fuji Photo film Co.), and the amount of Pr55gag in each of the different fractions was expressed as a percentage of the total (the sum of all fractions). Influenza virus NP expressed from SFV-C/NP genome (56), transferrin receptor (TR) expressed from SFV-C/TRΔ2 (49), and CD55 expressed from SFV1-CD55 were used as controls. Raft association of influenza virus NP was analyzed after a 15-min pulse and 1-h chase, and that of TR was analyzed after a 60-min pulse and 120-min chase. Cell lysates from SFV1-CD55-infected cells were prepared 5 h postinfection, and proteins in gradient fractions were precipitated with 10% trichloroacetic acid and analyzed by SDS-PAGE followed by Western blotting with anti-CD55 antibody (H-319) from Santa Cruz Biotechnology, Inc. Lck was analyzed from gradient fractions by Western blotting with anti-Lck antibody (3A5) from Santa Cruz Biotechnology, Inc.
(ii) Membrane-associated Pr55gag-crude cell homogenates.
Jurkat cells (107) were infected with SFV-C/HIVgag and metabolically labeled with [35S]methionine as described above. After chase, cells were placed on ice, washed once with ice-cold PBS (with MgCl2 and CaCl2), and homogenized in buffer containing 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 20 μg of phenylmethylsulfonyl fluoride per ml, and 1 μg of CLAP per ml with a tight-fitting Dounce homogenizer. Immediately after homogenization, NaCl was added to 150 mM, and the homogenate was diluted with 100% Nycodenz (Nycomed Pharma A/S) to give a final 60% Nycodenz concentration. The sample (2 ml) was overlaid with 1.5 ml of 50% Nycodenz and 1 ml of homogenization buffer (with 150 mM NaCl), and the gradient was centrifuged in an SW50.1 rotor at 63,000 × g for 45 min at 4°C. The total membrane fraction at the buffer-50% Nycodenz interphase was collected. About 200 to 400 μl of this membrane fraction was mixed with Triton X-100 (final concentration, 1%) and after 30 min of incubation at 0 or 37°C, the solubilized membranes were analyzed on iodixanol step gradients as described above. TR was analyzed after a 60-min pulse and 30-min chase, influenza virus NP was analyzed after a 60-min pulse, and CD55 and Lck were analyzed by Western blotting as described above.
Subconfluent 293T cells grown on 10-cm-diameter plates were transfected with 28 μg of pCMV-HIVgag plasmid by a calcium phosphate precipitation technique. The pCMV-HIVgag plasmid was derived from pCMVΔR8.91 (a generous gift from Didier Trono) (59) by deleting a BclI-EcoRI fragment from the Pol region. The transfected cells were collected ∼ 22 to 24 h posttransfection and processed as described above for the SFV-C/HIVgag-infected Jurkat cells. Total Pr55gag in gradient fractions was visualized by Western blotting using anti-p24 antibody from Biogenesis.
(iii) Membrane-associated Pr55gag postnuclear supernatants.
Jurkat cells (107) were infected with SFV-C/HIVgag and metabolically labeled with [35S]methionine as described above. Transfected 293T cells were analyzed ∼22 to 24 h posttransfection. After chase, cells were placed on ice, washed once with ice-cold PBS (with MgCl2 and CaCl2), and homogenized in buffer containing 25 mM HEPES-KOH (pH 7.4), 0.25 M sucrose, 4 mM MgCl2, 20 μg of phenylmethylsulfonyl fluoride per ml, and 1 μg of CLAP per ml by using a tight-fitting Dounce homogenizer. The crude cell homogenates were centrifuged at 1,400 × g for 4 min at 4°C to pellet nuclei and cell debris. About 70 to 80% of total labeled Pr55gag was found in the postnuclear supernatant (PNS), which was subsequently fractionated on the Nycodenz step gradient described above to separate membranes from cytosolic material (gradient solutions were prepared in 25 mM HEPES-KOH, pH 7.4, 150 mM NaCl and 1 mM EDTA). The total membrane fraction from the buffer-50% Nycodenz interphase was mixed with Triton X-100 (final concentration, 1%) and protease inhibitors, and after 30 min of incubation at 0°C, or at 37°C, the solubilized membranes were analyzed on iodixanol step gradients as described above. For Brij98 extraction, the total membranes were mixed with protease inhibitors and 0.5% Brij98, and after a 5-min incubation at 37°C, the extracts were mixed with 3.5 ml of 56% iodixanol and overlaid with 2 ml of 30% iodixanol, 2 ml of 25% iodixanol, 2 ml of 20% iodixanol, and 0.5 ml of 5% iodixanol (all iodixanol solutions were in 25 mM HEPES-KOH, pH 7.4, 150 mM NaCl, 1 mM EDTA). Gradients were centrifuged in an SW41 rotor at 77,100 × g for 14 h at 4°C.
(iv) VLPs.
Jurkat cells (107) were infected with SFV-C/HIVgag and metabolically labeled with [35S]methionine for 3 h as described above. Culture media were collected and clarified by centrifugation at 200 × g for 4 min. VLPs from clarified culture media were purified by a two-step gradient centrifugation. The clarified culture supernatant was first applied to a linear 5 to 20% iodixanol gradient (21). After centrifugation in an SW41 rotor at 160,000 × g for 1 h at 4°C, fractions were collected from the top, and Pr55gag-containing fractions were pooled, 1 ml of pooled fractions was mixed with 3 ml of 56% iodixanol, and this mixture was overlaid with 4 ml of 27% iodixanol, 3 ml of 20% iodixanol, and 0.5 ml of 5% iodixanol. The iodixanol solutions were made in a mixture of 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 1 mM EDTA (Triton X-100 extraction) or a mixture of 25 mM HEPES-KOH (pH 7.4), 150 mM NaCl, and 1 mM EDTA (Brij98 extraction). The gradients were centrifuged in a SW41 rotor at 77,100 × g for 14 h at 4°C, and VLPs from the 20 to 27% iodixanol interphase were collected and extracted with Triton X-100 or Brij98 as described above and analyzed on iodixanol step gradients as described above.
Production and analysis of [32P]orthophosphate- and [35S]methionine-labeled VLPs.
Jurkat cells were prelabeled with [32P]orthophosphate as described in reference 21. Prelabeled Jurkat cells (107) were infected with SFV-C/HIVgag and metabolically labeled with [35S]methionine for 3 h as described above. VLPs were harvested from clarified culture supernatants as described above. Material from SFV-C/NP-infected Jurkat cells prepared in a similar way was used as a control to estimate contamination of VLPs by microvesicles. Triton X-100- or Brij98-extracted VLPs or NP controls were fractionated on iodixanol step gradients as described above, except that gradients were centrifuged for 4.5 h. Buoyant Pr55gag complexes from the 30 to 40% iodixanol interphase (Triton X-100 extraction) or the 25% iodixanol zone (Brij98 extraction) were collected, diluted fourfold with a mixture of 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA (Triton X-100) or 25 mM HEPES-KOH (pH 7.4), 150 mM NaCl, 1 mM EDTA (Brij98), and pelleted by centrifugation in a SW41 rotor (160,000 × g for 1 h at 4°C). The pelleted Pr55gag complexes and the control intact, nonextracted VLPs were solubilized with 3% SDS at 70°C, mixed with SDS-PAGE buffer, and analyzed by SDS-PAGE with 20% polyacrylamide gels (21). Radioactivity in mixed SDS-lipid micelles was determined by using a Bas-III Image Plate and the Bio-Image analyzer system Bas 2000. [35S]methionine label in Pr55gag bands (analyzed by SDS-PAGE with 10% polyacrylamide gels) was used to ensure that similar amounts of Pr55gag from the intact particles and the buoyant detergent-treated Pr55gag complexes were loaded on 20% polyacrylamide gels.
Production and analysis of [3H]cholesterol-labeled VLPs.
Jurkat cells (107) were infected with SFV-C/HIVgag or with SFV-C/NP for 30 min. The virus inoculum was removed, and the cells were metabolically labeled with 10 μCi of [1α,2α(n)-3H]cholesterol per ml for 4 h in RPMI-BSA medium. The radioactive cholesterol label was removed and cells were incubated in RPMI-BSA medium for 3 h. VLPs were harvested from clarified culture supernatants by the two-step centrifugation procedure described above. After the last flotation gradient, the peak VLP fractions were pooled and then diluted threefold with buffer, and VLPs were pelleted by a 2-h centrifugation in an SW50.1 rotor at 173,000 × g (4°C). For Triton X-100 extraction, the pellets were placed into 160 μl of mixture containing 10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 20 μg of phenylmethylsulfonyl fluoride per ml, 1 μg of CLAP per ml, and 1% Triton X-100 and incubated at 0°C for 30 min, and the extracts were mixed with 1 ml of 56% iodixanol and overlaid with solutions of 400 μl of 40% iodixanol, 400 μl of 30% iodixanol, and 200 μl of 5% iodixanol. The gradients were centrifuged in TLS-55 rotors at 166,000 × g for 4 h at 4°C, 400-μl fractions were collected from the top, and [3H]cholesterol in fractions was quantified by scintillation counting. The counts from the NP control were subtracted from the VLP values, and the amount of [3H]cholesterol radioactivity in different fractions was expressed as a percentage of the total (the sum of all fractions). For Brij98 extraction, the pelleted VLPs were placed into a mixture containing 160 μl of 25 mM HEPES-KOH (pH 7.4), 150 mM NaCl, 1 mM EDTA, 20 μg of phenylmethylsulfonyl fluoride per ml, 1 μg of CLAP per ml, and 0.5% Brij98, incubated at 37°C for 5 min, and the extracts were mixed with 550 μl of 56% iodixanol and overlaid with 400 μl of 30% iodixanol, 800 μl of 20% iodixanol, and 200 μl of 5% iodixanol solutions. The gradients were centrifuged and collected as described for the Triton X-100 extraction.
Confocal fluorescence microscopy.
Subconfluent 293T cells seeded onto coverslips (3-cm-diameter plates) were transfected with 3 μg of pCMV-HIVgag. After 20 to 22 h, cells were placed on ice and washed twice with cold PBS (with MgCl2 and CaCl2) and then incubated for 20 min on ice with fluorescein isothiocyanate (FITC)-conjugated cholera toxin B subunit (10 μg/ml in PBS with MgCl2 and CaCl2; Sigma). After two washes in PBS, cells were fixed in 4% formaldehyde for 30 min at room temperature (unpatched cells) or incubated on ice in PBS (with MgCl2 and CaCl2) containing anti-cholera toxin antibodies (Calbiochem) for 30 min and subsequently transferred to 37°C for 30 min for patching. After two washes in PBS, cells were fixed in 4% formaldehyde as described above, permeabilized with 0.3% Triton X-100, and stained with mouse anti-Pr55gag antibodies (EF-7 or 38:9; generous gifts from Jorma Hinkula, Department of Virology, SMI/KI, Sweden) and tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-mouse immunoglobulin G (IgG) antibodies (Southern Biotechnology Associates, Inc.). Preparations were mounted in glycerol-PBS (pH 8.0) containing p-phenyldiamine and analyzed with the Carl Zeiss LSM 510 confocal system. The system is based on the compact scanning module, which has been fitted to an inverted (Axiovert 200 M) microscope. The LSM system is equipped with Zeiss software (LSM modes) and is controlled via a standard high-end Pentium PC. Two lasers were selected—one with a wavelength of 488 nm and the other with a wavelength of 514 to 543 nm. In most cases, 40 to 50 optical horizontal sections with intervals about 0.25 μm were taken through the whole preparation (Z-stack). Selected single optical sections were then analyzed, and fluorescence images were saved and processed in Photoshop 6.0. In some cases, the Z-stack was analyzed in the pseudo-three-dimensional LSM mode with orthogonal function allowing us to obtain a membrane-bound intracellular section through the whole cell.
RESULTS
Analyses of raft association of Pr55gag by using Triton X-100.
We chose to use SFV-driven expression of Pr55gag in Jurkat cells to analyze raft association of the protein. In the recombinant SFV genome SFV-C/HIVgag, the coding sequence of Pr55gag is fused to the C gene of SFV, and the resulting C-Pr55gag fusion protein is cotranslationally processed into C and Pr55gag proteins by the autoproteolytic activity of SFV C protein (1, 34). [3H]myristic acid labeling of cells indicated that Pr55gag expressed in SFV-C/HIVgag-infected cells was efficiently myristoylated (data not shown). We also tested kinetics and efficiency of membrane association of newly synthesized Pr55gag. Cell homogenates of SFV-C/HIVgag-infected Jurkat cells were prepared after 15 min of metabolic labeling with [35S]methionine and a 0- or 30-min chase, and membranes were separated from cytosolic material by fractionation on a flotation gradient. The results obtained indicated that, already by the 0-min chase point, ∼34% of labeled Pr55gag was membrane associated, and the membrane-bound pool increased to ∼50% after a 60-min chase (data not shown). Thus, the newly synthesized Pr55gag in SFV-C/HIVgag-infected cells becomes membrane associated with similar kinetics to Pr55gag expressed from proviral constructs (52).
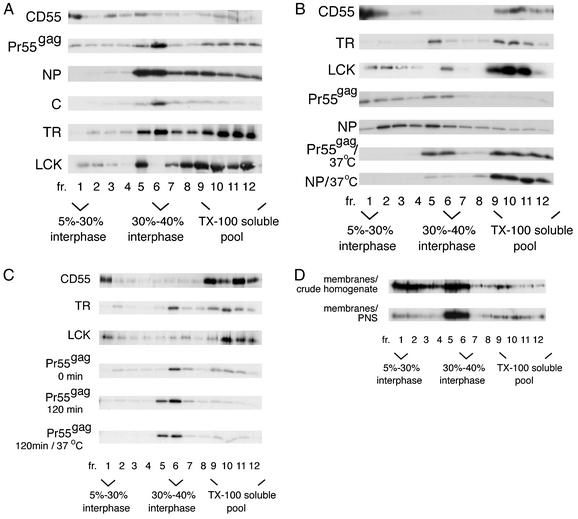
Pr55gag has been previously assigned to Triton X-100-resistant rafts based on results from a biochemical flotation assay that isolates lipid rafts on the basis of their insolubility in this detergent at low temperatures (32, 35, 36). However, as shown in Fig. 1, the assay is problematic, since the flotation phenotype of Pr55gag varies, depending on which cellular fraction is used as the starting material for Triton X-100 extraction. Figure 1A shows the results from crude cell lysates. Recombinant SFV-infected Jurkat cells expressing Pr55gag or control marker proteins were extracted with 1% Triton X-100 at 0°C for 30 min, and the extracts were analyzed on an iodixanol step gradient, which consisted of a 56% loading zone overlaid with 40, 30, and 5% iodixanol solutions. After ultracentrifugation, the Triton X-100-insoluble rafts float to the 5 to 30% iodixanol interphase (fraction 1), whereas detergent-soluble proteins stay in the loading zone (fractions 9 to 12) (4). Our positive control for a raft-associated protein, the glycosylphosphatidylinositol (GPI)-anchored protein CD55 (16, 46), efficiently floated to fraction 1. In contrast, e.g., after a 15-min pulse with [35S]methionine and a 60-min chase, only minor amounts of labeled Pr55gag were found in fraction 1, but a clear concentration of the protein was seen at the 30 to 40% iodixanol interphase in fraction 6. About 37% of labeled Pr55gag was in the Triton X-100-soluble pool (fractions 9 to 12), which could represent cytosolic Pr55gag, or be of both cytosolic and membranous origins. The Pr55gag complexes at the 30 to 40% interphase correspond to the “barges” described by Lindwasser and Resh (32). Barges have been interpreted to represent raft-like membranes that have a higher density than classical Triton X-100-resistant rafts due to the presence of multimeric Pr55gag complexes in barges (32). However, our control experiments indicated that both the influenza virus NP, a predominantly nuclear protein (56), and the cytosolic nucleocapsid protein C of SFV (50, 51) efficiently floated to the 30 to 40% iodixanol interphase after cold Triton X-100 extraction (Fig. 1A). Significant amounts of the non-raft membrane marker TR ended up in the 30 to 40% iodixanol interphase as well (Fig. 1A). We also analyzed the distribution of the endogenous Src family kinase Lck, which like Pr55gag associates with the cytoplasmic leaflet of the plasma membrane. As previously reported (24), the majority of Lck was found in the Triton X-100-soluble pool (fractions 9 to 12). Some Lck was in fraction 1, but significant amounts of the protein concentrated in fraction 5. Taken together, these controls demonstrated that different types of proteins in crude Triton X-100 cell lysates float to the 30 to 40% iodixanol interphase, and thus this flotation phenotype does not necessarily signify association of a protein with raft-like membranes. The extreme viscosity of the SDS-solubilized fraction 5 and 6 samples suggested that this gradient region contained large amounts of chromatin. This raised the possibility that flotation of some of the marker proteins, and possibly also that of Pr55gag, to the 30 to 40% iodixanol interphase region could actually be a postlysis artifact caused by unspecific trapping of proteins to nuclear remnants or chromatin.
FIG. 1.
Cold Triton X-100 extraction of cells yields variable complexes for Pr55gag, depending on which cellular fraction is used as a starting material for the detergent treatment. (A) Analyses of crude cell lysates. Pr55gag, CD55, influenza virus NP, the nucleocapsid protein C of SFV, and the TR were expressed in Jurkat cells from recombinant SFV genomes. Cells were extracted with 1% Triton X-100 (TX-100) at 0°C, and lysates were subjected to flotation on iodixanol step gradients. Aliquots of gradient fractions were analyzed by SDS-PAGE. The Pr55gag, NP, and C proteins were analyzed after a 15-min pulse with [35S]methionine and a 60-min chase, and TR was analyzed after a 60-min pulse and 120-min chase, whereas CD55 and the endogenous Lck were detected by Western blotting. (B) Analyses of the total membrane fraction from Jurkat crude cell homogenates. Pr55gag was analyzed after a 15-min pulse with [35S]methionine and a 60-min chase, NP was analyzed after a 60-min pulse, and TR was analyzed after a 60-min pulse and 30-min chase. Total CD55 and Lck were visualized by Western blotting. (C) Analyses of the total membrane fraction from the PNS of Jurkat cells. Pr55gag was analyzed after a 15-min pulse with [35S]methionine and a 0- or 120-min chase, and TR was analyzed after a 90-min pulse. Total CD55 and Lck were visualized by Western blotting. (D) Analyses of membrane-bound Pr55gag from transfected 293T cells. Total Pr55gag in gradient fractions was detected by Western blotting.
Since the results from crude cell lysates were difficult to interpret, we also tested alternative cellular fractions in the assay. Figure 1B shows results from experiments in which the total membrane fraction of cells was used as the starting material for Triton X-100 extraction. Crude cell homogenates were first fractionated on a flotation gradient to separate membrane-associated proteins from cytosolic components. The total membrane fraction was extracted with 1% Triton X-100 at cold temperatures, and the extracts were analyzed on the iodixanol step gradient described above. Our positive control CD55 still efficiently floated to fraction 1 in this modified raft fractionation scheme. The flotation phenotype of TR was otherwise unchanged as well, except that the buoyant TR had now shifted to fraction 5. In the case of Lck, there was a clear reduction in the relative amount of the protein in the middle of the gradient compared to the crude cell lysates, and the buoyant Lck complexes in this gradient region were now concentrated in fraction 6. Our total membrane fraction apparently still contained significant amounts of unspecified nuclear remnants, since ∼64% of intracellular influenza virus NP cofractionated with cellular membranes in the crude cell homogenates (data not shown). When this “membrane-associated” NP was extracted with Triton X-100 at cold temperatures, this led to scattering of the protein along the gradient, although with some concentration in fractions 2 to 5. Also the small amounts of the cytosolic nucleocapsid protein of SFV that copurified with membranes were predominantly found in fractions 2 to 5 (data not shown). Analyses of membrane-bound Pr55gag after a 15-min pulse with [35S]methionine and a 60-min chase indicated that labeled Pr55gag was predominantly found in buoyant complexes, but in contrast to the crude cell lysates, these buoyant complexes were now scattered in fractions 1 to 6. The Pr55gag panel in Fig. 1B suggests also that relatively more Pr55gag ended up in fraction 1 in this fractionation scheme than in crude cell lysates. However, the relative amounts of labeled Pr55gag in fraction 1 versus fraction 6 varied significantly between different experiments (data not shown), thus suggesting that fraction 1 material in fact could be a postlysis artifact of fraction 6 complexes, or vice versa. Furthermore, when comparing the gradient profiles of Pr55gag and NP in Fig. 1B, it is obvious that the buoyant complexes of Pr55gag are strikingly similar to those of NP. Since NP and Pr55gag are both nucleic acid binding proteins, these heterogeneous buoyant structures could represent yet another type of postlysis artifact induced by nuclear remnants. In an attempt to determine whether the Pr55gag buoyant complexes were artifacts or rafts, we tested the sensitivity of the complexes to Triton X-100 at 37°C. Rafts are solubilized with Triton X-100 if the extraction is performed at 37°C (6). As expected, extraction at 37°C shifted essentially all CD55 to the detergent-soluble pool (fractions 9 to 12) (data not shown). In the case of Pr55gag, floating of the protein to fractions 1 to 4 was suppressed, and increased amounts of labeled Pr55gag were found in the Triton X-100-soluble pool (Fig. 1B). However, the material in fractions 5 and 6 was largely resistant to extraction at 37°C. But since the buoyant NP complexes were efficiently solubilized at 37°C as well (Fig. 1B), the Triton X-100 solubility of the Pr55gag complexes in fractions 1 to 4 at 37°C cannot be taken as proof for these complexes representing rafts. In addition to solubilization of rafts, 37°C could also activate an enzymatic function or functions that destabilize the buoyant Pr55gag and NP complexes.
As a third approach to assess association of Pr55gag with Triton X-100-resistant rafts, we used membranes from PNS as the starting material for the detergent extraction. In experiments described above, we had used EDTA in the cell homogenization buffer, but the NP control indicated that nuclear remnants in EDTA-homogenized cell extracts were not quantitatively pelleted by the low-speed spin used to produce PNS (data not shown). We therefore replaced EDTA in the homogenization buffer with Mg2+, which is known to promote nuclear integrity during homogenization and thus to improve the removal of nuclear material by low-speed spin. PNS was first fractionated on a flotation gradient to separate membranes from cytosol, and the total membrane fraction was extracted with 1% Triton X-100 at cold temperatures, and the extracts were analyzed on the iodixanol step gradient. Under these conditions, no NP cofractionated with cellular membranes (data not shown). As shown in Fig. 1C, the relative amount of CD55 in fraction 1 was somewhat reduced compared to that of the membranes from crude cell homogenate, whereas the gradient profile of TR was essentially unchanged. Relatively more Lck was now found in fraction 1 than that shown in Fig. 1B, but most significantly, only trace amounts of Lck were present in fractions 5 and 6. In contrast, the majority of Pr55gag was concentrated in the middle of the gradient. After a 15-min pulse with [35S]methionine and a 0-min chase, about equal amounts of labeled Pr55gag were found in fraction 6 and the Triton X-100-soluble pool (fractions 9 to 12), whereas after a 60-min chase (data not shown) or 120-min chase, the protein was predominantly in fractions 5 and 6 (Fig. 1C). Similar results were obtained if the detergent concentration was reduced to 0.25% (data not shown). As shown in the lowest panel in Fig. 1C, the Pr55gag complexes at the 30 to 40% iodixanol interphase were resistant to Triton X-100 at 37°C.
Taken together, the results in Fig. 1A to C demonstrate that cold Triton X-100 extraction yields variable buoyant complexes for Pr55gag depending on which cellular fraction is used in the experiment. To ascertain that the variability is not an artifact of the SFV expression system, we also analyzed the Triton X-100 resistance of Pr55gag expressed in transfected 293T cells. Western blotting of the gradient fractions was used to visualize total Pr55gag. Also in these transfected cells, the different cellular fractions yielded variable buoyant complexes for Pr55gag: Triton X-100 extraction of membranes of crude homogenates yielded buoyant Pr55gag complexes that were scattered in fractions 1 to 6, whereas the membranes of PNS gave Pr55gag complexes that were concentrated into fractions 5 and 6 (Fig. 1D). Thus, the variability is a feature of Pr55gag, and is not due to the expression system used.
Triton X-100-treated buoyant Pr55gag complexes from VLPs are not physically associated with a raft-like membrane.
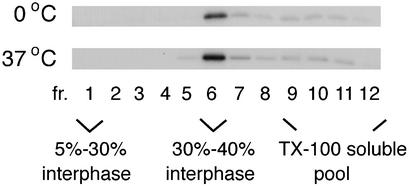
The technical problems associated with the flotation assay made it difficult to determine with certainty what was a genuine Triton X-100-resistant complex and what was an artifact in the case of the intracellular Pr55gag. However, one factor common to all of the fractionation schemes used in Fig. 1 was that some Pr55gag consistently floated to the 30 to 40% iodixanol interphase (fraction 6). In agreement with previously published results (32), we found that extraction of VLPs with Triton X-100 at either 0 or 37°C also yielded Pr55gag complexes that floated to fraction 6 (Fig. 2). Thus, the Pr55gag complexes at the 30 to 40% iodixanol interphase most likely are not artifacts but represent genuine Triton X-100-resistant complexes. Figure 2 shows analyses of VLPs produced from SFV-C/HIVgag-infected Jurkat cells, but VLPs from transfected 293T cells gave similar results (data not shown).
FIG. 2.
Triton X-100 extraction of VLPs at 0°C or 37°C yields buoyant Pr55gag complexes that float to the 30 to 40% iodixanol interphase. VLPs were collected from SFV-C/HIVgag-infected Jurkat cells which had been metabolically labeled with [35S]methionine for 180 min. VLPs were extracted with 1% Triton X-100 (TX-100) and fractionated on iodixanol step gradients.
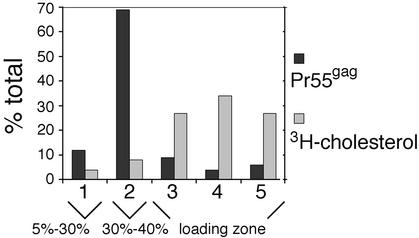
Since VLPs can be obtained in a relatively pure form, this prompted us to determine whether floating of VLP-derived Pr55gag to the 30 to 40% iodixanol interphase indeed is a result of these complexes being physically associated with Triton X-100-resistant membranes as has been previously suggested (32). To determine whether envelope cholesterol was retained in the Triton X-100-extracted Pr55gag complexes, we labeled SFV-C/HIVgag-infected Jurkat cells with [3H]cholesterol and purified VLPs from the culture media by a two-step gradient centrifugation as described in Materials and Methods. Since cultured cells are known to shed microvesicles, which have densities similar to that of retrovirus particles, we first controlled how much of the total 3H radioactivity in our VLP sample was of microvesicular origin. To this end, we collected medium from [3H]cholesterol-labeled Jurkat cells infected with a recombinant SFV encoding influenza virus NP and subjected that medium to the same purification procedure used for VLPs. The 3H counts in VLPs and the NP control samples produced from parallel experiments were determined by liquid scintillation counting, and comparison of the two values indicated that the radioactivity in the NP control amounted to only ∼10% of that of the VLPs (data not shown). Thus, the vast majority of the radioactive cholesterol in our VLP sample was likely to reside on the VLP envelope. To determine whether this envelope-associated radioactive cholesterol was retained on Pr55gag complexes after extraction with Triton X-100, the [3H]cholesterol-labeled VLPs and control NP samples were treated with Triton X-100 at 0°C, and the extracts were analyzed on an iodixanol step gradient otherwise similar to that in Fig. 1, except the gradient used was shorter. [35S]methionine-labeled VLPs were used as a control to track the position of Pr55gag complexes on the gradient. Five fractions were collected from the top of the gradient, and aliquots of fractions were analyzed by SDS-PAGE ([35S]methionine-labeled VLPs) or by scintillation counting ([3H]cholesterol-labeled VLPs and the NP control; the counts from the NP sample were subtracted from the VLP values). As already demonstrated in Fig. 2, the [35S]methionine-labeled Pr55gag complexes from the Triton X-100-extracted VLPs floated to the 30 to 40% iodixanol interphase (fraction 2, Fig. 3). In contrast, the majority of the radioactive cholesterol counts (88% of total) were found in the Triton X-100-soluble pool (fractions 3 to 5). These results indicated that Triton X-100 extraction efficiently stripped envelope cholesterol from the VLP-Pr55gag complexes.
FIG. 3.
Triton X-100 extraction efficiently strips envelope cholesterol from VLP-Pr55gag. VLPs were collected from SFV-C/HIVgag-infected Jurkat cells that had been metabolically labeled with [35S]methionine or [3H]cholesterol. VLPs were extracted with 1% Triton X-100 at 0°C, and the extracts were fractionated on iodixanol step gradients. Aliquots of gradient fractions were analyzed by SDS-PAGE ([35S]methionine-labeled VLPs) or by scintillation counting ([3H]cholesterol-labeled VLPs). Labeled Pr55gag and [3H]cholesterol in gradient fractions are expressed as percentages of the total (sum of all fractions).
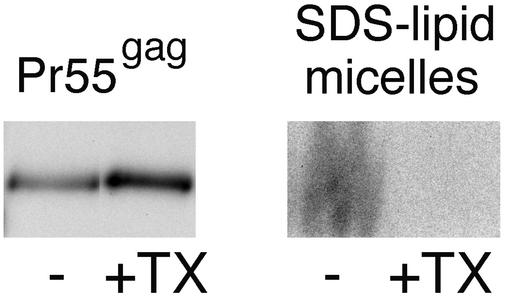
We used VLPs containing radioactively labeled phospholipids to determine whether the Triton X-100-treated Pr55gag complexes retain envelope phospholipids. The VLPs were collected from SFV-C/HIVgag-infected Jurkat cells that had been metabolically labeled with [32P]orthophosphate. The [32P]orthophosphate label is incorporated into both glycerophospholipids and sphingomyelin of VLPs. The VLPs were purified from the culture media by the same two-step centrifugation procedure used in the [3H]cholesterol experiments described above. Material collected from [32P]orthophosphate-labeled Jurkat cells infected with the recombinant SFV encoding influenza virus NP was used as a control for microvesicular contamination. The radioactively labeled phospholipids were quantitated by solubilization of samples with excess hot SDS and by analyzing the solubilized material by SDS-PAGE. On 20% polyacrylamide gels, the mixed SDS-lipid micelles are separated from other 32P-labeled components (proteins and RNA) (21), and this enables easy quantification of the glycerophospholipid and sphingomyelin contents of samples. We first estimated what percentage of the radioactivity in the VLP-derived SDS-lipid micelles truly originated from VLPs by comparing the radioactive signal of VLP sample to that of the control NP sample from a parallel experiment. Surprisingly, the SDS-lipid micelles from both the VLP and the control NP samples contained considerable amounts of radioactivity (data not shown), thus suggesting that Jurkat cells shed large amounts of microvesicles that were rich in phospholipids, but apparently poor in cholesterol, since the [3H]cholesterol-experiments described above had indicated only ∼10% microvesicular contamination for our VLP sample. The microvesicles were not an artifact of the SFV expression system used, since similar amounts of vesicles were also shed from noninfected Jurkat cells labeled with [32P]orthophosphate (data not shown). The exact degree of contamination of the VLP samples by microvesicles was difficult to establish, because the relative ratios of the VLP- and control NP-associated radioactivities differed between different experiments. From three different experiments though, we estimated that the microvesicle contamination accounted for 30 to 51% of the total phospholipid signal in VLP samples. To test what percentage of the VLP-envelope phospholipids were retained on Pr55gag complexes after extraction with Triton X-100, we collected VLPs from cells that had been metabolically labeled with both [32P]orthophosphate and [35S]methionine. The VLPs were extracted with Triton X-100 at 0°C, and extracts were fractionated on the iodixanol step gradient described in the legend to Fig. 1. Material from the 30 to 40% iodixanol interphase was recovered and concentrated by pelletation. The pellet was solubilized in hot SDS, and equal volumes of the solubilized sample were analyzed on 10 and 20% gels along with intact VLPs. A representative experiment is shown in Fig. 4. [35S]methionine label in Pr55gag bands (10% gels) was used to quantitate the amount of Pr55gag in the two samples. In the experiment shown in Fig. 4, the Triton X-100-treated sample contained ∼30% more Pr55gag than the intact VLP sample. However, as shown by the right-hand panel in Fig. 4, the two samples significantly differed in their phospholipid contents: the radioactive lipid signal was readily detected in the intact VLP sample, whereas the lipid signal was very weak in the Triton X-100-extracted sample. Assuming that 49 to 70% of the total phospholipid signal from the intact VLP sample was from VLP envelopes, quantitation of the Pr55gag and lipid signals of the two samples suggested that only 12 to 17% of the envelope phospholipids were retained on the Triton X-100-extracted buoyant Pr55gag complexes. Taken together, these results and the [3H]cholesterol results described above strongly suggest that cold Triton X-100 extraction solubilizes the bulk of VLP envelope lipids. Thus floating of Triton X-100-treated VLP-Pr55gag to the 30 to 40% iodixanol interphase is not due to these complexes being associated with a Triton X-100-resistant raft-like membrane.
FIG. 4.
Triton X-100 extracts the bulk of envelope phospholipids from the VLP-Pr55gag. VLPs were collected from SFV-C/HIVgag-infected Jurkat cells that had been metabolically labeled with [35S]methionine and [32P]orthophosphate. VLPs were extracted with 1% Triton X-100 at 0°C, and the lysate was fractionated on an iodixanol step gradient as shown in Fig. 1. Buoyant Pr55gag complexes from the 30 to 40% iodixanol interphase were concentrated by ultracentrifugation. The resulting pellet (+TX), as well as an intact, non-Triton X-100-extracted VLP sample (−), were solubilized with hot SDS and analyzed by SDS-PAGE. Equivalent amounts of samples were loaded on 10% (Pr55gag) and 20% (SDS-lipid micelles) polyacrylamide gels. On 20% polyacrylamide gels, mixed SDS-lipid micelles separate from other 32P-labeled material. Comparison of the 10 and 20% polyacrylamide gels demonstrates that although the intact VLP and the Triton X-100-treated samples contained similar amounts of Pr55gag complexes, the two samples significantly differed in their phospholipid contents. The radioactive signal in the mixed SDS-lipid micelles originates from both glycerophospholipids and sphingomyelin.
Analyses of raft association of Pr55gag by confocal fluorescence microscopy.
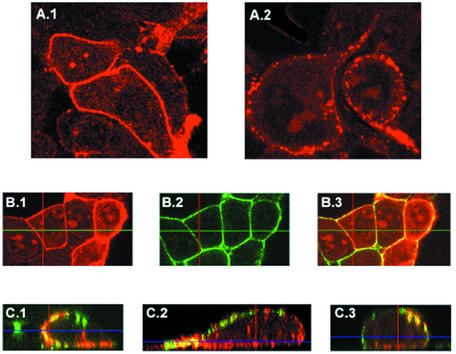
The results shown in Fig. 1 to 4 suggested that Pr55gag does not associate with Triton X-100-resistant rafts. However, the results do not exclude the possibility that Pr55gag is a raft-associated protein, since not all rafts are resistant to Triton X-100 (11, 41). We therefore probed raft association of Pr55gag by confocal fluorescence microscopy as well. Individual raft domains at the plasma membrane are small (∼50 nm in diameter) (40) and thus are below the detection level of standard light microscopes. However, cell surface rafts can be induced to form larger patches by antibody-mediated cross-linking of raft components (23). Since the raft-lipid marker GM1 has been reported to be incorporated into HIV-1 virions (35), we decided to cross-link rafts at the cell surface via GM1 and to test whether Pr55gag localizes to these cross-linked raft structures. Transfected 293T cells were used in the experiment. Pr55gag has been previously reported to localize at the cell surface in a punctate pattern (25). However, as shown in Fig. 5A, the staining pattern for plasma membrane-associated Pr55gag is dependent on the antibody used: the monoclonal EF-7 anti-Pr55gag antibody gave a smooth staining pattern (Fig. 5A.1), whereas a punctate pattern was obtained with another anti-Pr55gag monoclonal antibody (MAb 38:9; Fig. 5.A2). Since a possible raft localization of Pr55gag would be easier to score with a staining pattern that was initially smooth, we decided to use the EF-7 antibody in our raft cross-linking experiments. Cholera toxin B subunit specifically binds to GM1 (23), and the GM1-containing rafts were patched with this toxin and antibodies directed against it. The cells were subsequently fixed, permeabilized, and stained for Pr55gag. Figure 5B shows confocal analyses of nonpatched cells. Pr55gag (red, Fig. 5.B1) and GM1 (green, Fig. 5.B2) were both uniformly distributed on the plasma membrane of these nonpatched cells. Figure 5.C1 to C3 show three representative examples of cross-linked cells with the Pr55gag and GM1 signals superimposed. Both Pr55gag and GM1 were distributed in a patchy, dot-like pattern on these cells. Three different types of patches could be distinguished: patches that were positive only for GM1 (green), patches that were strongly positive for both GM1 and Pr55gag (yellow), and patches that appeared to be positive only for Pr55gag (red). However, from the nonoverlapped images, it was evident that the latter Pr55gag-containing patches were actually positive for GM1 as well, but the GM1 signal was weak in these patches (data not shown). Collectively, these results suggest that a significant fraction of Pr55gag at the cell surface might be localized to GM1-positive rafts, but according to the results from Fig. 1 to 4, these rafts apparently are Triton X-100 sensitive.
FIG. 5.
Confocal fluorescence microscopy of transfected 293T cells. (A) Different anti-Pr55gag antibodies give different staining patterns for Pr55gag at the cell surface. (A.1) MAb EF-7. (A.2) MAb 38:9. (B) Both Pr55gag and GM1 exhibit smooth staining patterns on transfected 293T cells if GM1-containing rafts have not coalesced. Live cells were first incubated with FITC-conjugated cholera toxin B subunit at a cold temperature to detect GM1, and the cells were subsequently fixed, permeabilized, and incubated with MAb EF-7 and TRITC-conjugated antimouse IgG antibodies to detect Pr55gag. (B.1) Pr55gag. (B.2) GM1. (B.3) Pr55gag and GM1 images superimposed. (C) Coalescence of GM1-positive rafts at the cell surface transforms the smooth staining pattern of Pr55gag into a patchy one. Live cells were first incubated with FITC-conjugated cholera toxin B subunit at a cold temperature, and the GM1-positive rafts were subsequently patched by brief incubation at 37°C in the presence of anti-cholera toxin antibodies. Cells were then fixed, permeabilized, and immunostained with MAb EF-7 and TRITC-conjugated antimouse IgG antibodies to detect Pr55gag. Three representative examples of patched cells (C.1 to C.3) are shown with Pr55gag (red) and GM1 (green) images superimposed. Yellow indicates extensive colocalization of Pr55gag and GM1. A single optical section is shown in panels A and B, whereas all optical sections (the Z-stack) were superimposed in panel C and tilted with the orthogonal function to show the view through the whole cell.
Intracellular membrane-bound Pr55gag, as well as VLP-associated Pr55gag, is largely resistant to extraction with Brij98.
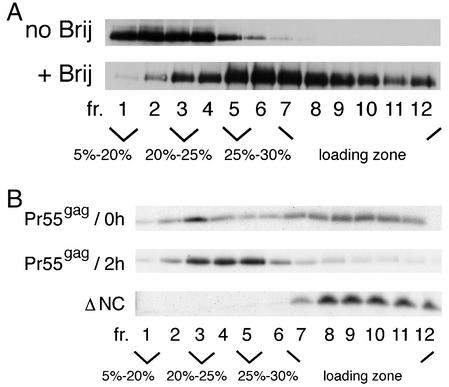
To obtain further evidence for localization of Pr55gag to distinct Triton X-100-sensitive rafts, we turned back to the biochemical assay and tested the sensitivities of membrane-bound Pr55gag to different detergents by using transfected 293T cells. Since we judged that chromatin and nuclear remnants were the main sources of artifacts in the biochemical assay, we chose the total membrane fraction of PNS as the starting material for detergent extractions. Our pilot assays indicated that the membrane-bound Pr55gag was partially resistant to Brij35, Brij56, Brij58, Brij98, Lubrol, and CHAPS (data not shown). We decided to focus on Brij98, since this is a detergent that detects rafts at physiological temperatures (11). Figure 6A shows analysis of Pr55gag-containing membranes either nonextracted or extracted with 0.5% Brij98 at 37°C for 5 min. The samples were analyzed on an iodixanol step gradient consisting of a 56% loading zone overlaid with 30, 25, 20, and 5% iodixanol solutions. Western blotting of the gradient fractions was used to visualize total Pr55gag. In the nonextracted sample, Pr55gag was predominantly found in fractions 1 to 4. After extraction with Brij98, the protein was scattered in fractions 2 to 12 (i.e., from the 20% iodixanol zone to the loading zone). Most importantly, significant amounts of the protein were found in the light-density fractions 2 to 5, thus suggesting that at steady state, a population of the membrane-bound Pr55gag was largely resistant to Brij98. To test the sensitivity of newly synthesized Pr55gag to Brij98, we turned to Jurkat cells and the SFV expression system. SFV-C/HIVgag-infected Jurkat cells were metabolically labeled with [35S]methionine for 15 min, and membrane-bound Pr55gag was analyzed after a 0- or 120-min chase. As shown in Fig. 6B, after a 0-min chase, labeled Pr55gag was broadly distributed along the gradient, with a minor concentration of the protein in fraction 3. After a 120-min chase, the amount of the protein in the loading zone was decreased, and a clear accumulation of the protein (∼61% of total) was observed in fractions 3 to 5. This suggested that Pr55gag accumulated in Brij98-resistant membrane domains of light density in a time-dependent fashion. In contrast to the wild-type Pr55gag, a carboxy-terminally-deleted mutant form of the protein consisting of MA-CA-p2-domains (ΔNC) remained sensitive to Brij98 even after 2 h of chase.
FIG. 6.
Intracellular membrane-bound Pr55gag is largely resistant to Brij98. (A) Analyses of transfected 293T cells. The total membrane fraction from PNS was extracted with Brij98 for 5 min at 37°C or left untreated, and the samples were subjected to flotation on iodixanol step gradients. Total Pr55gag in gradient fractions was visualized by Western blotting. (B) Brij98 resistance of newly synthesized Pr55gag. The wild type and ΔNC (MA-CA-p2) mutant of Pr55gag were expressed in Jurkat cells from recombinant SFV genomes. Cells were metabolically labeled with [35S]methionine for 15 min and chased for 0 min and 2 h (Pr55gag) or for 2 h (ΔNC). The total membrane fraction from PNS was extracted with Brij98 as described above, and the extracts were fractionated on iodixanol step gradients. Aliquots of gradient fractions were analyzed by SDS-PAGE.
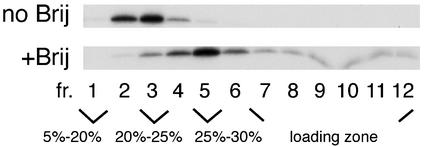
In order to determine whether the Brij98-resistant intracellular Pr55gag complexes were precursors for VLPs, we tested the sensitivity of VLPs to Brij98 extraction. Extracellular VLPs were extracted with 0.5% Brij98 at 37°C for 5 min, and the extracts were fractionated on an iodixanol step gradient similar to that shown in Fig. 6. As shown in Fig. 7, nonextracted VLPs produced from SFV-C/HIVgag-infected Jurkat cells floated through 25% iodixanol to fractions 2 and 3. After extraction with 0.5% Brij98 (5 min at 37°C), a slight shift in the density was evident, and Pr55gag peaked in fraction 5, thus overlapping the intracellular Brij98-resistant complexes. Similar results were obtained with VLPs produced from transfected 293T cells (data not shown). These results suggested that VLP envelope was largely resistant to Brij98, and the intracellular Brij98-resistant Pr55gag complexes banding at 25% iodixanol most likely represented precursors for VLPs.
FIG. 7.
Brij98-treated VLPs retain low buoyant density. VLPs were collected from SFV-C/HIVgag-infected Jurkat cells that had been metabolically labeled with [35S]methionine for 180 min. The VLPs were extracted with Brij98 and analyzed as described in the legend to Fig. 6. No Brij indicates intact, nonextracted VLPs.
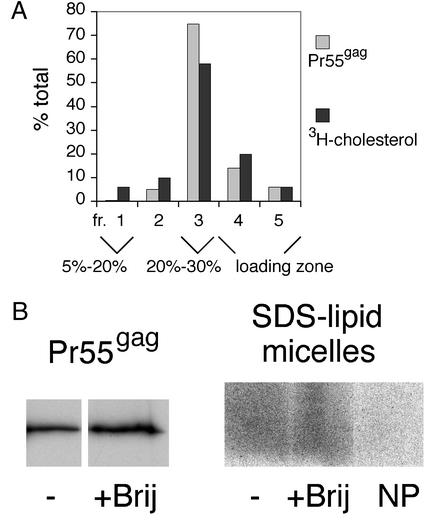
To confirm that the VLP envelope was indeed largely resistant to Brij98, we monitored the extractability of envelope lipids by the detergent. To determine whether envelope cholesterol was retained on Brij98-extracted Pr55gag complexes, we performed an assay similar to that described above for Triton X-100 extractions. [3H]cholesterol- or [35S]methionine-labeled VLPs were extracted with 0.5% Brij98 for 5 min at 37°C, and the extracts were analyzed on a iodixanol step gradient consisting of a 56% loading zone overlaid with 30, 20, and 5% iodixanol solutions. As shown in Fig. 8A, the majority of both the [35S]methionine-labeled Pr55gag complexes and the envelope [3H]cholesterol label (80 and 58% of the total, respectively) was found in fraction 3 at the 20 to 30% iodixanol interphase. This indicated that a large fraction of the envelope cholesterol was resistant to Brij98.
FIG. 8.
Large fraction of envelope phospholipids and cholesterol remains attached to Brij98-treated VLP-Pr55gag. (A) Brij98 extraction of [35S]methionine- or [3H]cholesterol-labeled VLPs produced from SFV-C/HIVgag-infected Jurkat cells. The VLPs were extracted with Brij98 for 5 min at 37°C, and the samples were processed as described in the legend to Fig. 3, with the exception that the iodixanol step gradient used was slightly different. (B) Brij98 extraction of [35S]methionine- and [32P]orthophosphate-labeled VLPs produced from SFV-C/HIVgag-infected Jurkat cells. The VLPs were extracted with Brij98 for 5 min at 37°C, and the extracts were fractionated on iodixanol step gradients as described in the legend to Fig. 6. The buoyant Pr55gag complexes from fractions 3 to 5 were pooled and concentrated by pelletation. The pellets, as well as intact, nonextracted VLP samples (−) were analyzed as described in the legend to Fig. 4. NP is a Brij98-treated control sample that was collected from [32P]orthophosphate-labeled Jurkat cells infected with recombinant SFV encoding influenza virus NP. The NP control demonstrates that the majority of phospholipid signal in the Brij98-treated VLP sample originates from VLPs, not from contaminating microvesicles.
To test the extractability of the envelope phospholipids by Brij98, we carried out an assay similar to that described above for Triton X-100 extractions. [35S]methionine- and [32P]orthophosphate-labeled VLPs and the [32P]orthophosphate-labeled NP control material were extracted with 0.5% Brij98 for 5 min at 37°C, and the extracts were fractionated on an iodixanol step gradient as described in the legend to Fig. 6. The buoyant Pr55gag complexes or the control NP material from fractions 3 to 5 was pooled and concentrated by pelletation, and the pellets were solubilized with excess hot SDS and analyzed on 10 and 20% polyacrylamide gels along with intact VLPs. As shown in Fig. 8B, the Brij98-treated Pr55gag complexes retained significant amounts of the envelope phospholipids. Comparison with the Brij-treated NP control indicated that majority of the Brij-resistant Pr55gag signal originated from VLPs. Assuming that microvesicles accounted for ∼30 to 51% of the total phospholipid signal in intact VLPs, quantitations of the Pr55gag and the SDS-lipid micelle signals from two separate experiments suggested that ∼37 to 53% of envelope phospholipids were retained on Brij98-treated Pr55gag complexes. Taken together, these results indicated that the VLP envelope was largely resistant to extraction with Brij98.
DISCUSSION
HIV-1 is one of the enveloped viruses whose assembly and budding has been proposed to be critically dependent on lipid rafts (32, 35, 36, 42). The strongest evidence for this raft connection has been the localization of Pr55gag, the protein that drives the assembly of virus, to Triton X-100-resistant rafts (32, 35, 36, 57). Localization of Pr55gag to the Triton X-100-resistant rafts was based on results from the commonly used biochemical raft fractionation assay, which involves extraction of cells with Triton X-100 at cold temperatures. Triton X-100-resistant rafts retain buoyant density after cold detergent treatment and float to a low density when the detergent extracts are fractionated on density gradients. However, our results indicate that the buoyancy of Triton X-100-treated Pr55gag complexes does not provide solid evidence for raft localization of Pr55gag.
Nguyen and Hildreth (35) have reported that the majority of [3H]myristic acid-labeled intracellular Pr55gag is associated with buoyant Triton X-100-resistant structures in cold cell lysates, and in a report by Zheng et al. (57), Nef was found to increase cofractionation of Pr55gag with Triton X-100-resistant buoyant structures. However, in these two reports, the buoyant structures were not further characterized. Lindwasser and Resh (32) reported that cold Triton X-100-extracted crude cell lysates, as well as Triton X-100-treated VLPs, yield buoyant Pr55gag complexes that band at the 30 to 40% iodixanol interphase. These buoyant Pr55gag complexes were of higher density than “classical” Triton X-100-resistant rafts, which float through 30% iodixanol. Ono and Freed (36) analyzed raft association of Pr55gag by using postnuclear supernatants of HIV-1-infected cell homogenates. They estimated that up to approximately half of membrane-bound Pr55gag associated with Triton X-100-resistant buoyant structures in cold lysates, and these structures were soluble in the detergent if the extraction was carried out at 37°C. In contrast to the study by Lindwasser and Resh (32), the buoyant Pr55gag complexes in the study by Ono and Freed (36) appeared to have a density similar to that of classical raft markers, but this discrepancy was most likely due to the different gradients used. Ono and Freed separated Triton X-100-sensitive and -resistant pools of proteins from each other by using sucrose gradients with a single, rather dense (65% [wt/vol] sucrose) separating layer. Nguyen and Hildreth (35), Lindwasser and Resh (32), Zheng et al. (57), and Ono and Freed (36) all interpreted that their buoyant Pr55gag complexes reflected association of the protein with rafts. However, our data presented in Fig. 1 indicate that, in the case of Pr55gag, the results from the biochemical raft fractionation assay have to be interpreted with caution, since the assay is very prone to artifacts. Figure 1 demonstrates that variable buoyant complexes are obtained for Pr55gag, depending on which cellular fraction is used as the starting material for the Triton X-100 treatment. Whole-cell lysates, as well as membranes from PNS, predominantly yielded buoyant Pr55gag complexes, which banded at the 30 to 40% iodixanol interphase, whereas membranes from crude cell homogenates gave buoyant complexes of more heterogeneous density. Furthermore, the influenza virus NP control (Fig. 1B) demonstrated that the sensitivity of a buoyant protein structure to Triton X-100 at 37°C does not necessarily mean that the structure represents rafts. Although it is difficult to determine with certainty which of the buoyant Pr55gag complexes reflect true in vivo Pr55gag structures and which are artifacts, our control experiments suggested that chromatin and other nuclear remnants could comprise a major source for artifacts. We therefore favor the interpretation that membranes from PNS are the best starting material for the detergent treatment, and, consequently, the Pr55gag complexes at the 30 to 40% iodixanol interphase represent the true intracellular Triton X-100-resistant structures of Pr55gag. This interpretation is further supported by the fact that similar complexes are obtained from Triton X-100-treated VLPs as well. Due to their higher relative purity, the VLP preparations are less likely to yield artifacts than the molecularly complex cell extracts.
Lindwasser and Resh coined the name “barges” for the Triton X-100-treated buoyant Pr55gag structures at the 30 to 40% iodixanol interphase (32). Since their results indicated that localization of Pr55gag to barges correlated with oligomerization of the protein, their interpretation was that barges represent Triton X-100-resistant rafts, which have an increased density due to the presence of high-valence Pr55gag multimers (32). However, they did not validate their interpretation by performing lipid analyses of the barge-associated Pr55gag. Our lipid analyses of the [32P]orthophosphate- and [3H]cholesterol-labeled VLPs do not support the notion that floating of the Triton X-100-treated Pr55gag to the 30 to 40% iodixanol-interphase reflects association of the protein with Triton X-100-resistant rafts. [32P]orthophosphate labeling of cells produces VLPs with radioactively labeled glycerophospholipids and sphingomyelin, and comparison of SDS-lipid micelles from intact VLPs and the Triton X-100-treated Pr55gag complexes indicated that the buoyant Pr55gag complexes were largely devoid of envelope glycerophospholipids and sphingomyelin. Furthermore, fractionation of [3H]cholesterol-labeled Triton X-100-extracted VLPs on an iodixanol step gradient indicated that the detergent extraction efficiently stripped envelope cholesterol from the VLP-Pr55gag. Since glycerophospholipids, sphingomyelin, and cholesterol together constitute the vast majority of HIV-1 envelope lipids (2, 3), these results strongly suggest that envelope lipids do not remain attached to the buoyant Triton X-100-treated Pr55gag complexes in large amounts. This conclusion is further supported by the fact that the Triton X-100 extraction causes a considerable shift in the density of membrane-bound Pr55gag (compare, e.g., Fig. 1D, PNS panel, with Fig. 6A, no-detergent-panel). The most plausible explanation for this shift is that the detergent extracts the bulk of membrane lipids from the Pr55gag complexes. Recently, Ding et al. (10) published a study demonstrating that only Pr55gag, but not classical raft markers, undergoes this clear shift in density upon cold Triton X-100 extraction. Thus, it is questionable whether the buoyant Triton X-100-resistant Pr55gag complexes can be interpreted to signify localization of the protein to rafts. Our finding that the buoyant Pr55gag complexes at the 30 to 40% iodixanol interphase, unlike classical raft markers, are insensitive to Triton X-100 at 37°C or that the buoyant Pr55gag complexes are insensitive to cholesterol depletion (10) provides a further argument against the buoyant Pr55gag structures representing Triton X-100-resistant raft membranes. At present, however, we have no explanation to offer for the floating phenotype of the Triton X-100-treated Pr55gag. Since we estimated that 12 to 17% of total membrane phospholipids could remain attached to the Triton X-100-treated Pr55gag, it is possible that these residual phospholipids, perhaps together with membrane lipids that were not scored in our assay (e.g., gangliosides), cause the buoyancy of the Pr55gag complexes. Our attempts to reliably measure the retention of GM1 on the Triton X-100-treated Pr55gag complexes were unsuccessful due to the low specific GM1 signal from VLPs (data not shown). Interestingly, only the unprocessed HIV-1 Pr55gag yielded buoyant Triton X-100-resistant complexes, whereas MA and CA of processed HIV-1 cores, as well as the unprocessed Gag precursor of Moloney murine leukemia virus VLPs, fractionated with the detergent-soluble pool (data not shown).
Triton X-100 has been the most commonly used detergent in raft-studies, but not all rafts are resistant to Triton X-100 (11, 41). We therefore probed the possible raft association of Pr55gag by confocal fluorescence microscopy and with detergents other than Triton X-100. The smooth cell surface staining pattern of Pr55gag in transfected 293T cells could be transformed into a patchy one by antibody-mediated coalescence of GM1-positive rafts. Furthermore, extraction of the total membrane fraction of PNS from transfected 293T cells suggested that at steady state, a population of membrane-bound Pr55gag was largely resistant to Brij98. In contrast to the Triton X-100-treated buoyant Pr55gag complexes, the Brij98-resistant complexes were of lighter density and banded at 25% iodixanol. Pulse-chase studies in Jurkat cells indicated that newly synthesized Pr55gag accumulated in these low-density Brij98-resistant structures in a time-dependent manner. In contrast, a mutant form of Pr55gag consisting of MA-CA-p2 remained sensitive to Brij98 even after extended chase periods. Since the MA-CA-p2-mutant is impaired in oligomerization (8; K. Holm and M. Suomalainen, unpublished data), the time-dependent accumulation of wild-type Pr55gag in the low-density Brij98-resistant structures could reflect oligomerization of the protein. The intracellular Brij98-resistant complexes most likely represent assembly intermediates, since Brij98 extraction of VLPs produced Pr55gag complexes of similar density. Analyses of [32P]orthophosphate- and [3H]cholesterol-labeled VLPs confirmed that the buoyant density of the floating Brij98 complexes was due to these complexes being associated with a membrane that was largely resistant to the detergent. We estimated that ∼37 to 53% of envelope phospholipids and ∼53% of cholesterol remained attached to the buoyant Brij98-treated VLP-Pr55gag. One possible explanation for this partial resistance is that the VLP membrane is a mosaic, consisting of both Brij98-resistant and Brij98-sensitive membrane domains. Taken together, these confocal and Brij98 results could indicate that Pr55gag associates with raft microdomains at the cell surface that are resistant to Brij98, and these microdomains form a part of the platform for Pr55gag-mediated assembly and budding of HIV-1. However, at present, we cannot exclude the possibility that oligomerization of Pr55gag induces a structural alteration in the membrane that results in increased Brij98 resistance (i.e., that the Brij98 resistance of membrane-bound Pr55gag has nothing to do with cellular rafts). Since Brij98-resistant rafts are newcomers in the raft field and are still poorly characterized, it is unknown at present whether any cellular rafts have a similar density to the Brij98-resistant Pr55gag structures described here. Of our three cellular marker proteins, only TR partially overlapped with Pr55gag complexes after Brij98 treatment, whereas CD55 floated through 20% iodixanol and Lck stayed in the loading zone (data not shown).
The growing list of viral raft-associated structural proteins has created a lot of excitement in the field of virus assembly. Even a partial localization of a viral structural protein to rafts has commonly been taken as evidence for rafts playing a critical role in the assembly of the virus in question. However, concrete evidence for rafts being mechanistically important for assembly and budding of enveloped viruses is actually scarce. In the case of HIV-1, it amounts to one observation—namely, that cholesterol-depleting agents inhibit particle release from HIV-1-infected cells (36). Cholesterol is essential for structural integrity of rafts (5, 45), and disruption of rafts by cholesterol depletion is expected to reduce virus production if assembly and budding of HIV-1 are dependent on rafts. However, since cholesterol depletion affects many different cellular processes (28, 29, 44, 45), it could bring about reduction in HIV-1 particle production entirely through indirect effects as well. The observations that viral spike proteins appear to localize (partially) to rafts and that cellular raft lipids and proteins are found in HIV-1 particles have been put forward as another piece of evidence for the importance of rafts in the assembly and budding of HIV-1 (35, 38, 42, 57). However, whether these observations indicate an obligatory linkage between rafts and the assembly and budding mechanisms of HIV-1 is unknown. With respect to the presence of host raft lipids and proteins in the HIV-1 envelope, it is worthwhile to remember that there are indications that rafts actually might be relatively abundant structures at the cell surface (12, 15, 22, 33, 39) and that retroviruses seem to be quite indiscriminate in choosing their envelope components (21, 37). Furthermore, in the case of viral spike proteins, the rafts in question appear to belong to the class of Triton X-100-resistant rafts. It is unclear how these rafts correlate with viral assembly intermediates, since according to our data, Pr55gag does not appear to have intrinsic affinity for Triton X-100-resistant rafts, and there are no data on Triton X-100 sensitivity or resistance of spike proteins in virus particles. In principle, it is possible that the Triton X-100-resistant rafts containing viral spike proteins and the Brij98-resistant Pr55gag membrane domains described here are similar types of rafts and that Pr55gag and viral envelope proteins only exhibit different sensitivities to Triton X-100 within these raft structures. However, we did not find any indications for Triton X-100-resistant raft-like lipid domains in the envelope of Pr55gag VLPs with cold Triton X-100 extractions: both the radioactive cholesterol (Fig. 3) and the bulk of radioactively labeled phospholipids (data not shown) in the VLP envelope were readily solubilized with cold Triton X-100. In summary, before it can be firmly concluded that rafts play a critical role in the assembly of HIV-1, direct evidence is needed that rafts or raft-associated host factors are important for some discrete step or steps in the viral assembly process.
Acknowledgments
We thank Henrik Garoff for many helpful discussions, Jose Casasnovas and Birgitta Lindqvist for providing SFV1-CD55, Didier Trono for providing pCMVΔR8.91, and Jorma Hinkula for providing antibodies.
This work was supported by a grant from the Swedish Research Council to M.S (K2001-06X-13052-03A) and by a grant from the European Union (FMRX-CT98-0225).
REFERENCES
- 1.Aliperti, G., and M. J. Schlesinger. 1978. Evidence for an autoprotease activity of Sindbis virus capsid protein. Virology 90:366-369. [DOI] [PubMed] [Google Scholar]
- 2.Aloia, R. C., F. C. Jensen, C. C. Curtain, P. W. Mobley, and L. M. Gordon. 1988. Lipid composition and fluidity of the human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 85:900-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aloia, R. C., H. Tian, and F. C. Jensen. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA 90:5181-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benting, J. H., A. G. Rietveld, and K. Simons. 1999. N-glycans mediate the apical sorting of a GPI-anchored, raft-associated protein in Madin-Darby kidney cells. J. Cell Biol. 146:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. A. 1998. Structure and origin of ordered lipid domains in biological membranes. J. Membr. Biol. 164:103-114. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 7.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, W. P., and J. C. Nickoloff. 1992. Site directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 200:81-88. [DOI] [PubMed] [Google Scholar]
- 10.Ding, L., A. Derdowski, J.-J. Wang, and P. Spearman. 2003. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drevot, P., C. Langlet, X.-J. Guo, A.-M. Bernard, O. Colard, J.-P. Chauvin, R. Lasserre, and H.-T. He. 2002. TCR signal initiation machinery is pre-assembled and activated in a subset of membrane rafts. EMBO J. 21:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edidin, M. 2001. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell Biol. 11:492-496. [DOI] [PubMed] [Google Scholar]
- 13.Ehrlich, L. S., S. Fong, S. Scarlata, G. Zybarth, and C. Carter. 1996. Partioning of HIV-1 Gag and Gag-related proteins to membranes. Biochemistry 35:3933-3943. [DOI] [PubMed] [Google Scholar]
- 14.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 15.Fridriksson, E. K., P. A. Shipkova, E. D. Sheets, D. Holowka, B. Baird, and F. W. McLafferty. 1999. Quantitative analysis of phospholipids in functionally important membrane domains from RBL-2H3 mast cells using tandem high-resolution mass spectrometry. Biochemistry 38:8056-8063. [DOI] [PubMed] [Google Scholar]
- 16.Friedrichson, T., and T. V. Kurzchalia. 1998. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature 394:802-805. [DOI] [PubMed] [Google Scholar]
- 17.Garoff, H., R. Hewson, and D.-J. E. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gheysen, D., E. Jacobs, F. D. Foresta, C. Thiriart, M. Francotte, D. Thines, and M. D. Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 19.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, B. H., G. M. Shaw, S. K. Arya, M. Popovic, R. C. Gallo, and F. Wong-Staal. 1984. Molecular cloning and characterization of the HTLV-III virus associated with AIDS. Nature 312:166-169. [DOI] [PubMed] [Google Scholar]
- 21.Hammarstedt, M., K. Wallengren, K. Winther Pedersen, N. Roos, and H. Garoff. 2000. Minimal exclusion of plasma membrane proteins during retroviral envelope formation. Proc. Natl. Acad. Sci. USA 97:7527-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao, M., S. Mukherjee, and F. R. Maxfield. 2001. Cholesterol depletion induces large scale domain segregation in living cell membranes. Proc. Natl. Acad. Sci. USA 98:13072-13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harder, T., P. Scheiffele, P. Verkade, and K. Simons. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawash, I. Y., K. P. Kesavan, A. I. Magee, R. L. Geahlen, and M. L. Harrison. 2002. The lck SH3 domain negatively regulates localization to lipid rafts through interaction with c-Cbl. J. Biol. Chem. 277:5683-5691. [DOI] [PubMed] [Google Scholar]
- 25.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton, R., H. Hunt, S. Ho, J. Pullen, and L. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 28.Ikonen, E. 2001. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13:470-477. [DOI] [PubMed] [Google Scholar]
- 29.Incardona, J. P., and S. Eaton. 2000. Cholesterol in signal transduction. Curr. Opin. Cell Biol. 12:193-203. [DOI] [PubMed] [Google Scholar]
- 30.Karacostas, V., K. Nagashima, M. A. Gonda, and B. Moss. 1989. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc. Natl. Acad. Sci. USA 86:8964-8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liljeström, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 32.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayor, S., and F. R. Maxfield. 1995. Insolubility and redistribution of GPI-anchored proteins at the cell surface after detergent treatment. Mol. Biol. Cell 6:929-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melancon, P., and H. Garoff. 1987. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J. Virol. 61:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen, D. H., and J. E. K. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott, D. E. 1997. Cellular proteins in HIV virions. Med. Virol. 7:167-180. [DOI] [PubMed] [Google Scholar]
- 38.Pickl, W. F., F. X. Pimentel-Muiños, and B. Seed. 2001. Lipid rafts and pseudotyping. J. Virol. 75:7175-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierini, L. M., and F. R. Maxfield. 2001. Flotillas of lipid rafts fore and aft. Proc. Natl. Acad. Sci. USA 98:9471-9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pralle, A., P. Keller, E.-L. Florin, K. Simons, and J. K. H. Hörber. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148:997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Röper, K., D. Corbeil, and W. B. Huttner. 2000. Retention of prominin in microvilli reveals distinct cholesterol-based lipid microdomains in the apical plasma membrane. Nat. Cell Biol. 2:582-592. [DOI] [PubMed] [Google Scholar]
- 42.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 97:13523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 44.Simons, K., and E. Ikonen. 2000. How cells handle cholesterol. Science 290:1721-1726. [DOI] [PubMed] [Google Scholar]
- 45.Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell. Biol. 1:31-39. [DOI] [PubMed] [Google Scholar]
- 46.Solomon, K. R., M. A. Mallory, and R. W. Finberg. 1998. Determination of the non-ionic detergent insolubility and phosphoprotein associations of glycosylphosphatidylinositol-anchored proteins expressed on T cells. Biochem. J. 334:325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spearman, P., R. Horton, L. Ratner, and I. Kuli-Zade. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spearman, P., J.-J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suomalainen, M., and H. Garoff. 1994. Incorporation of homologous and heterologous proteins into the envelope of Moloney murine leukemia virus. J. Virol. 68:4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suomalainen, M., K. Hultenby, and H. Garoff. 1996. Targeting of Moloney murine leukemia virus Gag precursor to the site of virus budding. J. Cell Biol. 135:1841-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suomalainen, M., P. Liljeström, and H. Garoff. 1992. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 66:4737-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vincent, S., D. Gerlier, and S. N. Manié. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yon, J., and M. Friend. 1989. Precise gene fusion by PCR. Nucleic Acids Res. 17:4895.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, H., M. Ekström, and H. Garoff. 1998. The M1 and NP proteins of influenza A virus form homo- but not hetero-oligomeric complexes when coexpressed in BHK-21 cells. J. Gen. Virol. 79:2435-2446. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, Y.-H., A. Plemenitas, T. Linnemann, O. T. Fackler, and B. M. Peterlin. 2001. Nef increases infectivity of HIV via lipid rafts. Curr. Biol. 11:875-879. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]