Abstract
In general, enveloped viruses use two different entry strategies and are classified accordingly into pH-dependent and pH-independent viruses. Different members of the retrovirus family use one or the other strategy. Little is known about the uptake of foamy viruses (FV), a special group of retroviruses, into the target cells. In this study, we examined the pH dependence of FV entry by analyzing FV envelope glycoprotein (Env)-mediated infection of target cells with murine leukemia virus or FV vector pseudotypes in the presence of various lysosomotropic agents. Similar to vesicular stomatitis virus glycoprotein G (VSV-G)-mediated uptake, FV Env-mediated entry was inhibited by various lysosomotropic agents, suggesting a pH-dependent endocytic pathway. However, in contrast to its effect on VSV-G pseudotypes, chloroquine failed to reduce the infectivity of FV Env pseudotypes, implying that the pathway is different from that of VSV-G. Glycoproteins of various other FV species showed inhibition profiles similar to that of the prototype FV (PFV) Env. Analysis of the pH dependence of the FV Env-mediated fusion process in a cell-to-cell fusion assay revealed an induction of syncytium formation by a short exposure to acidic pH, peaking around pH 5.5. Interestingly, of all FV Env species analyzed, only the PFV Env had a significant fusion activity at neutral pH. Taken together, these data suggest a pH-dependent endocytic pathway for infection of target cells by FV.
An essential step in the life cycle of enveloped viruses is the fusion of viral and cellular lipid membranes, mediated by the viral glycoproteins, to release the viral capsid into the cytoplasm of the host cell. Although the details of the mechanisms of viral entry pathways are poorly understood, there are in general two different entry strategies used by various viruses. Influenza A is the prototype of a virus that requires a low-pH environment in the endosomal compartment to trigger membrane fusion (30). Viruses that use a similar entry strategy are vesicular stomatitis virus (VSV) and Semliki Forest virus (6, 34). In contrast, other viruses, such as Sendai virus, use a pH-independent mechanism (5). For these viruses, fusion occurs at the plasma membrane but may occur in the endosomes as well.
The different members of the family of retroviruses use one or the other type of entry pathway. To date, human immunodeficiency virus and amphotropic murine leukemia virus (MuLV-A) are mammalian retroviruses classified as pH independent (15, 16, 32). Infection is thought to occur by direct fusion at the plasma membrane. However, a recent study suggests that MuLV-A might employ a pH-independent receptor-mediated endocytic pathway for infection (9). Other mammalian retroviruses, such as ecotropic MuLV and mouse mammary tumor virus, are thought to use a pH-dependent mode of entry (16, 24). For mouse mammary tumor virus, a moderate induction of cell fusion by low-pH treatment is observed (24), whereas for ecotropic MuLV, a cell-line-dependent inhibition of infectivity by lysosomotropic agents has been reported (16). Avian leukosis virus (ALV) has previously been classified as pH independent (3). Only recently, it was shown to require a low-pH step for infection that acts downstream of receptor binding (19).
Foamy viruses (FV) are a group of retroviruses showing many unique features in their replication cycles that set them apart from all other retroviruses (14). Because of this distinction, FV have recently been clustered together into a separate subgroup of retroviruses, the spumaretrovirinae, with only one genus, the foamy viruses. All other retroviral genera constitute the subgroup of orthoretrovirinae (33). Research on FV over the last years has shown that they are at the crossroad between retroviruses and pararetroviruses, and some of their special features have been characterized (for a review, see reference 14). However, there are several steps of the FV replication cycle that are not or are only poorly understood. In particular, little is known about the early steps, the binding to a yet unknown receptor, the penetration into the host cell, the intracellular transport to the nucleus, and the disassembly of the viral particle. There is only one report demonstrating that nuclear targeting of incoming FV Gag proteins involves a centriolar step (28).
In this study, we attempted to clarify which of the two principal entry pathways is used by FV.
MATERIALS AND METHODS
Cells.
The human kidney cell line 293T (2) and the human fibrosarcoma cell lines HT1080 (23) and HT1080 NLS-LacZ (21) were cultivated in Eagle's minimal essential medium or Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum and antibiotics. The quail fibroblast cell line QT-6 (18) was cultivated in DMEM supplemented with 10% fetal calf serum, 2% chicken serum, and antibiotics.
Expression constructs.
The MuLV Gag/Pol expression construct pHIT60 (31), the replication-deficient MuLV vector pczCFG2fEGN (12), and the replication-deficient prototype FV (PFV; formerly known as human FV) vector pMH118 (C. Leurs, M. Jansen, K. Pollok, M. Heinkelein, M. Schmid, D. Lindemann, C. von Kalle, A. Rethwilm, D. William, and H. Hanenberg, submitted for publication) were described earlier.
The glycoprotein expression constructs used in this study are shown schematically in Fig. 1. The MuLV-A envelope expression construct pHIT456 (1), the VSV glycoprotein G (VSV-G) Env expression plasmid pcVG-wt (20), the PFV Env expression construct pczHFVenvEM02 (13), and the chimeric PFV Env expression construct pczHFVenvΔ 2MuLV (11) containing a MuLV cytoplasmic domain (CyD) were described earlier.
FIG. 1.
Schematic illustration of the glycoprotein expression constructs. The coding regions of the different envelope expression constructs used in this study are shown schematically. The viral origins of the glycoproteins (virus) and the construct names (construct) are indicated to the left of the schematic outline. The individual constructs are described in detail in Materials and Methods. Ampho, amphotropic; LP, leader or signal peptide; SU, surface domain; TM, transmembrane domain; MSD, membrane-spanning domain.
The envelope expression constructs of PFV, simian FV type 1 (SFV-1), feline FV (FFV), bovine FV (BFV), and equine FV (EFV) are based on the pCDNA3.1zeo vector (Invitrogen) unless otherwise stated. The SFV-1 Env expression construct pczSFV-1env was created by inserting a 2,999-bp PCR amplicon containing the complete SFV-1 env open reading frame (ORF), with the infectious SFV-1 provirus clone pSFV-1 (17) as a template, into the XbaI site of the pcDNA3.1zeo vector. Subsequently, a 2,539-bp PCR-derived XcmI/Bpu1102I fragment of the SFV-1 Env expression construct was replaced by the corresponding fragment of the pSFV-1 plasmid. The pciSFV-1env expression construct was generated by inserting an EcoRI/PmeI fragment of pczSFV-1env into pCI (Promega). The pciSFV1-envSM01 construct generated by recombinant PCR expresses a chimeric FV envelope protein and contains amino acids (aa) 1 to 59 of FFV Env and aa 62 to 989 of SFV-1 Env. The FFV Env expression construct pczFFVenv was created by recombinant PCR by using the FFV subgenomic proviral clones FFVclone#94 and FFVenv1 (7) as templates, generating an amplicon of 2,973 bp containing the complete FFV env ORF, and cloning the amplicon into the XbaI site of pCDNA3.1zeo. Subsequently, a 1,740-bp PCR-derived KspAI/Eco91I fragment of the FFV Env expression construct was replaced by the corresponding fragment of the FFVenv-1 plasmid. The BFV Env expression construct pczBFVenv was created by cloning a 2,994-bp PCR amplicon containing the complete BFV env ORF, with the infectious BFV provirus lambda phage clone BSV-11 (26) as a template, into the Bsp120I and EcoRV sites of pCDNA3.1zeo. The EFV Env expression construct pciEFVenv was generated by subcloning a 3,024-bp EcoRI/SmaI fragment of pSGEFVenv (10) containing the complete EFV env ORF into pCI. The MuLV-A Env expression construct pcziMEA was generated by inserting a 1,225-bp HindIII/XbaI fragment of pHIT456 (1) containing the MuLV env ORF into pczi, a pcDNA3.1+zeo-derived vector containing the cytomegalovirus immediate-early intron A of pHIT60 (31). The R peptide-truncated variant, pcziMEA-R−, was generated by recombinant PCR by introducing a translation stop after aa 638. All PCR-derived inserts were completely sequenced to verify the integrity of the constructs.
Generation of viral supernatants and analysis of vector transduction efficiency.
Supernatants containing recombinant viral particles were generated essentially as described earlier (11, 13, 31). FV supernatants were generated by cotransfection of 293T cells with the Gag/Pol-expressing vector pMH118 and an Env expression plasmid as indicated in the figure legends. MuLV particles were obtained by cotransfection of 293T cells with the MuLV Gag/Pol expression vector pHIT60, the MuLV retroviral vector pczCFG2fEGN, and an Env expression vector as indicated in the figure legends. Transductions were performed by infection of 2 × 104 cells plated 24 h in advance in 12-well plates for 4 h with 1 ml of viral supernatant. The number of enhanced green fluorescent protein (EGFP)-positive cells was determined by fluorescence-activated cell sorter (FACS) analysis 48 h after infection.
Analysis of infectivity in the presence of lysosomotropic agents.
Stock solutions of ammonium chloride (cell culture grade; Sigma), chloroquine (Applichem), and methylamine (Fluka) were prepared in distilled H2O. Stock solutions of concanamycin A (Sigma) and bafilomycin A1 (Sigma) were prepared in dimethyl sulfoxide, and a stock solution of nigericin (Sigma) was prepared in ethanol. Target cells (2 × 104), plated 24 h in advance in 12-well plates, were preincubated for 30 min to 2 h with media containing lysosomotropic agents at the concentrations indicated in the figures. Subsequently, the cells were infected for 4 h with pretitrated supernatants. After infection, the cells were washed once and further incubated for an additional 30 min to 2 h in media containing lysosomotropic agents at the concentrations indicated in the figures. The transduction efficiency was determined by FACS analysis for EGFP-positive cells 48 h after infection. The percentages of EGFP-positive cells in cultures transduced with the respective pseudotypes in the absence of lysosomotropic agents were in the range of 15 to 50%. The relative infectivities for the cultures treated with the different lysosomotropic agents were calculated with regard to cells transduced with the respective pseudotype supernatants in the absence of lysosomotropic agents.
Transfections and pH-dependent cell fusion assay.
The pH dependence of the fusion activity of the different FV Env proteins was analyzed by using a cell-to-cell fusion assay. HT1080 cells were transiently transfected by using the Fugene 6 transfection reagent (Roche). Fugene 6 (6 μl) was mixed with the respective envelope protein expression construct (1.5 μg) and 92.5 μl of serum-free and antibiotic-free DMEM. The mixture was incubated at room temperature for 20 min. Subsequently, it was added to 2 × 105 HT1080 cells plated 24 h in advance in 6-well plates in 2 ml of medium. Twenty-four hours posttransfection, the cells were detached from the culture plate by trypsin treatment, mixed with an equal amount of HT1080 NLS-LacZ cells (HT1080 cells stably expressing a beta-galactosidase marker protein with a nuclear localization signal), and reseeded onto the plate. Four hours later, after attachment to the culture plate, the cells were exposed to different pHs. The cells were washed once with phosphate-buffered saline (PBS), pH 7.2, and then incubated for 30 s to 15 min with PBS at different pHs, ranging from 5.0 to 7.2. Subsequently, the cells were washed once with culture medium and fresh culture medium was added. Syncytia were allowed to form overnight. Next, the cells were fixed and histochemically stained for beta-galactosidase as described previously (29). Fusion activity was quantified by counting syncytia containing three or more nuclei. The fusion activity was described as the number of syncytia per square centimeter of culture plate.
RESULTS
FV Env-mediated transduction is inhibited by various lysosomotropic agents.
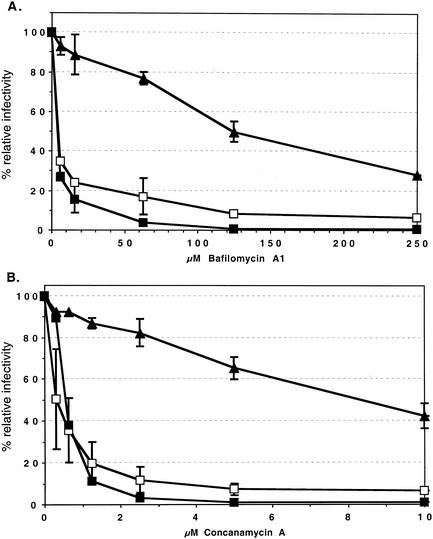
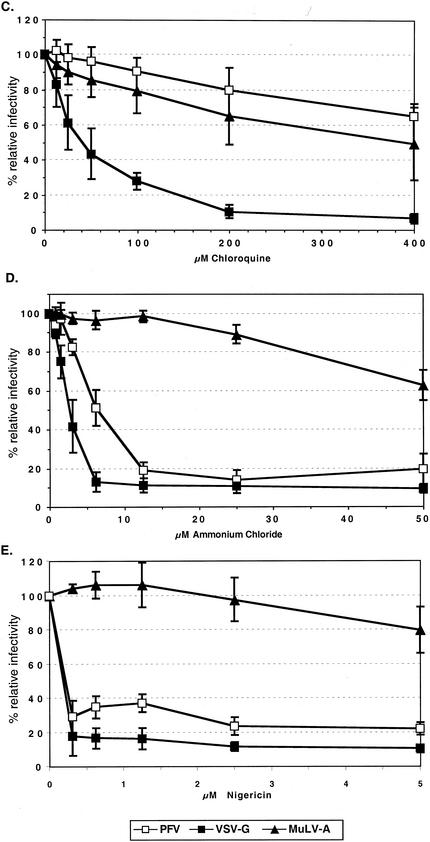
FV vectors cannot be pseudotyped by heterologous viral glycoproteins (20). Therefore, we predominantly analyzed the influence of lysosomotropic agents on the infectivity of PFV Env-pseudotyped MuLV vectors to determine whether FV Env-mediated entry involves a pH-dependent step. These were chosen over FV vectors as their use allowed the inclusion in these experiments of valid envelope controls, the MuLV-A envelope and the VSV-G protein, which are known to mediate pH-independent and pH-dependent entry processes, respectively. HT1080 target cells were incubated with various lysosomotropic agents at different concentrations for the time periods indicated in the figures before, during, and after transduction with EGFP-expressing MuLV pseudotypes. Subsequently, the infectivities relative to those for mock-treated controls were determined by FACS analysis. As expected for MuLV-A Env pseudotypes, only a minor inhibition of infectivities (1.5- to 3-fold) by bafilomycin A1, concanamycin A, chloroquine, ammonium chloride, and nigericin at the highest concentrations tested could be observed (Fig. 2). In contrast, VSV-G pseudotype infectivities were strongly reduced (10- to 50-fold) by all of these substances (Fig. 2). The inhibition patterns of PFV Env pseudotypes were comparable to those of the pH-dependent VSV-G pseudotypes for all lysosomotropic agents, with one exception, namely, chloroquine (Fig. 2). This substance (Fig. 2C) had no significant influence on the infectivity of the PFV Env pseudotypes, whereas their infectivity was inhibited by bafilomycin A1 (Fig. 2A), concanamycin A (Fig. 2B), ammonium chloride (Fig. 2D), nigericin (Fig. 2E), and methylamine (data not shown). Similar results with some of these substances were obtained on the quail cell line QT-6 and on mouse NIH3T3 cells, indicating that the results are neither cell nor species specific (data not shown). To analyze whether the observed phenotype of PFV Env-pseudotyped MuLV vectors is dependent on the viral capsid used, we also examined the influence of concanamycin A and chloroquine on the infectivity of PFV vectors. As shown in Fig. 3, PFV vectors displayed an inhibition characteristic in HT1080 cells similar to that of the corresponding PFV Env-pseudotyped MuLV vectors. In addition, inhibitor studies using replication-competent PFV showed a comparable pattern (data not shown).
FIG. 2.
Inhibition of MuLV pseudotype infectivity by different lysosomotropic agents. HT1080 target cells plated 24 h in advance were infected with pretitrated, EGFP-expressing MuLV pseudotype supernatants in the presence of different concentrations of bafilomycin (A), concanamycin A (B), chloroquine (C), ammonium chloride (D), and nigericin (E) as indicated. Lysosomotropic agents were present for 0.5 to 2 h before, 4 h during, and 0.5 to 2 h after infection. Forty-eight hours later, the percentages of EGFP-expressing cells were determined by FACS and the relative infectivities were calculated with respect to cells transduced with the respective pseudotype supernatants in the absence of lysosomotropic agents. The means and standard deviations of results from 3 to 10 experiments for each of the indicated concentrations of the lysosomotropic agents are shown. Pseudotypes were generated by cotransfection of 293T cells with the MuLV retroviral vector pczCFG2fEGN, the MuLV Gag/Pol expression vector pHIT60, and the respective Env expression construct as indicated: PFV, pczHFVenvΔ2MuLV; VSV-G, pcVG-wt; MuLV-A, pHIT456.
FIG. 3.
Inhibition of MuLV and PFV capsid infectivities by different lysosomotropic agents. HT1080 cells were infected with pretitrated, EGFP-expressing MuLV or PFV vectors pseudotyped by PFV glycoproteins in the presence of different concentrations of concanamycin A (A) or chloroquine (B) as indicated. Incubation with lysosomotropic agents, determination of the percentages of EGFP-expressing cells, and calculation of relative infectivities were done as described in the legend for Fig. 2. The means and standard deviations of results from 3 to 10 experiments for each of the indicated concentrations of the lysosomotropic agents are shown. Pseudotypes were generated by cotransfecting 293T cells with the following expression constructs: MuLV, pczCFG2fEGN plus pHIT60 plus pczHFVenvΔ2MuLV; PFV, pMH118 plus pczHFVenvEM02.
Taken together, these data suggest that the productive PFV Env-mediated infection by MuLV or PFV capsids involves a pH-dependent step that is strongly inhibited by incubation with various lysosomotropic agents. However, the lack of inhibition by chloroquine indicates that this entry process is at least to some extent different from that mediated by VSV-G.
Glycoproteins of different FV species are inhibited by lysosomotropic agents.
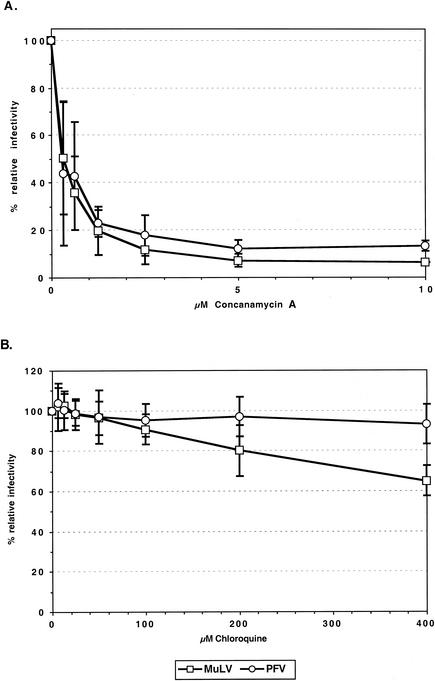
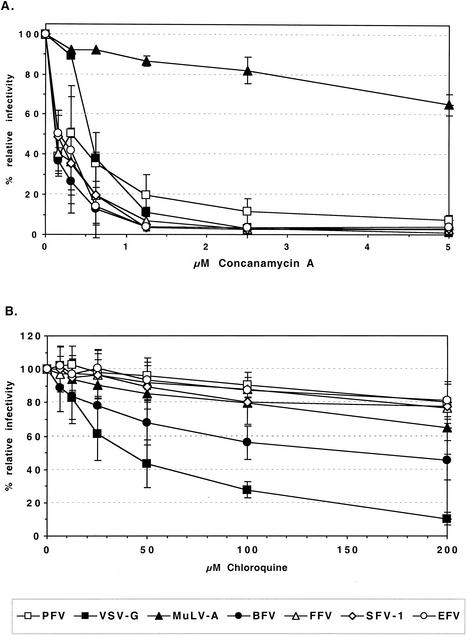
To determine whether the pattern of inhibition by lysosomotropic agents was unique to PFV Env, functional envelope glycoprotein expression constructs of various FV species, such as SFV-1, FFV, BFV, and EFV, were generated and analyzed with respect to their capacities for pseudotyping various retroviral capsids. For all FV species, individual envelope expression constructs that efficiently pseudotype MuLV particles could be identified (G. Jarmy, M. Picard-Maureau, and D. Lindemann, unpublished data). Subsequently, the infectivities of MuLV pseudotypes containing these FV Env proteins were tested with regard to their inhibition by chloroquine and concanamycin A. The results are summarized in Fig. 4. All of the FV Env MuLV pseudotypes were efficiently inhibited by concanamycin A, even more than the PFV Env pseudotypes (Fig. 4A). Similar to PFV Env pseudotypes, all of the FV Env MuLV pseudotypes except that containing BFV Env showed only a marginal reduction in infectivity by chloroquine (Fig. 4B). The BFV Env-mediated infectivity was slightly more inhibited by chloroquine than those mediated by the other FV Envs or MuLV-A Env, although the inhibition was clearly not as strong as that of the VSV-G pseudotypes (Fig. 4B).
FIG. 4.
Inhibition of infectivities of MuLV vectors pseudotyped with various FV Env species by different lysosomotropic agents. HT1080 cells were infected with pretitrated, EGFP-expressing MuLV pseudotyped by various viral glycoproteins in the presence of different concentrations of concanamycin A (A) or chloroquine (B) as indicated. Incubation with lysosomotropic agents, determination of the percentages of EGFP-expressing cells, and calculation of relative infectivities were done as described in the legend for Fig. 2. The means and standard deviations of results from 3 to 10 experiments for each of the indicated concentrations of the lysosomotropic agents are shown. Pseudotypes were generated by cotransfection of 293T cells with the MuLV retroviral vector pczCFG2fEGN, the MuLV Gag/Pol expression vector pHIT60, and the respective Env expression construct as indicated: PFV, pczHFVenvΔ2MuLV; VSV-G, pcVG-wt; MuLV-A, pHIT456; BFV, pczBFVenv; FFV, pczFFVenv; SFV-1, pciSFV-1envSM01; EFV, pciEFVenv.
Taken together, these results indicate that the inhibitory effect of certain lysosomotropic agents, such as concanamycin A, is common to all FV species. Furthermore, the general failure of chloroquine to reduce the infectivity of MuLV vectors pseudotyped with glycoproteins of different FV species suggests that FV use a common pH-dependent entry pathway different from that mediated by VSV-G.
FV Env-mediated fusion is pH dependent.
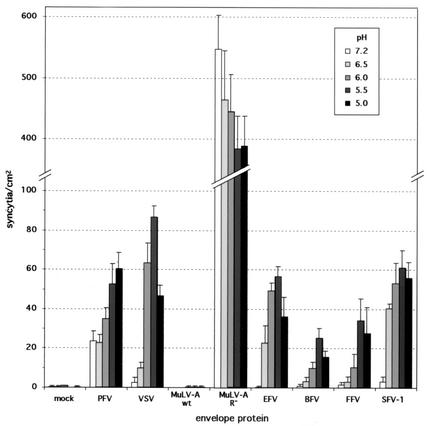
The data presented above demonstrate that the FV Env-mediated infectivity of MuLV and PFV pseudotypes involves a pH-dependent step. However, the experiments did not reveal whether the FV Env-mediated fusion process itself or another step in the replication cycle, between the binding of the viral particles to the cell surface and the expression of the EGFP marker genes, is pH dependent. Therefore, we examined the pH dependency of the FV Env fusion process in a cell-to-cell fusion assay. HT1080 cells were transfected with various FV Env expression constructs and mixed with NLS-LacZ-expressing HT1080 cells, and after adherence to the culture dishes, the cells were exposed to PBS at different pHs. Subsequently, syncytium formation was quantified after histochemical beta-galactosidase staining. The data are summarized in Fig. 5. The MuLV Env fusion activity is not controlled by pH but by a posttranslational processing event of the CyD (22). Only upon removal of the C-terminal 16 aa (R peptide) of the CyD through proteolytic cleavage by the viral protease does the MuLV Env reach a fusion-competent state (22, 25). In agreement with these data, no syncytium formation could be observed upon expression of the MuLV-A Env, regardless of the pH. Cells expressing an R peptide-truncated MuLV Env mutant showed very strong fusion activities that were similar at the different pHs tested. In contrast, VSV-G-mediated syncytium formation was very low at a neutral pH but was induced ∼30-fold by exposure to decreasing pHs (35). Under sensitive assay conditions, PFV Env expression alone showed a significant basal fusion activity at neutral pH (Fig. 5), similar to what has been reported previously (4, 21). However, a reproducible two- to threefold increase in the number of syncytia could be obtained by short-term exposure of PFV Env-expressing cells to acidic pH. Interestingly, all other FV Envs tested had very low basal fusion activities around background levels under these sensitive assay conditions but were strongly induced (∼20- to 180-fold) by low-pH treatment (Fig. 5). Maximal activities were observed in most cases at pH 5.5.
FIG. 5.
Induction of FV Env-mediated syncytium formation by low-pH treatment. HT1080 cells were transfected with the indicated expression constructs and 24 h later mixed with HT1080 NLS-LacZ cells. Four hours later, after attachment to the culture plate, the cells were exposed to different pHs as indicated. Syncytia were allowed to form overnight and subsequently quantified after histochemical beta-galactosidase staining. The means and standard deviations of results from three experiments for each of the indicated pHs are shown. Mock, pcDNA3.1+zeo; PFV, pczHFVenvEM02; VSV, pcVG-wt; MuLV-A wt, pcziMEA-wt; MuLV-A-R−, pcziMEA-R−; BFV, pczBFVenv; FFV, pczFFVenv; SFV-1, pciSFV-1; EFV, pSGEFVenv.
Taken together, these data suggest that the FV Env-mediated fusion process is induced by exposure to acidic pH. The PFV Env was the only FV Env tested that had significant fusion activity at neutral pH. It is noteworthy that in line with this finding, of all FV Envs tested, the PFV Env was the least inhibited by lysosomotropic agents.
A short exposure to low pH is sufficient for FV Env fusion induction.
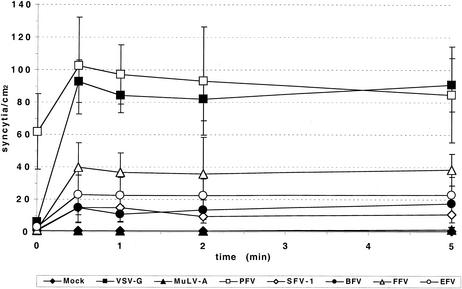
One unique feature of the pH-dependent fusion process of ALV described recently is the requirement for a prolonged exposure to acidic pH for induction of giant cell formation in a cell-to-cell fusion assay (19). We therefore analyzed the influence of the time period of pH shift on the induction of FV Env-mediated syncytium formation. As shown in Fig. 6, exposure of all FV Envs tested to pH 5.5 for as little as 30 s was sufficient to induce fusion activity. This indicates that the pH-dependent FV fusion process deviates from that of ALV and resembles more closely that of influenza A and VSV.
FIG. 6.
Time course of pH-dependent FV Env-mediated syncytium formation. HT1080 cells were transfected with different expression constructs as indicated below and 24 h later mixed with HT1080 NLS-LacZ cells. Four hours later, after attachment to the culture plate, the cells were exposed to pH 5.5 for different time periods as indicated. Syncytia were allowed to form overnight and subsequently quantified after histochemical beta-galactosidase staining. The means and standard deviations of results from three experiments for each of the indicated pHs are shown. Mock, pcDNA3.1+zeo; PFV, pczHFVenvEM02; VSV-G, pcVG-wt; MuLV-A, pHIT456; BFV, pczBFVenv; FFV, pczFFVenv; SFV-1, pczSFV-1; EFV, pSGEFVenv.
DISCUSSION
The first steps of the FV replication cycle are poorly characterized. The results from this study indicate that FV probably enter target cells by using a pH-dependent endocytic pathway. Heterologous viral glycoproteins are not packaged into FV particles (20). However, the comparison of MuLV FV Env pseudotypes with FV virions in our study and other studies (11, 12, 20) suggests that MuLV pseudotypes accurately mimic the entry process of FV. A major advantage of using FV Env MuLV pseudotypes for this study was the possibility of using viral particles differing only in their incorporated glycoproteins and then directly comparing the FV Env to viral glycoproteins known to mediate pH-dependent or pH-independent virus entry. The incubation of target cells with various lysosomotropic agents and subsequent analysis of the infectivities of various MuLV pseudotypes in an EGFP marker gene transfer assay showed that the inhibition of PFV Env pseudotypes by these agents was similar to that of VSV-G control pseudotypes. However, one difference from the VSV-G pseudotypes could be observed, namely, a lack of inhibition of PFV Env pseudotypes by chloroquine. The reason for this is currently unclear, but it may suggest that the FV Env-mediated entry process is different at least in some aspects from that of VSV-G. FV Env-mediated fusion may take place in a different endosomal compartment from the VSV-G-mediated fusion process. It is noteworthy that a similar phenomenon has been observed for the penetration of Sindbis virus in mosquito cells (8). Unlike Sindbis virus entry into vertebrate cells, the entry of this virus into mosquito cells is inhibited by ammonium chloride but not chloroquine, although the acidification of endosomes seems to be blocked by both substances (8). One possible explanation suggested by Hernandez et al. is that these cells possess two classes of endosomes, one sensitive to both substances and another sensitive only to NH4Cl (8). The fact that the inhibition of FV Env-mediated entry by the different tested lysosomotropic agents, except chloroquine, could be observed with either MuLV or PFV capsids indicates that the phenomenon is not a special feature of the MuLV pseudotypes and also not dependent on the retroviral capsid but is solely mediated by FV envelope glycoproteins. Furthermore, these results seem to exclude a postfusion effect of the lysosomotropic agents resulting in the observed inhibition of marker gene transfer and expression. Similar inhibition characteristics of the different tested lysosomotropic substances were observed in cells of different origin, suggesting that a unique entry pathway mediated by the FV Env may be common to different species.
FV Env proteins from different species showed similar characteristics of inhibition by the tested lysosomotropic agents. However, PFV Env pseudotypes were somewhat less inhibited by concanamycin A. This observation is in good agreement with the pH dependence of the different FV Envs, with PFV being the only FV Env having a significant fusion activity at neutral pH. This suggests that it may allow for the entering of target cells by a pH-independent mechanism also, at least to some extent. Recently, the acquirement of a pH-independent fusion activity by VSV-G during intracellular transport in a polarized endometrial cell line was described (27). This activity was efficiently inhibited by treatment with lysosomotropic agents. It seems unlikely that a similar phenomenon is responsible for the basal fusion activity observed for the PFV Env at neutral pH in our fusion assays with HT1080 cells, because syncytium formation was not reduced by the addition of lysosomotropic agents during the assay (data not shown). Nevertheless, it would be of interest to examine FV Env fusion induction in HEC cells, especially that of FV Env proteins with low basal activities at neutral pH, to determine whether FV Env is transported to the cell surface by a route similar to that of VSV-G in these cells. It is currently unclear why the BFV Env pseudotypes seem to be affected slightly more by chloroquine in HT1080 cells than the other FV Env proteins and whether this is an indication for a somewhat different entry pathway of BFV.
Recently, the inhibition of ecotropic MuLV and MuLV-A infectivities by lysosomotropic agents was reexamined (9). The results in this study suggest that the inhibitory effect of these agents on viral titers reflects the stability of viral particles during the course of the experiment rather than the necessity for an acidic environment. We were unable to detect a direct effect of either ammonium chloride or concanamycin A on the particle stability of the different pseudotypes (data not shown). In contrast to the data reported by Katen et al. (9), VSV-G and MuLV-A pseudotypes showed similar stabilities in our assay system. However, the use of different target cells and assay systems for the determination of relative infectivities makes a direct comparison difficult.
Neither ecotropic MuLV nor MuLV-A Env-expressing cells are induced to undergo syncytium formation upon the artificial lowering of the extracellular pH, indicating that indeed the MuLV Env-mediated fusion process itself is pH independent (22). Therefore, it was striking to observe that FV Env-expressing cells could be induced to form giant cells at low-pH exposure, similar to the results that have recently been reported for the ALV Env (19). This confirmed our assumption that the FV Env fusion process itself is pH controlled. Unlike the Avian sarcoma/leukosis virus Env, which requires an extended exposure to low pH, FV Env requires an exposure of only 30 s for fusion induction. With respect to this characteristic, FV Env more closely resembles classical pH-dependent viral glycoproteins, such as influenza hemagglutinin and VSV-G, where fusion is induced immediately after exposure to low pH (35). Interestingly, at neutral pH only the PFV Env showed a significant basal fusion activity, which could still be enhanced two- to threefold by low-pH exposure. All other FV Env proteins tested at neutral pH had low or undetectable fusion activities, which were strongly induced by lowering the extracellular pH. These results indicate that PFV Env is an exception with respect to the fact that it may also use a pH-independent pathway for entry. This is in agreement with the inhibitory effect of concanamycin A on FV Env pseudotypes and the somewhat lower sensitivity of PFV Env. Why PFV is different with respect to its basal fusion activity is currently unclear. One possible explanation might be an in vitro adaptation, since the PFV proviral sequence from which the PFV Env expression construct is derived was isolated only after extensive in vitro passage of the virus. It might therefore be interesting to analyze the Env proteins of closely related primary chimpanzee virus isolates from which the PFV isolate most probably was derived.
Acknowledgments
We thank A. Saïb for pSGEFVenv, A. Mergia for pSFV-1, C. R. Helps and D. A. Harbour for FFV clone#94 and Env1, and R. W. Renshaw and J. W. Casey for BSV-11.
This work was supported by grants from the Bayerische Forschungsstiftung, DFG (Li621/2-1, Li621/2-3, SFB479, Re627/6-1, and Europäisches Graduiertenkolleg “Gene regulation in and by microbial pathogens”), and EU (QLK5-CT-1999-51410).
REFERENCES
- 1.Cannon, P. M., N. Kim, S. M. Kingsman, and A. J. Kingsman. 1996. Murine leukemia virus-based Tat-inducible long terminal repeat replacement vectors: a new system for anti-human immunodeficiency virus gene therapy. J. Virol. 70:8234-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du Bridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbert, J. M., D. Mason, and J. M. White. 1990. Fusion of Rous sarcoma virus with host cells does not require exposure to low pH. J. Virol. 64:5106-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goepfert, P. A., K. L. Shaw, G. D. Ritter, Jr., and M. J. Mulligan. 1997. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J. Virol. 71:778-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haywood, A. M., and B. P. Boyer. 1982. Sendai virus membrane fusion: time course and effect of temperature, pH, calcium, and receptor concentration. Biochemistry 21:6041-6046. [DOI] [PubMed] [Google Scholar]
- 6.Helenius, A., M. Marsh, and J. White. 1982. Inhibition of Semliki forest virus penetration by lysosomotropic weak bases. J. Gen. Virol. 58:47-61. [DOI] [PubMed] [Google Scholar]
- 7.Helps, C. R., and D. A. Harbour. 1997. Comparison of the complete sequence of feline spumavirus with those of the primate spumaviruses reveals a shorter gag gene. J. Gen. Virol. 78:2549-2564. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez, R., T. Luo, and D. T. Brown. 2001. Exposure to low pH is not required for penetration of mosquito cells by Sindbis virus. J. Virol. 75:2010-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katen, L. J., M. M. Januszeski, W. F. Anderson, K. J. Hasenkrug, and L. H. Evans. 2001. Infectious entry by amphotropic as well as ecotropic murine leukemia viruses occurs through an endocytic pathway. J. Virol. 75:5018-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lecellier, C. H., M. Neves, M. L. Giron, J. Tobaly-Tapiero, and A. Saib. 2002. Further characterization of equine foamy virus reveals unusual features among the foamy viruses. J. Virol. 76:7220-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindemann, D., M. Bock, M. Schweizer, and A. Rethwilm. 1997. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J. Virol. 71:4815-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindemann, D., and A. Rethwilm. 1998. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J. Virol. 72:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClure, M. O., M. Marsh, and R. A. Weiss. 1988. Human immunodeficiency virus infection of CD4-bearing cells occurs by a pH-independent mechanism. EMBO J. 7:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClure, M. O., M. A. Sommerfelt, M. Marsh, and R. A. Weiss. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71:767-773. [DOI] [PubMed] [Google Scholar]
- 17.Mergia, A., and M. Wu. 1998. Characterization of provirus clones of simian foamy virus type 1. J. Virol. 72:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moscovici, C., M. G. Moscovici, H. Jimenez, M. M. Lai, M. J. Hayman, and P. K. Vogt. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11:95-103. [DOI] [PubMed] [Google Scholar]
- 19.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679-689. [DOI] [PubMed] [Google Scholar]
- 20.Pietschmann, T., M. Heinkelein, M. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietschmann, T., H. Zentgraf, A. Rethwilm, and D. Lindemann. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J. Virol. 74:4474-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasheed, S., W. A. Nelson-Rees, E. M. Toth, P. Arnstein, and M. B. Gardner. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027-1033. [DOI] [PubMed] [Google Scholar]
- 24.Redmond, S., G. Peters, and C. Dickson. 1984. Mouse mammary tumor virus can mediate cell fusion at reduced pH. Virology 133:393-402. [DOI] [PubMed] [Google Scholar]
- 25.Rein, A., J. Mirro, J. Gordon Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renshaw, R. W., M. A. Gonda, and J. W. Casey. 1991. Structure and transcriptional status of bovine syncytial virus in cytopathic infections. Gene 105:179-184. [DOI] [PubMed] [Google Scholar]
- 27.Roberts, P. C., T. Kipperman, and R. W. Compans. 1999. Vesicular stomatitis virus G protein acquires pH-independent fusion activity during transport in a polarized endometrial cell line. J. Virol. 73:10447-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saib, A., F. Puvion Dutilleul, M. Schmid, J. Peries, and H. de The. 1997. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J. Virol. 71:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt, M., and A. Rethwilm. 1995. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology 210:167-178. [DOI] [PubMed] [Google Scholar]
- 30.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 31.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein, B. S., S. D. Gowda, J. D. Lifson, R. C. Penhallow, K. G. Bensch, and E. G. Engleman. 1987. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell 49:659-668. [DOI] [PubMed] [Google Scholar]
- 33.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2002. Virus taxonomy: the classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 34.White, J., J. Kartenbeck, and A. Helenius. 1980. Fusion of Semliki forest virus with the plasma membrane can be induced by low pH. J. Cell Biol. 87:264-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White, J., K. Matlin, and A. Helenius. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J. Cell Biol. 89:674-679. [DOI] [PMC free article] [PubMed] [Google Scholar]