Abstract
The importance of the small envelope (E) protein in the assembly of coronaviruses has been demonstrated in several studies. While its precise function is not clearly defined, E is a pivotal player in the morphogenesis of the virion envelope. Expression of the E protein alone results in its incorporation into vesicles that are released from cells, and the coexpression of the E protein with the membrane protein M leads to the assembly of coronavirus-like particles. We have previously generated E gene mutants of mouse hepatitis virus (MHV) that had marked defects in viral growth and produced virions that were aberrantly assembled in comparison to wild-type virions. We have now been able to obtain a viable MHV mutant in which the entire E gene, as well as the nonessential upstream genes 4 and 5a, has been deleted. This mutant (ΔE) was obtained by a targeted RNA recombination method that makes use of a powerful host range-based selection system. The ΔE mutant produces tiny plaques with an unusual morphology compared to plaques formed by wild-type MHV. Despite its low growth rate and low infectious titer, the ΔE mutant is genetically stable, showing no detectable phenotypic changes after several passages. The properties of this mutant provide further support for the importance of E protein in MHV replication, but surprisingly, they also show that E protein is not essential.
The coronavirus virion consists of a nucleocapsid core surrounded by an envelope containing three integral membrane proteins that are common to all members of the genus (19). The nucleocapsid is a helical ribonucleoprotein made up of the positive-strand RNA genome with monomers of a single nucleocapsid (N) protein bound along its length (20). It has been suggested that this structure is further compacted into a shell with spherical, possibly icosahedral, symmetry (31, 32). The viral envelope that packages the nucleocapsid is derived from the membrane of the intracellular budding site, which is the endoplasmic reticulum-Golgi intermediate compartment or cis-Golgi network (16, 34, 40). Embedded in the envelope membrane are two major structural proteins. The first of these is the spike (S) glycoprotein, which forms morphologically characteristic projections on the virion surface that mediate both attachment to species-specific host cell receptors and fusion with the plasma membrane. The second is the membrane (M) glycoprotein, the most abundant virion component, which is a triple-spanning integral membrane protein with a short ectodomain and a large carboxy-terminal endodomain (33). The carboxy terminus of M has been shown by both biochemical (10, 26, 27, 38) and genetic (18) criteria to interact with the nucleocapsid, specifically, with the carboxy terminus of the N protein (18).
The third critical membrane-bound constituent of the virion is the small envelope (E) protein. E was only belatedly recognized as a viral structural protein, owing to its size (ca. 10 kDa) and its very low abundance relative to the M, N, and S proteins (13, 21, 45). E proteins are well conserved within each of the three groups of coronaviruses, but they show very limited homology across the different groups. All E proteins have the same general structure: a short (7- to 9-amino-acid) hydrophilic amino terminus preceding a large (21- to 29-amino-acid) hydrophobic region, followed by a large hydrophilic carboxy terminus making up one-half to two-thirds of the mass of the molecule (21). Earlier work suggested an external topology for the carboxy-terminal domain of the E protein of transmissible gastroenteritis virus (TGEV) (13). However, more recent studies of the E proteins of mouse hepatitis virus (MHV) and infectious bronchitis virus (IBV) have demonstrated that E is an integral membrane protein displaying its carboxy terminus on the cytoplasmic face of the endoplasmic reticulum or Golgi, which corresponds to the interior of assembled virions (3, 30, 42). Interestingly, it is the carboxy-terminal tail of the IBV E protein, in the absence of the membrane-bound domain, which specifies Golgi targeting for that molecule (4). The disposition of the amino terminus of the E molecule is less well established. A lumenal (or virion-exterior) orientation of this domain has been inferred for the IBV E protein (3), while for the MHV E protein, the amino terminus has been proposed to be buried within the membrane near the cytoplasmic face (23).
The case for a major role for the E protein in coronavirus assembly was solidified by fundamental observations by Vennema and coworkers (42) and Bos and coworkers (2) that cellular expression of just the M and E proteins of MHV leads to formation and release of virus-like particles (VLPs) that are morphologically identical to spikeless virions. These results were subsequently generalized to TGEV (1), bovine coronavirus (BCoV) (1), and IBV (3). Thus, the coronavirus envelope can form itself in the absence of either S protein or nucleocapsid, although work with constructed MHV M protein mutants indicates that the nucleocapsid stabilizes virion structure (7, 18). Further support for the importance of the E protein came from the construction of clustered charged-to-alanine mutants of the MHV E protein (12). Two of these mutants were partially temperature sensitive and markedly thermolabile, and electron microscopic examination of the more impaired mutant revealed virions with dramatic deviations from the more uniformly spherical virions of the wild type.
Collectively, these results would imply that the E protein is essential for coronavirus replication, and indeed, this has recently been reported for TGEV (5, 29) and for equine arterivirus (EAV), a related member of the order Nidovirales (37). Surprisingly, however, when we rigorously tested this assumption for MHV, we were able to isolate mutants in which the E gene was totally deleted. These viruses, although severely debilitated, were nevertheless viable and infectious. We therefore conclude that the E protein is critical, but not essential, for MHV replication.
MATERIALS AND METHODS
Cells and viruses.
Wild-type MHV-A59 and MHV recombinants were propagated in mouse 17 clone 1 (17Cl1) or L2 cells; plaque assays and plaque purification were performed with mouse L2 cells. The interspecies chimeric virus fMHV (17) was grown in feline AK-D fetal lung cells.
Plasmid constructs.
For the purpose of deleting the E gene in this study, plasmid pLK70 was constructed from the transcription vector pMH54, which has been described previously (17). The deletion mutation, which encompasses genes 4 and 5a as well as the E gene, was derived from a PCR fragment that was generated using primers LK87 and LK10 (Table 1) with pMH54 as the template. After restriction digestion with Sse8783I and BssHII, the PCR product was transferred into pMH54, replacing the Sse8783I-BssHII fragment of that plasmid. The resulting construct, pLK70, was 1,023 bp shorter than the pMH54 parent but was otherwise identical to pMH54.
TABLE 1.
Primers used in this study
| Primer | Gene | Sense | Sequence |
|---|---|---|---|
| LK87a | ΔE junction | + | 5′CCGAAGCCTGCAGGAAAGACAGAAAATCTAAACATTATGAGTAGTACTACTCAG3′ |
| LK10 | M | − | 5′CAACAATGCGGTGTCCGCGCCAC3′ |
| CK4 | MHV S | + | 5′TCGAGAAGTTAAATGTTA3′ |
| PM147 | M | − | 5′AGAAAATCCAAGATACAC3′ |
| PM145 | N | − | 5′CACACTCCCGGAGTTGGG3′ |
| LK59 | MHV S | + | 5′GCTGCACAGGTTGTGGCT3′ |
| LK58 | FIPV S | + | 5′CTGGACTGTACCTGAATT3′ |
| LK17 | M | − | 5′CCGAACTGTAGTATGATAG3′ |
| LK50 | FIPV S | + | 5′TGCACTGGCTATGCTACC3′ |
| LK67 | FIPV S | − | 5′GATATACAACCAATAGGA3′ |
| LK57 | FIPV S | + | 5′GTTAATGAATGTGTTAGG3′ |
| LK60 | FIPV S | − | 5′ATGGCAAGTTCTACTGTA3′ |
| CM82 | 5a | + | 5′AACAGGCCGTTAGCAAGT3′ |
| PM152 | E | − | 5′ATGGTGACCATCAAACAC3′ |
| PM160 | E | + | 5′ATTCCTTACAGACACAGT3′ |
| FF42 | E | − | 5′CATCCACCTCTAATAGGG3′ |
| CM80 | 5a | + | 5′GGTTACGCCGCACGCGGG3′ |
| CM95 | 3′ UTR | + | 5′GAGAATGAATCCTATGTC3′ |
| PM112 | 3′ UTR | − | 5′CCATGATCAACTTCATTC3′ |
Primer used to generate the deletion in pLK70 (see Materials and Methods). Underlined bases are the Sse8387I site and the new TRS for the M gene, respectively.
Targeted RNA recombination.
To incorporate the ΔE mutation into the MHV genome, targeted RNA recombination was performed using fMHV as the recipient virus, as described previously (18, 25). In brief, feline AK-D cell monolayers were infected with fMHV at a multiplicity of roughly 1 PFU per cell for 4 h at 37°C; the infected cells were then suspended by treatment with a low concentration of trypsin. Capped donor RNA transcripts were synthesized with the T7 mMessage mMachine transcription kit (Ambion) using PacI-truncated pLK70 or pMH54 template following the manufacturer's instructions. Synthetic donor RNA (5 to 10 μg) was transfected into 1.2 × 107 fMHV-infected AK-D cells by two pulses with a Gene Pulser electroporation apparatus (Bio-Rad) at settings of 960 μF and 0.3 kV. The infected and transfected AK-D cells were then overlaid onto murine 17Cl1 cell monolayers. Progeny virus released into the supernatant medium was harvested when syncytia or cytopathic effect was apparent in the murine cells, typically 48 h postinfection at 37°C. Both large- and small-plaque recombinant candidates were purified through two rounds of plaque titration on murine L2 monolayers at 37°C.
Genomic analysis of candidate recombinants.
Each purified recombinant candidate was used to infect a 10-cm2 monolayer of L2 cells at 37°C. Total cellular RNA was extracted either 24 (for large-plaque candidates) or 72 (for small-plaque candidates) h postinfection using Ultraspec reagent (Biotecx). Reverse transcription (RT) of RNA was carried out using a random hexanucleotide primer (Boehringer Mannheim) with avian myeloblastosis virus reverse transcriptase (Life Sciences) under standard conditions (35). To confirm the presence, absence, or alteration of different genomic regions, PCR amplifications of cDNA were performed using various oligonucleotide primer pairs. PCR was carried out with AmpliTaq polymerase (Perkin-Elmer) under standard conditions of 1 min at 94°C, 1 min at 49°C, and 2 min at 72°C, repeated for 30 cycles. The resulting products were analyzed directly by agarose gel electrophoresis or were purified with Quantum-prep columns (Bio-Rad) prior to automated sequencing.
Radiolabeling of viral RNA.
Metabolic labeling of virus-specific RNA was carried out under conditions similar to those reported previously (25). Monolayers of 17Cl1 cells in 20-cm2 dishes were infected with the ΔE mutant Alb291 or the wild-type MHV recombinant Alb240 at a multiplicity of 0.125 PFU per cell, and incubation was continued at 33°C. When infections had progressed to the extent of ∼50% syncytium development (at ∼12 h), the cells were starved for 2 h at 37°C in Eagle's minimal essential medium containing 5% dialyzed fetal bovine serum and 1/10 of the normal phosphate concentration. Labeling was then carried out for 2 h at 37°C by incubation of the starved cells in phosphate-free medium containing 100 μCi of [33P]orthophosphate (ICN) per ml, 5% dialyzed fetal bovine serum, and 20 μg of actinomycin D (Sigma) per ml. Total cytoplasmic RNA was purified by a gentle NP-40 lysis procedure. RNA samples containing equal amounts of radioactivity were denatured with formaldehyde and formamide, analyzed by electrophoresis through 1% agarose containing formaldehyde, and visualized by fluorography.
Northern blot hybridization.
For the analysis of viral RNA by Northern blotting, total cytoplasmic RNA was harvested and purified from 17Cl1 cells infected with MHV recombinants or wild-type MHV. After fractionation in a 1% agarose gel containing formaldehyde, the RNA was blotted and then UV cross-linked onto a Nytran Supercharge membrane (Schleicher and Schuell). Alternatively, RNA denatured with formaldehyde and formamide was directly dot blotted onto a membrane by filtration through a vacuum manifold followed by UV cross-linking. A DNA probe corresponding to the 5′ 283 nucleotides of the MHV 3′ untranslated region (UTR), generated with PCR primers CM95 and PM112 (Table 1), was used to detect all virus-specific RNA species. As a probe for E gene-specific sequences, a DNA fragment of 233 bp, covering almost the entire E open reading frame (ORF), was generated with the PCR primers PM160 and FF42 (Table 1). The DNA fragments were directly labeled during synthesis with [α-32P]dCTP in PCR using the respective primers (35). Hybridization was carried out at 65°C (16 to 20 h) in a buffer containing 0.5 M sodium phosphate, pH 7.2, 7% sodium dodecyl sulfate, and 1 mM EDTA. After hybridization, the blots were washed at 65°C three times in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% sodium dodecyl sulfate (35), and the radioactivity was visualized by fluorography.
RESULTS
Construction of a viable MHV recombinant with the E gene deleted.
Targeted RNA recombination has been successfully employed as a method to carry out reverse genetics on the 3′-most 10 kb of the 31-kb MHV genome (15, 17, 24, 25), a region that encompasses all of the structural genes of this coronavirus. This technique exploits the recombinational propensity of the viral polymerase to obtain crossover events between a recipient virus and a transfected synthetic donor RNA. Recombinant viruses are then recovered based on a counterselectable property of the recipient virus. A particularly strong selection is afforded by the host cell species restriction of a chimeric mutant, fMHV, that contains the ectodomain of the S protein of feline infectious peritonitis virus (FIPV) in place of its MHV counterpart (17). As a result of this substitution, fMHV grows in feline cells and has lost the ability to grow in murine cells. We have recently shown that the use of fMHV as the recipient virus in targeted RNA recombination allows the recovery of extremely defective MHV mutants based on selection for the reacquisition of the MHV S protein ectodomain and the concomitant ability to grow in murine cells (18).
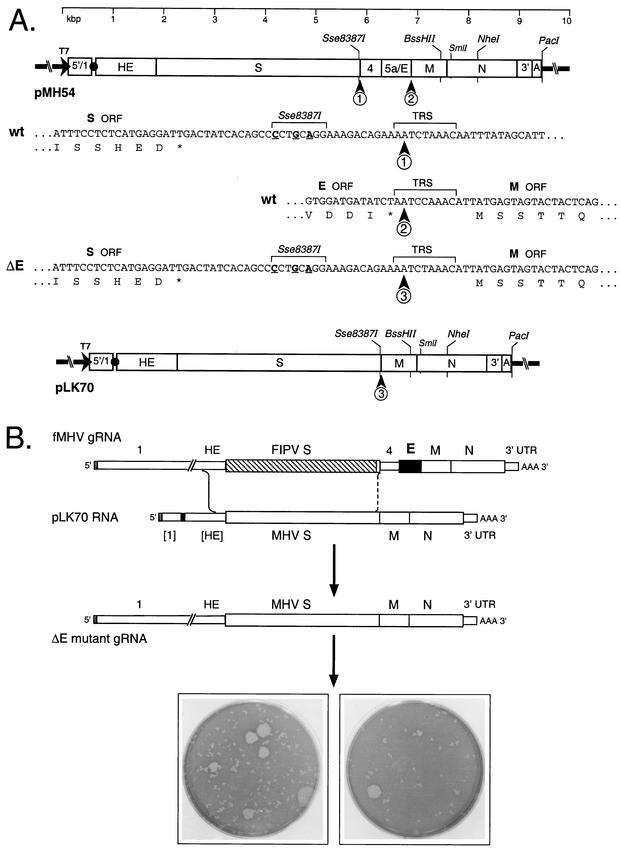
To pursue the fundamental question of whether the E protein is essential for MHV replication, we deleted the E gene from a transcription vector from which donor RNA could be synthesized. Plasmid pLK70 (Fig. 1A) was constructed from the previously described wild-type pMH54 (17) via PCR-facilitated removal of the entirety of genes 4, 5a, and E. A number of prior reports have established that genes 4 and 5a are nonessential in MHV (8, 11, 28, 43, 44). The 1,023-bp deletion in pLK70 ran from the transcription-regulating sequence for gene 4 (TRS4) through the TRS for the M gene (TRS6); the canonical TRS4 core sequence (AATCTAAAC) was retained instead of the slightly variant (underlined) TRS6 core sequence (AATCCAAAC) (Fig. 1A).
FIG.1.
Selection of the ΔE mutant. (A) Construction of a transcription vector for synthesis of donor RNA. Plasmid pLK70 was derived from pMH54 (17), as detailed in Materials and Methods. In each plasmid schematic, the arrow indicates the T7 promoter, and the solid circle represents the linker between cDNA segments corresponding to the 5′ and 3′ ends of the MHV genome. The restriction sites shown are those relevant to plasmid construction (Sse8387I, BssHII, and NheI), in vitro transcription (PacI), or mutant analysis (SmlI). The DNA sequences shown are as follows: 1 and 2, the boundaries of the region that was deleted from the wild type (wt); and 3, the newly created junction in the deletion mutant. The underlined nucleotides are the three base changes made in pMH54 that generated an Sse8387I site (17). Also indicated are the TRSs governing synthesis of sgRNA4 and the M mRNA. Shown beneath the DNA sequences are translations of the S, M, and E ORFs (asterisks indicate stop codons). (B) Scheme for generation of the ΔE mutant by targeted RNA recombination between the interspecies chimera fMHV (17) and donor RNA transcribed from plasmid pLK70. fMHV contains the ectodomain-encoding region of the FIPV S gene (hatched rectangle) and is able to grow in feline cells but not in murine cells. A single crossover (solid line), within the HE gene, should generate a recombinant that has simultaneously reacquired the MHV S ectodomain and the ability to grow in murine cells and has also incorporated the deletion of the E gene. A potential second crossover (broken line), in the distal portion of the S gene, would produce a recombinant retaining the E gene. At the bottom are shown the mixed progeny of two independent targeted recombination experiments, forming tiny and large plaques on mouse L2 cells. The monolayers were stained with neutral red 75 h postinfection and were photographed 19 h later.
Synthetic donor RNAs transcribed from pLK70, or from the control, pMH54, were independently transfected into feline AK-D cells that had been infected with fMHV (17) (Fig. 1B), and recombinants were selected in murine cells. When the titers of these were determined on mouse L2 cells, the pLK70-derived recombinants generated a mixed population of very tiny plaques and much larger, wild-type-sized plaques (Fig. 1B). By contrast, only wild-type-sized plaques were observed with RNA from the parental plasmid, pMH54 (not shown). Based on our previous experience with this selection scheme (18), we expected that the small plaques represented ΔE recombinants, produced by a single crossover upstream of the S gene, while the large plaques were expected to be wild-type recombinants resulting from an additional downstream crossover event. (These expectations were subsequently verified.) It is likely that the small plaques outnumbered the large ones because the elimination of genes 4 and 5a, in addition to the E gene, in pLK70 provided a minimal (233-nucleotide) region of homology downstream of the S ectodomain in which a productive second crossover could occur (Fig. 1B).
Plaque phenotype of the ΔE mutant.
Multiple plaques resulting from independent infection-transfection experiments with pLK70-derived donor RNA were purified by plaque titration. The respective phenotypes of the small and large plaques did not change throughout two rounds of plaque purification or following additional passages in murine cells. Infections begun from the small-plaque ΔE mutants produced the hallmark syncytia and subsequent cytopathic effect of MHV in cell monolayers, but they did so much more slowly than the wild type, and ΔE stocks reached optimal titers of only 1 × 105 to 2 × 105 PFU/ml, at least 3 orders of magnitude lower than those typically observed for the wild type. The large-plaque recombinants, on the other hand, were indistinguishable from wild-type MHV in growth rate, plaque size, and virus titers.
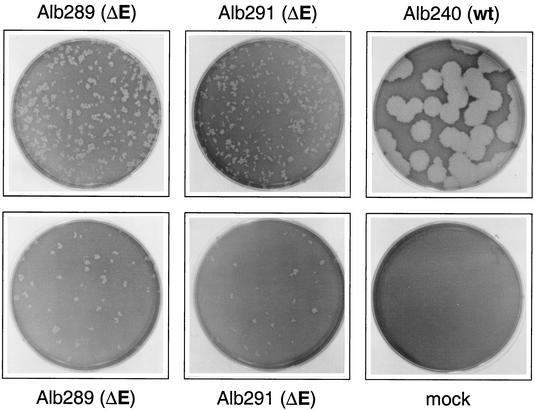
In addition to their drastically reduced size, plaques of the ΔE mutants also exhibited an intriguing morphology. Figure 2 shows two dilutions of two independent ΔE mutants to demonstrate this feature. In contrast to the uniform, circularly symmetric plaques typically formed by wild-type MHV, plaques formed by ΔE mutants showed much more pronounced size variations, with some plaques of pinpoint size and others several times larger. The tiny plaques also had irregular shapes with jagged edges, unlike the usually smooth edges found in wild-type plaques. Notably, these peculiarities were not seen for wild-type plaques at early stages postinfection, i.e., at a comparably small plaque size. This suggests that the absence of E protein somehow affects the rate or the directionality of cell-to-cell spread of the virus during infection. Elucidating the basis for this unexpected observation will require much further work.
FIG. 2.
Titration of passage 2 stocks obtained from purified tiny plaques from independent infection-transfection experiments. Two dilutions each of the ΔE mutants Alb289 and Alb291 are shown (top, 10−2; bottom, 10−3) to emphasize the atypical morphology of the tiny plaques. Also shown, for comparison, are plaques of Alb240, a wild-type (wt) recombinant derived from pMH54 and fMHV (17), and mock-infected cells (mock). The titers of the viruses were determined on mouse L2 cell monolayers, which were stained with neutral red 75 h postinfection and photographed 19 h later.
Confirmation of the ΔE genotype.
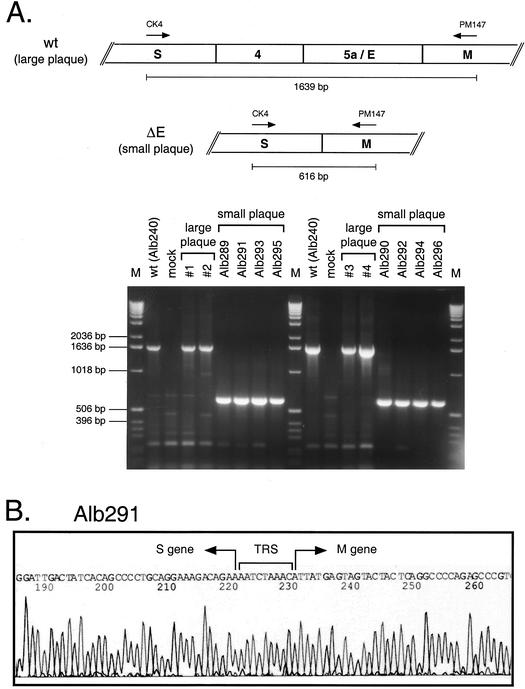
For genotypic analysis, we initially chose eight plaque-purified small-plaque recombinants (designated Alb289 through Alb296), which were collectively derived from four independent infection-transfection experiments. As controls, we used four large-plaque recombinants, which were derived from four independent infection-transfection experiments, and Alb240, a well-characterized wild-type recombinant that had previously been obtained from targeted recombination between fMHV and pMH54 RNAs. Total cellular RNA was isolated from monolayers of 17Cl1 cells infected with each of the recombinants or from mock-infected cells as an additional control. To examine the site of the ΔE deletion in each recombinant, the RNA was reverse transcribed with a random primer and then PCR was carried out with primers CK4 and PM147, which are specific for the regions of the MHV S and M genes, respectively, flanking the deleted genomic segment (Fig. 3A and Table 1). Each of the four independently generated large-plaque recombinants yielded a single product that was identical in size to that of the wild-type control, Alb240, and consistent with the predicted size of 1,639 bp (Fig. 3A). Direct sequencing of these PCR products showed that they corresponded exactly to the sequence of the wild type. Therefore, in accord with our earlier experience in isolating the extremely defective MΔ2 mutant (18), the large-plaque recombinants appeared to be regenerated wild-type viruses resulting from two crossover events during targeted recombination (Fig. 1A), and they were not analyzed further.
FIG. 3.
Analysis of large-plaque and small-plaque progeny from targeted recombination. (A) RNA was isolated from cells infected with plaque-purified recombinants, reverse transcribed with random primer p(N)6, and amplified with primers CK4 and PM147. The PCR products were analyzed by agarose gel electrophoresis. Large plaques (#1 to 4) were purified from independent infection-transfection experiments. The small-plaque recombinants Alb289 through Alb296 represent four independent sets of mutants in which consecutively numbered odd-even pairs are siblings. Lanes M, DNA fragment size markers. wt, wild type; mock, mock-infected cells. (B) Sequence of the S gene-M gene junction in the ΔE mutant Alb291. The junction sequences of Alb289, Alb290, and Alb292 to Alb296 were identical.
By contrast, each of the small-plaque recombinants produced a single PCR product that was much smaller than that of the wild type and was consistent with the size of 616 bp predicted for the ΔE mutant (Fig. 3A). This strongly suggested that the small-plaque recombinants lacked the 1,023-bp region encompassing genes 4, 5a, and E. Direct sequencing of these purified PCR fragments confirmed that all eight of them had the expected sequence for the entire amplified region. In particular, the junction between the S and M genes was exactly as had been constructed in the transcription vector pLK70. A representative sequence, that of the junction in Alb291, is shown in Fig. 3B. Because we were concerned about the possibility that MHV, in order to survive in the absence of the E protein, might require adaptive mutations in the M protein, we sequenced another PCR product, obtained with primers CK4 and PM145 (Table 1), that contained the entire M gene. This revealed that there were no mutations in the M genes of the small-plaque recombinants, except that Alb289 and Alb293 each had a point mutation in the last codon of the M ORF, resulting in the amino acid change T228I. Since the other six small-plaque recombinants had the entire wild-type M gene sequence, this mutation cannot be required for ΔE mutant viability. We have presented evidence elsewhere that the T228I mutation likely results from recombination between the MHV genome (g) RNA and subgenomic (sg) RNA7, the mRNA for N protein (18).
Analyses to rule out a heterologous source or location of the E gene.
It has previously been established by a number of criteria that the interspecies chimeric virus fMHV is incapable of growth in murine cells that lack feline aminopeptidase N, the receptor for FIPV (17). All of the ΔE mutants that we isolated from fMHV-based targeted RNA recombination were purified through two rounds of plaque titration on murine cells to ensure their homogeneity and were then propagated in murine cells, in some cases for as many as seven passages. Nevertheless, it remained conceivable that, if MHV were unable to survive in the absence of E protein, we might then have selected for an fMHV variant that was pseudotyped with MHV S protein that was somehow provided by a copackaged RNA. We tested for this possibility in two ways. First, exhaustive attempts were made to cultivate fMHV from passage 3 or 4 ΔE mutant stocks by serial passaging in feline AK-D cells. We were never able to detect signs of infection caused by ΔE virus, in sharp contrast to fMHV controls, which produced extensive syncytia and cytopathic effect in the same cell line.
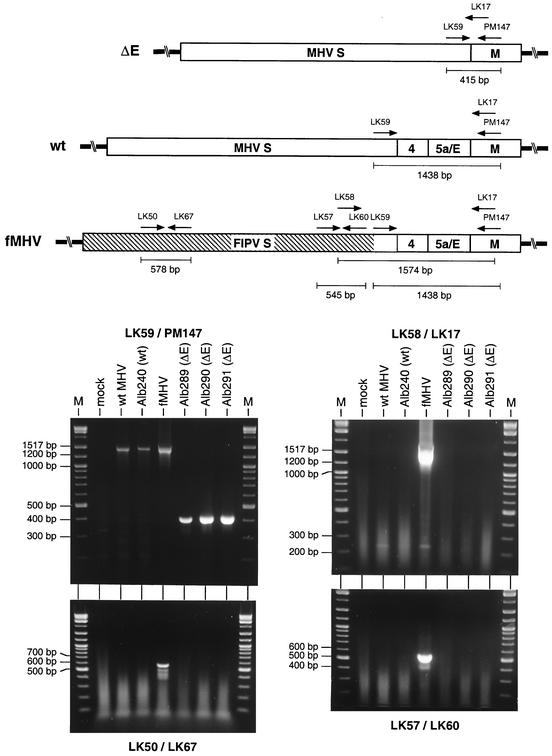
As a further test for the possible presence of fMHV in ΔE virus stocks, total cellular RNA was extracted from murine 17Cl1 monolayers that had been infected with passage 4 stocks of three ΔE mutants, Alb289, Alb290, and Alb291, or as controls, with wild-type MHV or the wild-type recombinant Alb240. Additional controls were provided by RNA isolated from mock-infected 17Cl1 cells and from fMHV-infected AK-D cells. Following random-primed RT, PCR was performed using several specific primer pairs (Table 1). With primer pair LK59-PM147, products of two alternative sizes were detected in the viral samples (Fig. 4). The upstream primer, LK59, resides in the region of the S gene encoding the S protein endodomain, which is common to both MHV and fMHV. For fMHV and the two MHV wild-type controls, we observed a PCR product in agreement with the expected size of 1,438 bp. For all three ΔE mutants, a much smaller product was detected, at a mobility consistent with the predicted size of 415 bp (Fig. 4), and there was no trace of the larger product that had been found with the fMHV or wild-type control. This further validated the previous RT-PCR result (Fig. 3) that had showed the deletion of genes 4, 5a, and E in the ΔE mutants. It also confirmed the integrity of the RNA and RT product for all viral samples, particularly those of the ΔE mutants. PCRs were then carried out with three additional primer pairs. Two of these, LK50-LK67 and LK57-LK60, were specific for sequences mapping entirely within the region encoding the FIPV S ectodomain. The third set paired an FIPV S-specific primer, LK58, with a downstream primer, LK17, located in the M gene. All three primer pairs yielded products only with fMHV, and these were consistent with the expected sizes of 578, 545, and 1,574 bp, respectively (Fig. 4). No signal was obtained with the ΔE mutants or with the wild-type or mock-infected controls. Collectively, these results ruled out the presence of fMHV in the ΔE mutant virus stocks.
FIG. 4.
RT-PCR analysis to rule out the presence of fMHV in purified ΔE recombinants. The random-primed RT product obtained with RNA isolated from infected cells was amplified with primer pairs either unique to fMHV or common to fMHV, wild-type (wt) MHV, and the ΔE mutant. In the genomic schematics at the top, the hatched region indicates the ectodomain-encoding portion of the FIPV S gene; primer positions are not drawn strictly to scale. The PCR products were analyzed by agarose gel electrophoresis. Lanes M, DNA fragment size markers; mock, mock-infected cells.
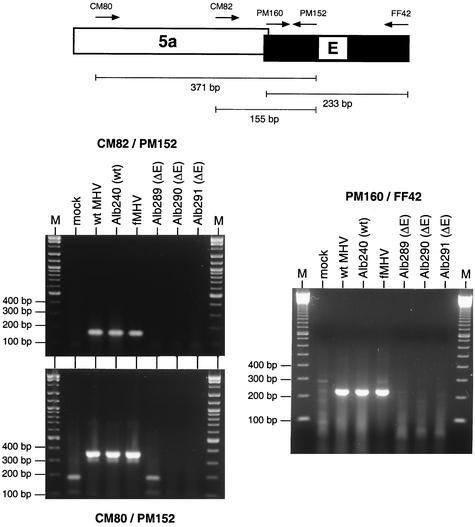
We next tested for any other potential extraneous source of the E gene or for the possible presence of the E gene at an ectopic location in the ΔE mutant genome. To accomplish this, we performed PCR with three different pairs of primers targeted to various regions of genes 5a and E (Fig. 5). Primer pairs CM82-PM152, CM80-PM152, and PM160-FF42 each gave strong, specific products of 155, 371, and 233 bp, respectively, with fMHV and the wild-type MHV controls. However, none of the ΔE mutants produced a detectable signal with these primers. Although a weak nonspecific band of roughly 200 bp can be seen for one of the ΔE mutants, Alb289, with the primer pair CM80-PM152, the same band was also present in the mock-infected control. Several other sets of primers mapping in genes 5a and E, in different combinations, were also used in additional RT-PCR and similarly produced products with the fMHV and wild-type MHV controls but not with any of the ΔE mutants (data not shown). It should be noted that all of these PCRs were carried out with the same ΔE mutant cDNA samples that yielded strong positive signals with the primer pair LK59-PM147 (Fig. 4) and also with an N gene-specific primer pair (data not shown).
FIG. 5.
RT-PCR analysis to rule out the presence of the E gene elsewhere in the genome. The random-primed RT product obtained with RNA isolated from infected cells was amplified with primer pairs internal to the wild-type (wt) E ORF or including the E ORF and the upstream gene 5a. The PCR products were analyzed by agarose gel electrophoresis. Lanes M, DNA fragment size markers; mock, mock-infected cells.
Direct analysis of intracellular RNA.
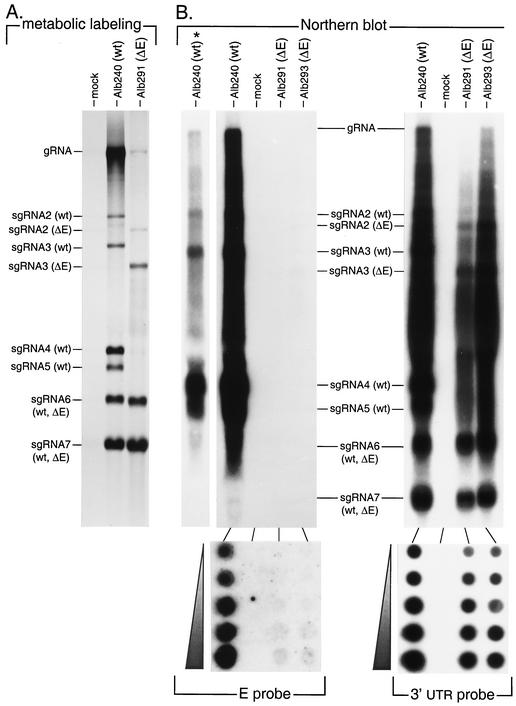
To obtain a more long-range picture of the MHV genome than had been achieved by RT-PCR, we metabolically labeled virus-specific RNA with [33P]orthophosphate in cells infected with the ΔE mutant Alb291 or with the wild-type recombinant Alb240 (Fig. 6A). During infection, MHV-A59 produces a 3′-nested set of six sgRNAs, each of which consists of a 70-nucleotide leader sequence joined at a TRS to all genomic sequence downstream of that TRS (36, 41). Deletion of a given region of the genome will thus affect not only gRNA but also all sgRNAs encompassing that region. The pattern of transcripts for the ΔE mutant, therefore, exactly conformed to that expected to result from the deletion of genes 4, 5a, and E and their two associated TRSs. The ΔE mutant entirely lacked sgRNAs 4 and 5, and sgRNAs 2 and 3 had mobilities consistent with having been shortened by 1 kb with respect to their wild-type counterparts. We would not have expected to be able to detect the size difference between the ΔE mutant gRNA (30 kb) and the wild-type gRNA (31 kb). The gRNA of the ΔE mutant appeared to be much less abundant than that of the wild type; however, this likely reflected variation in recovery of gRNA in this particular sample, since other independent labeling experiments did not show this contrast. As expected, there were no detectable differences in the sizes of mutant and wild-type sgRNAs 6 and 7, which are both governed by TRSs downstream of the deleted region. Notably, in the ΔE mutant the amount of sgRNA6, the mRNA for M protein, was not significantly different from that of its wild-type counterpart. This appears to preclude the possibility that the mutant compensates for the absence of E protein by overexpressing the M protein.
FIG. 6.
Analysis of RNA species synthesized by the ΔE mutant. (A) Virus-specific RNA was labeled with [33P]orthophosphate in the presence of actinomycin D in cells infected with wild-type (wt) recombinant Alb240 or ΔE mutant Alb291 or in mock-infected (mock) control cells. Purified RNA was analyzed in a formaldehyde-agarose gel, as detailed in Materials and Methods. (B) Northern blot analysis of unlabeled RNA isolated from cells infected with wild-type recombinant Alb240 or ΔE mutants Alb291 and Alb293 or from mock-infected control cells. Purified RNA was separated in a formaldehyde-agarose gel and transferred to a nylon filter. Alternatively, RNA was directly dot blotted onto nylon filters; each set of dots corresponds to serial twofold dilutions, starting with 5 μg of total cellular RNA. The RNA was hybridized with a 32P-labeled DNA probe specific either for the E gene or for the 3′ UTR of the MHV genome; bound probe was visualized by fluorography. The Alb240 lane marked with an asterisk is a short exposure of the adjacent, overexposed lane.
To corroborate the conclusion of the RT-PCR analysis in Fig. 5, we used Northern blotting to search for the presence of E gene-related sequences in RNA from ΔE mutant-infected cells. RNA isolated from cells infected with two ΔE mutants (Alb291 and Alb293) and the wild-type recombinant (Alb240) was electrophoretically separated, blotted onto nylon, and hybridized with a labeled probe encompassing almost the entire E ORF (Fig. 6B). As expected, for wild-type RNA the probe detected sgRNA5, the mRNA for E protein, and all species larger than sgRNA5. However, even after prolonged exposure of the blot, no E gene-specific hybridization signal was obtained with RNA from either of the ΔE mutants. As a control, a duplicate blot was hybridized with a labeled probe specific for the 3′ UTR. As expected, this probe hybridized to all viral RNAs in both wild-type and ΔE mutant samples. To guard against the possibility that potential hybridizing material was lost in the Northern blots owing to inefficient transfer of the largest of the MHV RNA species, we also carried out the same hybridizations in dot blots and obtained the same results (Fig. 6B, bottom). Overexposed blots using the E gene-specific probe failed to reveal any hybridizing material in the ΔE mutant RNA samples, while the same samples showed an extent of hybridization to the 3′ UTR probe comparable to that exhibited by wild-type RNA. These results strongly support our conclusion that in the ΔE mutants, the E gene has been deleted from its normal locus and is neither present elsewhere in the genome nor provided by an exogenous source.
DISCUSSION
Coronaviruses, arteriviruses, and toroviruses have been grouped together in the order Nidovirales principally on the basis of similarities in their RNA synthesis mechanisms, polymerase genes, and genome organizations (9). Despite considerable differences among their surface glycoproteins and nucleocapsids, all of these viruses have in common envelopes with a triple-spanning M protein as a major constituent. In addition, all coronaviruses and arteriviruses encode an E protein, and no naturally occurring strains or variants have been found that lack E. By contrast, no homolog for E has yet been identified in the torovirus genome.
Three prior reports have used reverse genetics to explicitly abrogate nidovirus E gene expression. For the arterivirus EAV, Snijder and coworkers (37) eliminated expression of the E gene by creating a point mutation in its start codon. The frugal genomic organization of EAV prevents complete elimination of the E ORF because the latter overlaps extensively with the ORF for an essential surface glycoprotein, GS. In addition, the E start codon shares 2 nucleotides with the stop codon of the upstream polymerase gene, and it is very close to the TRS that governs transcription of the mRNA from which both E and GS are translated. The EAV E knockout mutant RNA was found to be unable to produce infectious progeny virus, whereas there was no unintended effect on expression of either polymerase or GS in initially transfected cells. The authors did note, however, that in a quantitative transfection-infectious center assay, the E knockout mutant had an apparent reversion frequency at least 10-fold greater than those of other EAV start codon point mutants (37). It is conceivable that this higher apparent reversion rate was generated by a low level of replication of the E knockout mutant that occasionally led to breakaway plaque formation by an arising revertant. For the coronavirus TGEV, Curtis and coworkers (5) constructed a virus containing the gene for green fluorescent protein in place of the nonessential gene 3a. This was accomplished through manipulation of a set of six cloned cDNAs spanning the TGEV genome that were then ligated in vitro to form a template for the synthesis of infectious RNA. The authors next deleted the nonessential gene 3b and the first 10 nucleotides of the downstream E ORF, and they were not able to obtain infectious material upon passage of supernatant from cells transfected with this mutant RNA. Similarly, Ortego and coworkers (29) deleted the entirety of the TGEV genes 3a, 3b, and E from a full-length infectious cDNA and found that this resulted in complete loss of ability to recover infectious virus. Each of these TGEV E gene knockout mutants was demonstrated to function as a (noninfectious) RNA replicon that could be packaged in the presence of E protein exogenously provided by an alphavirus vector (5, 29) or by a cell line constitutively expressing E protein (29). In the latter case, TGEV particles produced by such complementation appeared to follow the same assembly pathway and exhibited the same morphology as the wild-type virus.
The discrepancy between our successful isolation of a viable E gene deletion mutant of MHV and the apparent lethality of E gene knockouts in EAV and TGEV may indicate basic differences in assembly dynamics among various nidoviruses. Alternatively, it may reflect the relative robustness of MHV growth and plaque formation in tissue culture. With a more defective MHV mutant, the truncated M protein mutant MΔ2, we have previously noted that in one cell line the kinetics of viral infection were sufficiently slow that monolayers were always overtaken by the growth of uninfected cells (18). It is possible that a similar phenomenon may obscure detection of ΔE mutants of some nidoviruses.
Our results show that the MHV ΔE mutant produces viable, independently replicating virus, despite lacking the entire E ORF. As yet, we have not been able to formally demonstrate the absence of E protein in the ΔE mutant by immunological criteria. Antibodies raised against the E protein of either the Utrecht laboratory strain of MHV-A59 (30) or MHV-JHM (45), in our hands, fail to detect E protein in cells infected with our laboratory strain of MHV-A59. It has been shown that this loss of recognition, in the case of anti-E (MHV-A59; Utrecht), is caused by a mere 2-amino-acid-residue difference between the Utrecht and Albany MHV-A59 E proteins (8); in the case of anti-E (MHV-JHM), similar strain-specific E protein amino acid differences may pertain. Nevertheless, by much more sensitive analyses using RT-PCR, RNA metabolic labeling, and Northern blotting, we have established that the E gene has been deleted from its normal position in the genome of the ΔE mutant, that it is not present at some ectopic genomic locus, and that it is not provided by some epigenetic mechanism in infected cells.
Multiple independent isolates of ΔE gave rise to small plaques having the same atypical morphology, the basis for which is currently unclear. Stocks of all ΔE isolates had low titers, suggesting that virion assembly in the mutant occurs with orders of magnitude lower efficiency than in the wild type. We also found that the ΔE mutant was stable upon repeated passaging of viral stocks, unlike extremely defective mutants of M protein (18) and N protein (unpublished results), for which wild-type-like revertants overrun higher-passage stocks. This likely indicates that there are no single second-site point mutations that can compensate for the loss of the entire E gene. It must be noted that the deletion in the ΔE mutant also removed the nonessential genes 4 and 5a. It has previously been shown that the deletion of genes 4 and 5a produced no detectable change in viral plaque size or morphology compared to the wild type and had only a minimal effect on viral growth kinetics in tissue culture (8). Thus, we believe that the observed phenotype of the ΔE mutant is due almost entirely to the loss of the E protein. Moreover, the tissue culture phenotype of the Δ45a mutant (8) may also be attributable to alteration of E protein expression, since gene 5a is thought to function as an internal ribosome entry site for translation of E (14, 39).
The dispensability of the E protein, at least for MHV, leads us to conclude that E protein greatly enhances virion envelope formation but is not essential for this process to proceed. Results from some previous studies suggested that the E protein acts independently of other viral components. It has been found that the expression of the MHV or IBV E protein, in the absence of other viral proteins, results in vesicles that are exported from cells (3, 22). Expression of the MHV E protein alone has also been shown to induce the formation of clusters of convoluted membranous structures highly similar to those seen in coronavirus-infected cells (6, 30). This may mean that the principal role of E is to induce membrane curvature in the budding compartment and that M-M monomer interactions drive the remainder of virion morphogenesis. By contrast, other experimental results implied that E must specifically act in concert with M. In particular, it was found that, although both TGEV and BCoV VLPs are efficiently generated by coexpression of homologous constituents, VLPs cannot be produced by expression of the M protein of TGEV with the E protein of BCoV. Conversely, VLPs cannot be produced by expression of the M protein of BCoV with the E protein of TGEV (1). This seems to indicate that virion envelope maturation is promoted by a direct interaction between M and E instead of, or in addition to, the effect of E on intracellular membranes.
One of the most pressing questions raised by the existence of the ΔE mutant is what do its virions look like? We are eager to learn the answer to this, but we have been thwarted so far by the low titers of the ΔE mutant. Similarly, we are curious whether the ΔE mutant buds at the same intracellular site as the wild type or whether it exploits some alternative pathway. Preliminary results from immunofluorescence experiments suggest that there is no significant difference in the intracellular localization of M protein in cells infected with the ΔE mutant and in those infected with the wild type (data not shown), but higher-resolution analyses will be required to resolve this question. We expect that further characterization of ΔE viruses will help to shed light on the functions of the E protein in wild-type MHV assembly.
Acknowledgments
We are grateful to Kelley Hurst for expert technical assistance. We thank Matthew Shudt, Heather Berry, Jolene Wilson, and Tim Moran of the Molecular Genetics Core Facility of the Wadsworth Center for oligonucleotide synthesis and for DNA sequencing, and we deeply regret the untimely loss of our friend and colleague Tim Moran.
This work was supported in part by Public Health Service grant AI 39544 from the National Institutes of Health.
REFERENCES
- 1.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos, E. C. W., W. Luytjes, H. van der Meulen, H. K. Koerten, and W. J. M. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi and direct release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 76:1273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David-Ferreira, J. F., and R. A. Manaker. 1965. An electron microscope study of the development of a mouse hepatitis virus in tissue culture cells. J. Cell Biol. 24:57-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Haan, C. A. M., L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72:6838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haan, C. A. M., P. S. Masters, X. Shen, S. Weiss, and P. J. M. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organization of the nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godet, M., R. L'haridon, J.-F. Vautherot, and H. Laude. 1992. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology 188:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jendrach, M., V. Thiel, and S. Siddell. 1999. Characterization of an internal ribosome entry site within mRNA5 of murine hepatitis virus. Arch. Virol. 144:921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koetzner, C. A., M. M. Parker, C. S. Ricard, L. S. Sturman, and P. S. Masters. 1992. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 66:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krijnse Locker, J., M. Ericsson, P. J. M. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laude, H., and P. S. Masters. 1995. The coronavirus nucleocapsid protein, p. 141-163. In S. G. Siddell (ed.), “The Coronaviridae”. Plenum Press, New York, N.Y.
- 21.Liu, D. X., and S. C. Inglis. 1991. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology 185:911-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, J., A. Maeda, and S. Makino. 1999. Release of E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda, J., J. F. Repass, A. Maeda, and S. Makino. 2001. Membrane topology of coronavirus E protein. Virology 281:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masters, P. S. 1999. Reverse genetics of the largest RNA viruses. Adv. Virus Res. 53:245-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masters, P. S., C. A. Koetzner, C. A. Kerr, and Y. Heo. 1994. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 68:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayanan, K., and S. Makino. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 29.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raamsman, M. J. B., J. Krijnse Locker, A. de Hooge, A. A. F. de Vries, G. Griffiths, H. Vennema, and P. J. M. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Risco, C., I. M. Antón, L. Enjuanes, and J. L. Carrascosa. 1996. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 70:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Risco, C., M. Muntión, L. Enjuanes, and J. L. Carrascosa. 1998. Two types of virus-related particles are found during transmissible gastroenteritis virus morphogenesis. J. Virol. 72:4022-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rottier, P. J. M. 1995. The coronavirus membrane glycoprotein, p. 115-139. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 34.Salanueva, I. J., J. L. Carrascosa, and C. Risco. 1999. Structural maturation of the transmissible gastroenteritis coronavirus. J. Virol. 73:7952-7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440: 215-219. [DOI] [PubMed] [Google Scholar]
- 37.Snijder, E., H. van Tol, K. W. Pedersen, M. J. B. Raamsman, and A. A. F. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 73:6335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiel, V., and S. G. Siddell. 1994. Internal ribosome entry in the coding region of murine hepatitis virus mRNA5. J. Gen. Virol. 75:3041-3046. [DOI] [PubMed] [Google Scholar]
- 40.Tooze, S. A., J. Tooze, and G. Warren. 1988. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J. Cell Biol. 106:1475-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Most, R. G., and W. J. M. Spaan. 1995. Coronavirus replication, transcription, and RNA recombination, p. 11-31. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 42.Vennema, H., G.-J. Godeke, J. W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D.-J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, S. R., P. W. Zoltick, and J. L. Leibowitz. 1993. The ns 4 gene of mouse hepatitis virus (MHV), strain A59 contains two ORFs and thus differs from ns 4 of the JHM and S strains. Arch. Virol. 129:301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokomori, K., and M. M. C. Lai. 1991. Mouse hepatitis virus S RNA sequence reveals that nonstructural proteins ns4 and ns5a are not essential for murine coronavirus replication. J. Virol. 65:5605-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu, X., W. Bi, S. R. Weiss, and J. L. Leibowitz. 1994. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology 202:1018-1023. [DOI] [PubMed] [Google Scholar]