Abstract
Here we provide evidence that EBNA2 is methylated in vivo and that methylation of EBNA2 is a prerequisite for binding to SMN. We present SMN as a novel binding partner of EBNA2 by showing that EBNA2 colocalizes with SMN in nuclear gems and that both proteins can be coimmunoprecipitated from cellular extract. Furthermore, in vitro methylation of either wild-type EBNA2 or a glutathione S-transferase-EBNA2 fusion protein encompassing the arginine-glycine (RG) repeat element is necessary for in vitro binding to the Tudor domain of SMN. The recently shown functional cooperation of SMN and EBNA2 in transcriptional activation and the previous observation of a severely reduced transformation potential yet strongly enhanced transcriptional activity of an EBNA2 mutant lacking the RG repeat indicate that binding of SMN to EBNA2 is a critical step in B-cell transformation by Epstein-Barr virus.
The Epstein-Barr virus (EBV) causes infectious mononucleosis and is associated with a variety of human tumors (reviewed in reference 25). The virus transforms B cells into continuously growing lymphoblastoid cell lines by expressing 12 genes (reviewed in reference 1). The nuclear antigen EBNA2 is the first viral gene expressed after infection and is essential for EBV-mediated transformation through the activation of viral and cellular genes like the viral EBNAs and the latent membrane proteins LMP1, LMP2A, and LMP2B as well as the cellular CD21, CD23, c-fgr, and c-myc (reviewed in reference 13) and AML-2 genes (26). EBNA2 does not bind directly but is tethered to promoters by interacting with cellular transcription factors like RBPJκ, Sp-1/Spi-B, hnRNP-D/AUF1, or ATF/CRE. Domains of EBNA2 critical for transformation of B cells and for activation of gene expression had been identified previously through mutational analysis (6, 28, 29). EBNA2 binds to RBPJκ with its conserved WWP325 (18) motif, the deletion of which results in a severe reduction of activation of the LMP1 promoter and a complete loss of transforming capacity (6). An adjacent arginine-glycine (RG) repeat element between amino acids (aa) 337 and 354 of EBNA2 was shown elsewhere to be critical but not essential for B-cell transformation in vitro, but the deletion of aa 337 to 354 increased the potential of EBNA2 to activate the LMP1 promoter (29). While most interacting partners bind to the C terminus of EBNA2, we had recently demonstrated that EBNA2 interacts through its N-terminal aa 121 to 216 with a putative helicase/ATPase termed DP103 (12), which in turn binds to SMN, the product of the spinal muscular atrophy gene (SMN) (4, 5, 30). The survival of motor neurons (SMN) gene is lost or mutated in spinal muscular atrophy (SMA) (17). SMN is found in different macromolecular complexes, and one of these has recently been shown to facilitate the assembly of spliceosomal U snRNPs by mediating the attachment of the Sm proteins onto snRNAs U1, U2, U4, and U5 (7, 20, 22). The function of the SMN complex involves direct binding of Sm proteins to SMN, an interaction that is strongly enhanced by modification of arginines in Sm proteins B/B′, D1, and D3 to symmetrical dimethylarginines (sDMAs) (2, 3, 9, 21). This modification is catalyzed by the PRMT5 complex (also termed the methylosome), which consists of the PRMT5 methyltransferase and the WD45 and pICln proteins (10, 21). In addition to its role in RNA metabolism, SMN appears to be also involved in transcriptional regulation (23, 27). It has previously been shown that SMN cooperates with EBNA2 in the activation of the viral LMP1 promoter (30).
EBNA2 binds to SMN in cell extracts.
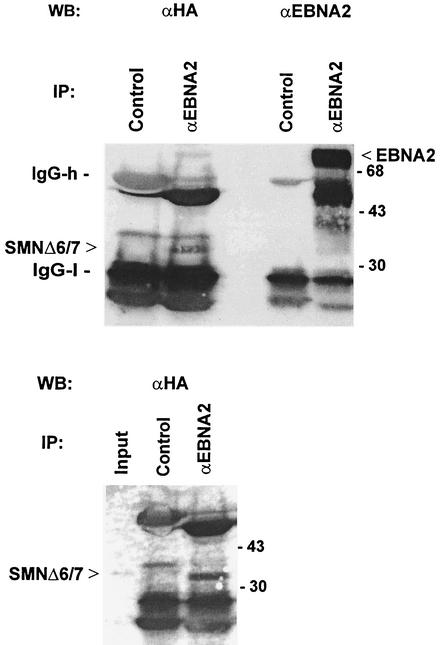
To test whether EBNA2 directly binds to SMN, we performed coimmunoprecipitation studies of EBV-transformed Raji cells with EBNA2-specific monoclonal antibody (MAb) 1E6 (14). Under the conditions employed, we could not observe coprecipitation of SMN and EBNA2, probably because only a small subfraction of EBNA2 is present in nuclear gems while a major part of EBNA2 is localized in the nucleoplasm (11, 24), where SMN is not abundant. However, our previous study had indicated that the deletion of exons 6 and 7 (SMNΔ6/7) resulted in its redistribution to the nucleoplasm, where it strongly colocalized with EBNA2. The SMNΔ6/7 mutant also showed an increased coactivation potential, probably because binding to DP103 was inhibited (30). We therefore cotransfected EBNA2wt and hemagglutinin (HA)-tagged SMNΔ6/7 in HeLa cells and precipitated EBNA2 with specific MAb 1E6 (14) and nonspecific rat MAbs as the negative control. The bound proteins were subsequently analyzed by Western blotting with either anti-HA MAb to visualize SMNΔ6/7 or EBNA2-specific MAb R3 (14). As shown in Fig. 1, SMNΔ6/7 was coprecipitated with EBNA2. In contrast, we did not observe a signal of HA-tagged SMNΔ6/7 when control MAb was used. Precipitation of tagged SMNΔ6/7 could also be achieved by using HA-specific antibody but not the control MAb; the precipitated SMNΔ6/7 migrated to the same position as did the protein from the whole-cell extract. Likewise, the EBNA2 from the transfected cells comigrated with EBNA2 from B95.8 cell extract (data not shown). Taken together, these experiments suggest that SMN interacts with EBNA2 in vivo.
FIG. 1.
EBNA2 binds to SMN in vivo. (A) Coimmunoprecipitation of EBNA2 and SMN from transfected HeLa cell extract. HeLa cells were transiently transfected by the calcium phosphate method with pSG5-EBNA2wt and pSG5-SMNΔ6/7 (30) expressing full-length EBNA2 and HA-tagged SMN with a deletion of aa 242 to 293 encoded by exons 6 and 7 of SMN. The cell extract was bound to protein G-Sepharose (Pharmacia) preadsorbed with nonspecific MAb 5A10 (lanes designated “Control”) or the EBNA2-specific MAb 1E6 (14) (lanes designated “αEBNA2”). The immune complexes were separated by SDS-10% PAGE, transferred to a nitrocellulose membrane, and stained with HA-specific MAb 3F10 or EBNA2-specific MAb R3 (14). The positions of the precipitated EBNA2, SMNΔ6/7, and immunoglobulin G heavy (“IgG-h”) and light (“IgG-l”) chains as well as the position of coelectrophoresed molecular mass marker proteins (103 kDa) are indicated by arrows or bars. In the lower panel, the position of the input SMNΔ6/7 protein (lane designated “Input”) is shown as a reference for the precipitated SMNΔ6/7 protein. The input represents about 3% of the total amount used for precipitation. WB, Western blot; IP, immunoprecipitation.
EBNA2 is methylated in vivo.
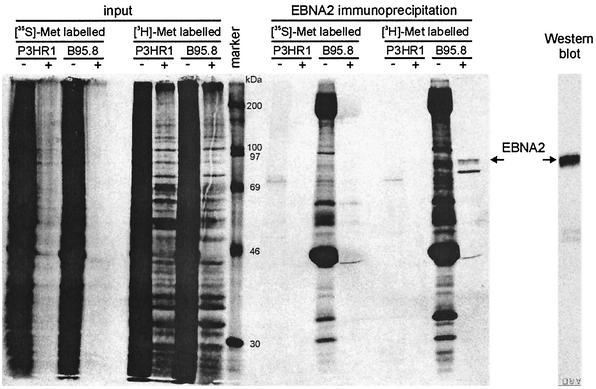
In vitro binding studies had shown that SMN preferentially binds to proteins that contain sDMAs. We therefore analyzed whether the interaction of EBNA2 and SMN was likewise influenced by this type of posttranslational modification. First, we tested in vivo whether EBNA2 can acquire methylated residues. The methylation state of EBNA2 was determined in vivo by metabolic labeling of EBNA2 in EBV-transformed B95.8 cells by using l-[methyl-3H]methionine as a donor for methyl groups essentially as described previously (15, 19). The EBV-positive, EBNA2-deficient P3HR1 cell line served as a negative control. De novo protein synthesis was inhibited by cycloheximide and chloramphenicol to exclude metabolic incorporation of 3H-labeled methionine into EBNA2. EBNA2 was immunoprecipitated from cell extracts with MAb R3 (14) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography. As shown in Fig. 2, de novo protein synthesis was efficiently blocked in both cell lines (lanes designated “[35S]-Met”) by cycloheximide-chloramphenicol (lanes designated “+”) while incorporation of 3H-methyl groups was still observed (lanes designated “[3H]-Met”). Correspondingly, no 35S-labeled EBNA2 protein could be immunoprecipitated. In contrast, an EBNA2-specific 3H-labeled band was clearly precipitated from the [3H]methionine-labeled cell extract, demonstrating incorporation of 3H-methyl groups into EBNA2. We noted an additional band of a 3H-labeled protein in the lane with the precipitated EBNA2 protein. In the control experiment the results of which are shown in the rightmost lane in Fig. 2, the efficient precipitation of EBNA2 from B95.8 cell extract was demonstrated in a Western blot analysis; under the conditions employed, we observed no degradation of EBNA2; we therefore assume that the additional band did not represent degraded EBNA2 protein but was probably derived from a coprecipitated, methylated protein. In conclusion, the experiments show that EBNA2 is methylated in vivo.
FIG. 2.
In vivo methylation of EBNA2. B95.8 cells (EBNA2 positive) and P3HR1-1 cells (EBNA2 negative) were labeled in vivo with either [35S]methionine or [3H-methyl]methionine in the presence (+) or absence (−) of the protein synthesis inhibitors cycloheximide and chloramphenicol. The cell extracts were subjected to immunoprecipitation with the EBNA2-specific MAb R3 (14) and analyzed by SDS-PAGE and autoradiography (lanes designated “EBNA2 immunoprecipitation”). The unprecipitated cell extracts (lanes designated “input”) were also analyzed to demonstrate that protein de novo synthesis was efficiently inhibited. The rightmost lane, designated “Western blot,” shows that MAb R3 efficiently precipitated the EBNA2 protein. Arrows indicate the positions of the precipitated EBNA2 protein; the lane designated “marker” shows the positions of coelectrophoresed 14C-labeled molecular mass marker proteins (Amersham).
Methylation of EBNA2 at its RG repeat is a prerequisite for binding to SMN in vitro.
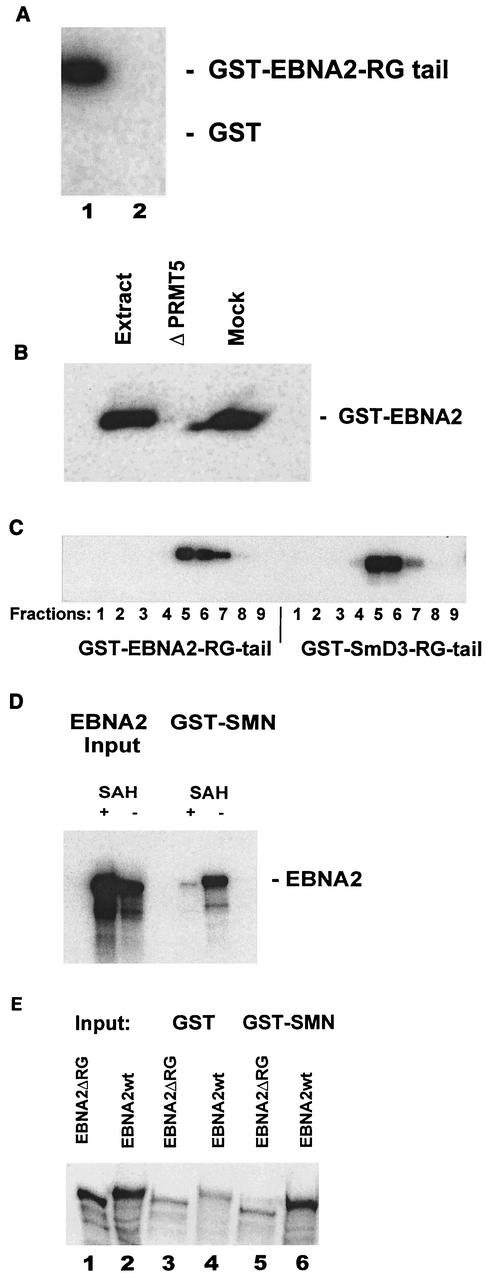
Next, the in vitro binding of EBNA2 to SMN was analyzed. In particular, we asked whether methylation of EBNA2 was a prerequisite for binding. In a first experiment, we assayed the methylation of a glutathione S-transferase (GST)-EBNA2 fusion protein encoding aa 301 to 400 (GST-EBNA2-tail), encompassing the arginine-glycine repeat of EBNA2 at aa 337 to 354, by using a HeLa cytosolic extract which contains the PRMT5 complex. This complex had previously been demonstrated to catalyze sDMA modifications in Sm proteins that contain RG repeat motifs (9, 21). As shown in Fig. 3A, the extract contained an activity which efficiently transferred methyl groups from 3H-labeled S-adenosylmethionine to the GST-EBNA2 tail but not to the GST protein (Fig. 3A, lanes 1 and 2, respectively). To obtain further evidence that indeed the PRMT5 complex was responsible for methylation of the GST-EBNA2 substrate, the HeLa cytosolic extract was either untreated, depleted with a PRMT5-specific polyclonal antibody, or treated with a nonspecific control antiserum (Fig. 3B, lanes designated “Extract,” “ΔPRMT5,” and “Mock,” respectively) as described previously (21). Clearly, the depleted extract lost most of its methyltransferase activity, while the mock-depleted extract was still active. To further support the findings shown above, the HeLa cell extract was separated by gel filtration and individual fractions were tested for methylation of EBNA2. The GST-EBNA2 fusion protein was incubated with the PRMT5-containing fractions obtained from gel filtration, which were assayed in parallel with GST-SmD3 fusion protein, a known substrate for this enzyme (21). As can be seen in Fig. 3C, the fractions containing the PRMT5 methyltransferase that efficiently methylated the GST-SmD3 fusion protein also showed labeling of the GST-EBNA2 protein. These experiments indicate that the RG-rich region of EBNA2 can be methylated by the PRMT5 methyltransferase complex in vitro. We then tested whether EBNA2, when translated in vitro under conditions that either allowed or inhibited methylation, would bind to SMN. The commercially available in vitro transcription-translation extracts contain methyltransferases including the PRMT5 complex which efficiently methylate proteins cotranslationally (21). To generate methylated and unmethylated EBNA2, full-length EBNA2 was translated in vitro with [35S]methionine as a label in the presence or absence of S-adenosyl-homocysteine (SAH), an inhibitor of protein methylation. As shown in Fig. 3D, EBNA2 was efficiently synthesized under either condition (lanes designated “EBNA2 Input”). We then tested the binding of the EBNA2 proteins to GST-SMN protein immobilized on glutathione beads. As shown in the right part of Fig. 3D (lanes designated “GST-SMN”), only the methylated EBNA2 was retained by the beads (lane designated “− SAH”) while unmethylated EBNA2 did not bind (lane designated “+ SAH”). In the experiments the results of which are shown, a GST-SMN fusion protein containing aa 1 to 150 of SMN was employed. A fusion protein containing aa 90 to 150, which encompasses just the Tudor domain of SMN, yielded the same results (data not shown), indicating that the Tudor domain of SMN is responsible for binding to EBNA2. As previous experiments had shown that SmD3 is methylated on its arginine-glycine (RG) repeat element (9, 21), we tested the EBNA2ΔRG mutant, which features a deletion of the RG repeat (29), for methylation and subsequently for binding to SMN (see below). As shown in Fig. 3E, both EBNA2ΔRG and EBNA2wt were efficiently labeled to the same extent during in vitro translation in the presence of [35S]methionine (lanes 1 and 2, respectively, designated “Input”). The proteins were then applied to glutathione beads containing either GST alone (lanes 3 and 4) or GST-SMN fusion protein (lanes 5 and 6). As can be seen, the GST-SMN beads retained the EBNA2wt (lane 6) protein but significantly less of the EBNA2ΔRG mutant (lane 5). The lanes designated GST show the amount of EBNA2wt or EBNA2ΔRG that was nonspecifically retained under the conditions employed.
FIG. 3.
In vitro methylated EBNA2 binds to SMN. (A) Methylation of EBNA2 by the PRMT5 complex. GST-EBNA2 (aa 301 to 400, “GST-EBNA2-tail”) (lane 1) or GST-wt (lane 2) bound to a column was incubated with a HeLa cytosolic extract in the presence of S-[3H-methyl]adenosylmethionine as a donor for methyl groups The labeled proteins were separated by SDS-PAGE, and the labeled proteins were visualized by autoradiography as described previously (21). (B) Depletion of the PRMT5 complex reduces methyl incorporation into GST-EBNA2-tail. The HeLa cell extract was either untreated (lane designated “Extract”), mock treated (lane designated “Mock”), or depleted of PRMT5 (lane designated “ΔPRMT5”) by nonspecific or PRMT5-specific rabbit serum, respectively (21). (C) Methylation of GST-EBNA2 (aa 300 to 400) by using partially purified PRMT5 complex. HeLa cell cytosolic extract was purified by gel filtration, and the fractions obtained were incubated with either GST-EBNA2 or GST-SmD3 and analyzed as described for panel A. (D) Binding of methylated and unmethylated EBNA2 to SMN. EBNA2 was in vitro transcribed-translated (lanes designated “EBNA2 Input”) by using [35S]methionine to label the newly synthesized EBNA2 protein in the presence (lanes designated “+SAH”) or absence (lanes designated “−SAH”) of SAH, a known inhibitor of methylation. The labeled EBNA2 proteins were then passed through a column containing immobilized GST-SMN (aa 1 to 150) (lanes designated “GST-SMN”). The inputs as well as the protein retained by the column were analyzed by SDS-PAGE and fluorography as described previously (21). (E) EBNA2 with a deletion of the RG repeat (EBNA2ΔRG) does not bind to SMN. EBNA2wt and EBNA2ΔRG were in vitro transcribed-translated in the presence of [35S]methionine (lanes designated “Input”). The labeled proteins were then passed through a column that contained either only GST or GST-SMN (aa 1 to 150). Bound proteins were visualized by SDS-PAGE and fluorography.
Although we have no formal proof that EBNA2 indeed contains sDMAs, several observations strongly suggest that EBNA2 is modified in this manner: (i) the same fractions obtained by purification through gel filtration that generated sDMAs on the known target SmD3 also transferred methyl groups to EBNA2 and (ii) depletion of a HeLa cell cytosolic extract with antibodies specific for the PRMT5 complex efficiently removed the type II (i.e., sDMA-generating) methyltransferase activity for EBNA2. So far, up to six different protein arginine methyltransferases have been detected (8). Of those, only the PRMT5 complex has been shown to generate sDMAs (21); the deletion of the RG repeat within EBNA2 results in its inability to bind to SMN in vitro. In summary, the in vitro binding assays strongly suggest that binding to the Tudor domain of SMN is mediated through the methylated RG repeat of EBNA2.
SMN costimulates EBNA2ΔRG in the activation of the LMP1 promoter.
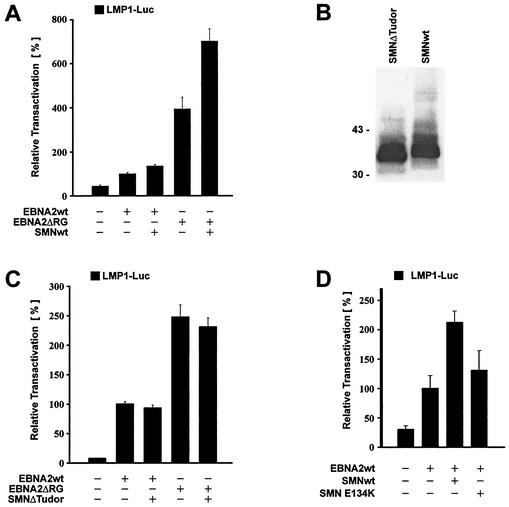
It was previously demonstrated that the deletion of the arginine-glycine repeat of EBNA2 (EBNA2ΔRG) resulted in an approximately four- to fivefold increase in the activation of the viral LMP1 promoter; this mutant, however, exhibited a severely reduced transforming potential (29). When we compared EBNA2ΔRG with EBNA2wt by using the LMP1 reporter (16, 30), we also observed a fourfold stimulation as shown in Fig. 4A. In this experimental setting, we found an increase in activity when both proteins were coexpressed with SMNwt. In particular, the EBNA2ΔRG mutant was further activated by approximately 1.5-fold through coexpression of SMN. This indicates that SMN can cooperate with EBNA2 in the activation of the LMP1 promoter regardless of the presence of the RG repeats.
FIG. 4.
Activation of LMP1 promoter by combinations of EBNA2 and SMN mutants (A) Coactivation of the LMP1 promoter by SMN and EBNA2ΔRG. The EBNA2wt and EBNA2ΔRG expression vectors were transfected with or without the SMNwt expression plasmid and tested for activation of the LMP1 promoter (−327/+40) in BJAB cells as described in the work of Voss et al. (30). (B) Expression of HA-tagged SMNwt and an HA-tagged SMN expression construct with a deletion of the Tudor domain (SMNΔTudor) in HeLa cells after transient transfection, separation by SDS-10% PAGE, and detection with HA-specific MAb 3F10. Numbers at left are molecular masses in kilodaltons. (C) SMNΔTudor with a deletion of the Tudor domain fails to coactivate the LMP1 promoter. The SMNΔTudor mutant was expressed with EBNA2wt or EBNA2ΔRG in BJAB cells and tested for activation of the LMP1 promoter construct. (D) No significant coactivation of the LMP1 promoter by EBNA2 and an SMA patient-derived point mutant in the Tudor domain of SMN (SMN E134K). The EBNA2wt expression vector was transfected with SMNwt or SMN E134K and tested for activation of the LMP1 promoter in BJAB cells. The experiments the results of which are shown in panels A and C were carried out five times each in duplicate; the experiments the results of which are shown in panel D were carried out three times in duplicate.
The deletion of its Tudor domain eliminates the coactivation potential of SMN.
The in vitro binding studies had shown that the binding site for EBNA2 on SMN was located within the Tudor domain encompassing aa 90 to 140 of SMN. We therefore generated an HA-tagged SMN expression construct with a deletion of aa 90 to 140 (SMNΔTudor) to be tested for its ability to costimulate EBNA2wt and EBNA2ΔRG. The expression of SMNwt and the SMNΔTudor mutant is shown in Fig. 4B. The cotransfection of this mutant together with EBNA2wt or the SMN-EBNA2ΔRG mutant did not result in a significant increase in stimulation of the LMP1 promoter compared to either EBNA2wt or EBNA2ΔRG alone (Fig. 4C). These data provide further evidence for a functional cooperation and binding of EBNA2 to SMN, which is mediated by the Tudor domain of SMN. To analyze the importance of the Tudor domain for binding to EBNA2 in more detail, we tested the coactivation of EBNA2 by the E134K point mutant of SMN derived from a patient with SMA (31). As can be seen in Fig. 4D, the point mutant showed only a small coactivation of the LMP1 promoter. A statistical analysis (by t test) revealed that the increase was not significant. We conclude that even a small perturbation within the Tudor domain that should otherwise leave the three-dimensional structure of the protein intact affects the coactivation potential of SMN.
We also analyzed the colocalization of the EBNA2ΔRG and the SMN protein and, conversely, of EBNA2wt and SMNΔTudor to determine whether the deletion of the contact domain for SMN on EBNA2 and vice versa resulted in a different subnuclear localization and/or association with SMN. The proteins clearly showed an overlap in nuclear gems as described previously (30). The deletion of the RG repeat did not result in an altered subnuclear localization of the EBNA2 or the SMN protein; the merged images likewise did not reveal any differences from the wild-type proteins. In the experiment involving EBNA2wt and SMNΔTudor, we also did not observe a grossly altered subnuclear localization of either protein. However, we observed only a partial overlap of the EBNA2wt-SMNΔTudor combination in the gems (data not shown), supporting the results of the reporter gene assays described above.
In light of our previous observation that the coexpression of SMN leads to an increase in the activation of the LMP1 promoter by EBNA2, we were not surprised by the observation that the non-EBNA2-binding SMNΔΤudor protein was inactive in all settings, failing to coactivate EBNA2 or EBNA2ΔRG. However, we were puzzled by the fact that the expression of SMN with EBNA2ΔRG still resulted in a highly reproducible 1.5-fold increase in stimulation over expression with EBNA2ΔRG alone. One possible explanation for the unexpected costimulation of EBNA2ΔRG by SMN might be (i) that there was residual binding of SMNwt to EBNA2ΔRG not revealed by the in vitro assays and (ii) that the RG motif of EBNA2 might otherwise be targeted by another, negatively regulating factor(s) which is now unable to bind and repress EBNA2 or (iii) that the RG region constitutes only part of a larger binding region of EBNA2 for SMN while the Tudor domain, in contrast, appears to represent the complete binding region for EBNA2 on SMN. The mechanism of the EBNA2-SMN cooperation remains enigmatic. Nevertheless, the interaction of EBNA2 with SMN has strong implications for the transformation by EBNA2. The elucidation of the mechanism of the EBNA2-SMN cooperation will shed new light on the transformation by EBV and might also reveal novel aspects of the function of SMN in addition to its role in RNA processing.
Acknowledgments
We thank Ruth Nord and Maria Kemele for expert technical assistance. The expression vectors pSG5-EBNA2wt and pSG5-EBNA2ΔRG were kindly provided by E. Kieff, Boston, Mass.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG), grant Gr950/9-1 to F.A.G.
REFERENCES
- 1.Bornkamm, G. W., and W. Hammerschmidt. 2001. Molecular virology of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:437-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brahms, H., L. Meheus, V. de Brabandere, U. Fischer, and R. Luhrmann. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branscombe, T. L., A. Frankel, J. H. Lee, J. R. Cook, Z. Yang, S. Pestka, and S. Clarke. 2001. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276:32971-32976. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, L., C. M. D. Hunter, P. Mohagheg, J. M. Tinsley, M. A. Brasch, and K. E. Davies. 2000. Direct interaction of SMN with dp103, a putative RNA helicase: a role for SMN in transcription regulation? Hum. Mol. Genet. 9:1093-1100. [DOI] [PubMed] [Google Scholar]
- 5.Charroux, B., L. Pellizzoni, R. A. Perkinson, A. Shevchenko, M. Mann, and G. Dreyfuss. 1999. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 147:1181-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, J. I., F. Wang, and E. Kieff. 1991. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J. Virol. 65:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer, U., Q. Liu, and G. Dreyfuss. 1997. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90:1023-1029. [DOI] [PubMed] [Google Scholar]
- 8.Frankel, A., N. Yadav, J. Lee, T. L. Branscombe, S. Clarke, and M. T. Bedford. 2002. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J. Biol. Chem. 277:3537-3543. [DOI] [PubMed] [Google Scholar]
- 9.Friesen, W. J., S. Massenet, S. Paushkin, A. Wyce, and G. Dreyfuss. 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7:1111-1117. [DOI] [PubMed] [Google Scholar]
- 10.Friesen, W. J., S. Paushkin, A. Wyce, S. Massenet, G. S. Pesiridis, G. Van Duyne, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grässer, F. A., P. Haiss, S. Göttel, and N. Mueller-Lantzsch. 1991. Biochemical characterization of Epstein-Barr virus nuclear antigen 2A. J. Virol. 65:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grässer. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136-19144. [DOI] [PubMed] [Google Scholar]
- 13.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 14.Kremmer, E., B. Kranz, A. Hille, K. Klein, M. Eulitz, G. Hoffmann-Fezer, W. Feiden, K. Herrmann, H.-J. Delecluse, G. Delsol, G. W. Bornkamm, N. Mueller-Lantzsch, and F. A. Grässer. 1995. Rat monoclonal antibodies differentiating between the Epstein-Barr virus nuclear antigens 2A (EBNA2A) and 2B (EBNA2B). Virology 208:336-342. [DOI] [PubMed] [Google Scholar]
- 15.Kzhyshkowska, J., H. Schutt, M. Liss, E. Kremmer, R. Stauber, H. Wolf, and T. Dobner. 2001. Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem. J. 358:305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laux, G., F. Dugrillon, C. Eckert, B. Adam, U. Zimber Strobl, and G. W. Bornkamm. 1994. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J. Virol. 68:6947-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefebvre, S., L. Burglen, S. Reboullet, O. Clermont, P. Burlet, L. Viollet, B. Benichou, C. Cruaud, P. Millasseau, M. Zeviani, et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155-165. [DOI] [PubMed] [Google Scholar]
- 18.Ling, P. D., J. J. Ryon, and S. D. Hayward. 1993. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J. Virol. 67:2990-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Q., and G. Dreyfuss. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 15:2800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meister, G., D. Buhler, R. Pillai, F. Lottspeich, and U. Fischer. 2001. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3:945-949. [DOI] [PubMed] [Google Scholar]
- 21.Meister, G., C. Eggert, D. Buhler, H. Brahms, C. Kambach, and U. Fischer. 2001. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11:1990-1994. [DOI] [PubMed] [Google Scholar]
- 22.Pellizzoni, L., B. Charroux, and G. Dreyfuss. 1999. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl. Acad. Sci. USA 96:11167-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellizzoni, L., B. Charroux, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 152:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petti, L., C. Sample, and E. Kieff. 1990. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology 176:563-574. [DOI] [PubMed] [Google Scholar]
- 25.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 26.Spender, L. C., G. H. Cornish, A. Sullivan, and P. J. Farrell. 2002. Expression of transcription factor AML-2 (RUNX3, CBFa-3) is induced by Epstein-Barr virus EBNA-2 and correlates with the B-cell activation phenotype. J. Virol. 76:4919-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strasswimmer, J., C. L. Lorson, D. E. Breiding, J. J. Chen, T. Le, A. H. Burghes, and E. J. Androphy. 1999. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum. Mol. Genet. 8:1219-1226. [DOI] [PubMed] [Google Scholar]
- 28.Tong, X., F. Wang, C. J. Thut, and E. Kieff. 1995. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J. Virol. 69:585-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong, X., R. Yalamanchili, S. Harada, and E. Kieff. 1994. The EBNA-2 arginine-glycine domain is critical but not essential for B-lymphocyte growth transformation; the rest of region 3 lacks essential interactive domains. J. Virol. 68:6188-6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voss, M. D., A. Hille, S. Barth, A. Spurk, F. Hennrich, D. Holzer, N. Mueller-Lantzsch, E. Kremmer, and F. A. Grässer. 2001. Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter. J. Virol. 75:11781-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wirth, B. 2000. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum. Mutat. 15:228-237. [DOI] [PubMed] [Google Scholar]