Abstract
Human CD46 is a cellular receptor for human herpesvirus 6 (HHV-6). Virus entry into host cells requires a glycoprotein H (gH)-glycoprotein L (gL) complex. We show that the CD46 ectodomain blocked HHV-6 infection and bound a complex of gH-gL and the 80-kDa U100 gene product, designated glycoprotein Q, indicating that the complex is a viral ligand for CD46.
Human herpesvirus 6 (HHV-6) was first isolated from the peripheral blood of patients with AIDS and lymphoproliferative disorders (31). HHV-6 isolates can be classified into two groups, variant A (HHV-6A) and variant B (HHV-6B). HHV-6B is the causative agent of exanthem subitum (40). The two variants can be differentiated on the basis of genomic polymorphism, antigenicity, and host cell tropism (1, 3, 5, 39). Human CD46 is reported to be a cellular receptor for HHV-6 (32). Recently, Mori et al. showed that HHV-6A, but not HHV-6B, mediates fusion from without in a variety of cells expressing human CD46 (20). Human CD46 is composed of four short consensus repeating units (SCRs), a Ser/Thr (ST)-rich domain, a 13-amino-acid sequence of unknown significance (UK), a transmembrane (TM) domain, and a cytoplasmic tail (CYT) (17) (Fig. 1A). In a study by Mori et al., HHV-6A-induced cell-cell fusion was not seen in Chinese hamster ovary cells expressing the CD46 deletion mutants ΔSCR2, ΔSCR3, and ΔSCR4, although it was seen in cells expressing ΔSCR1, ΔST, ΔUK, and ΔCYT, indicating that SCR domains 2, 3, and 4 of the CD46 ectodomain are essential for virus-induced cell-cell fusion (20). Recently, Greenstone et al. reported that SCR domains 2 and 3 are required for HHV-6 receptor activity (9). Moreover, Mori et al. found that monoclonal antibodies (MAbs) against glycoprotein B (gB) and gH of HHV-6A inhibited the fusion event, indicating that the cell-cell fusion induced by HHV-6A required gH and gB (20). However, it is as yet unknown which glycoprotein(s) of HHV-6 can associate with human CD46. It is important to identify the HHV-6 glycoprotein(s) that physically interacts with CD46 to better understand virally induced cell-cell fusion and entry.
FIG. 1.
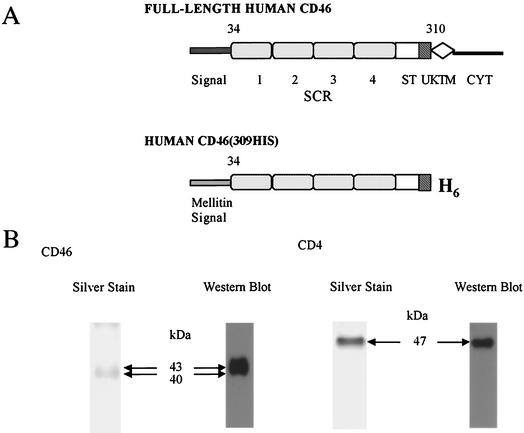
(A) Schematic representations of the human CD46 protein. Diagrams of the full-length human CD46 and the recombinant soluble form CD46(309his) are shown. The SCRs (indicated by numbers 1 to 4), ST-rich domain (ST), UK, TM domain (TM), and CYT are indicated. In CD46(309his), the natural signal peptide was replaced by the honeybee mellitin signal peptide encoded by the insertion vector. CD46(309his) was truncated at leucine 309 prior to the predicted transmembrane region (TM) and six additional histidine residues were added to the C terminus. (B) SDS-PAGE analysis of purified CD46(309his) and CD4(387his). Purified CD46(309his) and CD4(389his) were fractionated by SDS-12% PAGE under reducing conditions and visualized by silver staining and Western blotting with the His probe.
We recently found that the HHV-6A gH-gL complex interacts with one form of the U100 gene products (8), namely, an 80-kDa protein with N-linked oligosaccharides (21). Glycosidase digestion analysis showed that the U100 gene products existed as two forms of glycoproteins, an 80-kDa form containing complex N-linked oligosaccharides and a 74-kDa form containing immature, high-mannose N-linked oligosaccharides. Based on these characteristics, Mori et al. designated the U100 gene products glycoprotein Q (gQ) (21).
In this study, we identified the gH-gL-gQ complex as the viral ligand for human CD46. First, to search for the viral ligand(s) for human CD46, we produced a soluble form of the CD46 ectodomain from baculovirus-infected cells. To construct a recombinant baculovirus expressing a truncated and secreted form of human CD46, an 828-bp fragment of CD46, corresponding to amino acids 34 to 309 (in which amino acid 34 is the first amino acid after the predicted signal peptide sequence), was amplified by PCR from the plasmid pME18S CD46 STc/CYT 2 (10). The plasmid pME18S CD46 STc/CYT 2 was generously provided by Tsukasa Seya (Osaka Medical Center for Cancer and Cardiovascular Disease, Osaka, Japan). The amino-terminal primer, 5′-agatctTGCCTGTGAGGAGCCACCAAC-3′, was hybridized to CD46; it added a BglII site (indicated by the lowercase letters) just upstream from the codon for amino acid 34. The carboxy-terminal primer, 5′-ctgcagTTAGTGATGGTGATGGTGATGAAGTATTCCTTCCTCAGGTTTAG-3′, added six histidine codons after amino acid 309 of CD46, a stop codon, and a PstI site (lowercase letters). As a negative control, we used a secreted form of human CD4, which was a 1,092-bp fragment of CD4 that corresponded to amino acids 24 to 387. The amino-terminal primer, 5′-ggatccTCAGGGAAAGAAAGTGGTGCTG-3′, was hybridized to CD4; it added a BamHI site (lowercase letters) just upstream from the codon for amino acid 24. The carboxy-terminal primer, 5′-ctgcagTTAGTGATGGTGATGGTGATGCAGAACCTTGATGTTG-3′, added six histidine codons after amino acid 387 of CD4, a stop codon, and a PstI site (lowercase letters). The PCR-amplified product was digested with BglII (for CD46) or BamHI (for CD4) and PstI and inserted into the plasmid pFastBac-Msp-Fc (30), which had been digested with BamHI and PstI. pFastBac-Msp-Fc was generously provided by Yoshimi Takai (Osaka University, Osaka, Japan). The honeybee melittin signal peptide sequence replaced the CD46 or CD4 signal peptide. The recombinant baculoviruses were prepared according to the protocol of the manufacturer (Invitrogen) and were named bac-CD46(309his) and bac-CD4(387his). The recombinant proteins were designated CD46(309his) (Fig. 1A) and CD4(387his).
To purify CD46(309his) or CD4(387his), Hi5 cells grown in 20-ml cultures were infected with bac-CD46(309his) or bac-CD4(387his) at a multiplicity of infection (MOI) of 5. At 72 h postinfection (PI), the supernatant was clarified by centrifugation. Using a Centricon apparatus (Millipore), the supernatant was concentrated 10-fold and dialyzed against phosphate-buffered saline (PBS). The protein solution was mixed with 250 μl of Nexus IMAC resin (Valen Biotech, Inc.) preequilibrated with PBS and was incubated for 3 h at 4°C on a rotary shaker. The resin was pelleted (700 × g for 5 min at 4°C), resuspended in PBS, transferred to a column, and washed first with PBS and then with 5 bed volumes of washing buffer (50 mM sodium phosphate, 300 mM NaCl, pH 7.0) with 5 mM imidazole. The protein was eluted by adding elution buffer (50 mM sodium phosphate, 300 mM NaCl, 150 mM imidazole, pH 7.0). The eluate, containing most of the CD46(309his) or CD4(387his), was collected and dialyzed against PBS. The yield of purified CD46(309his) or CD4(389his) was 400 to 500 μg/20 ml of supernatant. The purified protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and examined by silver staining (Fig. 1B, first panel from left). Using an anti-rabbit polyhistidine antibody, His probe (Santa Cruz Biotechnology), the structure of CD46 or CD4 was confirmed by DNA sequencing and immunoblotting.
On immunoblots, broad bands ranging from 40 to 43 kDa for CD46(309his) reacted with the His probe (CD46; Fig. 1B, second panel from left).
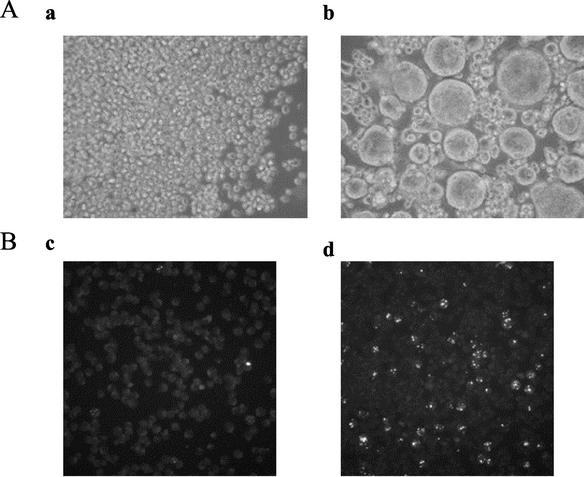
Santoro et al. reported that when recombinant vaccinia virus technology was used, soluble CD46 produced in mammalian cells inhibited HHV-6 binding and cell fusion (32).Using the recombinant baculovirus system in this study to determine whether the CD46(309his) produced in insect cells also acts as the cellular receptor, the ability of CD46(309his) to block HHV-6-mediated cell-cell fusion and virus entry was tested. The HHV-6A strain GS was incubated for 30 min at room temperature with 2 μM purified CD46(309his) or CD4(389his). SupT1 cells (1 × 105) were then spun with the treated virus preparation at an MOI of 1 for 1 h at 37°C, and the infection was monitored by light-microscopic examination of the extent of cell-cell fusion 2 h following infection with the virus (Fig. 2A). To further examine the blockage of HHV-6 entry into HSB-2 cells, strain GS was incubated with 2 μM purified CD46(309his) or CD4(387his) for 30 min at room temperature. HSB-2 cells (1 × 105) were then incubated with the treated virus preparation at an MOI of 0.1 for 1 h at 37°C. After incubation, the cells were treated with 0.1 M citrate buffer (pH 3.0) to inactivate extracellular virus. The cells were washed and grown at 37°C for 12 h. They were then fixed with acetone and stained with a MAb against the HHV-6A immediate-early protein, IE1 (8, 25, 34); this antibody is named AIE1-32 (Fig. 2B). The results showed that CD46(309his) was effective in blocking both HHV-6-mediated cell-cell fusion and virus entry (Fig. 2A, panel a, and Fig. 2B, panel c), whereas the CD4(387his) control had no effect on either function (Fig. 2A, panel b, and Fig. 2B, panel d). The CD46 expressed in insect cells from baculovirus recombinants had properties similar to those of CD46 reported by Santoro et al. (32). These results suggest that CD46(309his) produced by a recombinant baculovirus interacted with at least one virion component that is essential for virus-induced cell-cell fusion and entry.
FIG. 2.
Inhibition of HHV-6A-mediated cell-cell fusion and virus entry by CD46(309his). (A) HHV-6A strain GS was incubated with either CD46(309his) or CD4 (389his). SupT1 cells were then spun with the treated virus preparation, and the infection was monitored by light-microscopic examination of cell-cell fusion. Results for SupT1 cells infected with strain GS preincubated with CD46 (panel a) and SupT1 cells infected with strain GS preincubated with CD4 (panel b) are shown. The cell-cell fusion was inhibited in the cells infected with CD46-pretreated virus (panel a) but not in those infected with CD4-pretreated virus (panel b). (B) Strain GS was incubated with either CD46(309his) or CD4(389his). HSB-2 cells were then incubated with the treated virus preparation. At 12 h PI, the cells were fixed and stained with a MAb for HHV-6 IE1. The expression of IE1 in the nucleus was inhibited in the cells infected with CD46-pretreated virus (panel c) but not in those infected with CD4-pretreated virus (panel d).
To identify the viral ligands for human CD46, HSB-2 cells were infected with HHV-6A strain GS and the cells were harvested at 72 h PI, incubated for 30 min with lysis buffer (10 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1% Nonidet P-40) containing protease inhibitor cocktail (Roche), sonicated, and spun at 200,000 × g for 1 h and the lysates were collected. Using a Centricon apparatus (Millipore), the culture supernatants containing each secreted protein [CD46(309his) or CD4(387his), as described above] were concentrated 10-fold. The concentrated supernatant (300 μl) containing CD46(309his) or CD4(387his) was then incubated with immobilized cobalt chelate (ProFound pull-down PolyHis Protein: Protein Interaction Kit; Pierce) at 4°C for 2 h. After the resin was washed, it was incubated at 4°C for 4 h with the lysates from the HHV-6-infected cells. After extensive washing with washing buffer (ProFound pull-down PolyHis Protein: Protein Interaction Kit; Pierce), the proteins were eluted in elution buffer (washing buffer with 290 mM imidazole) and the eluted proteins were detected by silver staining. Broad bands of approximately 100 to 102 kDa were found in the samples eluted from the CD46(309his)-bound resin but not in those eluted from the CD4(387his)-bound resin (data not shown).
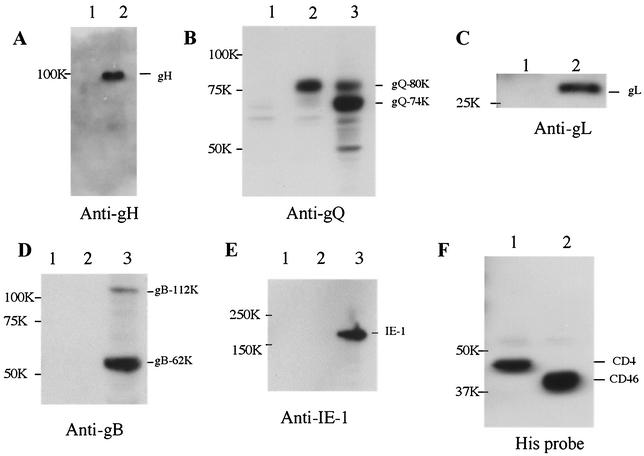
Thereafter, we searched for the gene responsible for these bands, and the most likely candidate was the U48 gene, which encodes gH, a glycoprotein consisting of 694 amino acid residues and containing 13 potential N-linked glycosylation sites (8) (Table 1). The mature form of HHV-6 gH is reported to have a molecular mass of 98 to 102 kDa (16, 23, 28, 36). Thus, the lysates were immunoblotted with an anti-HHV-6A gH mouse antiserum. The mouse antisera specific for HHV-6A gH and gL were generated as previously reported (21). Proteins of 100 to 102 kDa were labeled with anti-gH by immunoblotting (Fig. 3A, lane 2) but did not react with preimmune sera (data not shown), supporting the idea that the 100- to 102-kDa component corresponded to the product of the gH gene. These results showed that CD46(309his) associated with HHV-6A gH. In HHV-6 as well as the other herpesviruses, gH and gL interact to form a gH-gL complex (16) (Table 1). Furthermore, Mori et al. recently found that the gH-gL complex associated with the 80-kDa U100 gene product, which contains complex N-linked oligosaccharides, but not with a 74-kDa form of the U100 product that contains immature, N-linked oligosaccharides with high levels of mannose (21). Mori et al. have designated these two forms of the HHV-6A U100 gene product gQ (21); they form the gp105-82 complex (26, 27) in HHV-6A-infected cells.
TABLE 1.
Characteristics of predicted products of HHV-6 gH and gL genes
| Gene | No. of amino acids | No. of NXT/S sites | No. of cysteine residues |
|---|---|---|---|
| gH | 694 | 13 | 14 |
| gL | 250 | 2 | 6 |
FIG. 3.
Cosedimentation of HHV-6A with soluble CD46(309his). HSB-2 cells were infected with HHV-6A strain GS, and the cells were then lysed in lysis buffer. Strain GS-infected cell lysates were incubated with either CD46(309his)- or CD4(389his)-bound immobilized cobalt chelate resin, and the proteins were eluted with 290 mM imidazole. The eluted proteins were analyzed by SDS-8 or 10% PAGE followed by immunoblotting with anti-gH or gL antiserum, anti-gQ MAb, anti-gB MAb, anti-IE1 MAb, or His probe. gH (panel A, lane 2), the 80-kDa form of gQ (panel B, lane 2), and gL (panel C, lane 2) were detected in the eluate from the CD46-bound resin but not in that from the CD4-bound resin (panels A, B, and C, lanes 1). The 80-kDa and 74-kDa bands were detected by the anti-gQ MAb in strain GS-infected cell lysates (panel B, lane 3). The gB (panel D) and IE-1 (panel E) proteins were not detected in the eluate from the CD46-bound (panels D and E, lanes 2) or CD4-bound (panels D and E, lanes 1) resin. Either CD4 (panel F, lane 1) or CD46 (panel F, lane 2) was detected in each eluate by the His probe. The 112-kDa and 62-kDa bands were detected in strain GS-infected cell lysates by anti-gB MAb (panel D, lane 3), and the 190-kDa band was detected by anti-IE1 MAb (panel E, lane 3). Numbers beside panels indicate kilodaltons.
Therefore, to examine whether CD46 associates with the gH-gL-gQ-80-kDa complex, the eluted proteins from the CD46(309his)-bound resin were immunoblotted with the anti-HHV-6A gQ MAb or anti-HHV-6A gL antiserum. The MAb against HHV-6A gQ (designated AU100-119) was produced as previously described (21). As expected, the 80-kDa gQ (gQ-80K) and the 30-kDa gL protein were detected on the blot membrane with the anti-AU100-119 MAb and anti-gL antiserum, respectively (Fig. 3B, lane 2, and Fig. 3C, lane 2), but not with preimmune serum (data not shown), supporting the idea that the HHV-6A gH-gL-gQ-80K complex associates with CD46. As a negative control, the blot membrane was reacted with MAbs against HHV-6 gB (OHV1) (24) and with the MAb AIE1-32 against the nonstructural protein, HHV-6 IE1. The blot did not react with these MAbs (Fig. 3D, lane 2, and Fig. 3E, lane 2).
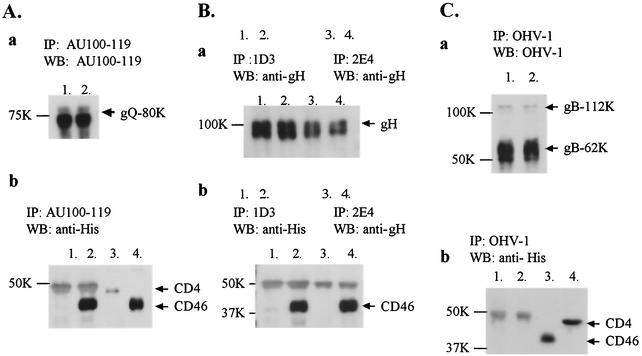
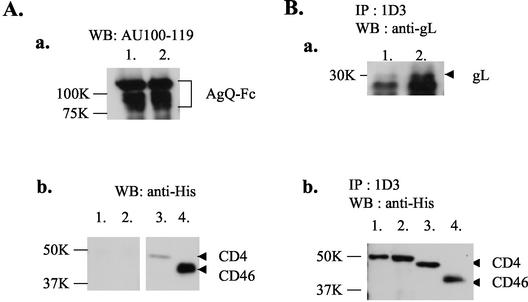
To confirm the interaction between CD46 and the glycoprotein complex, immunoprecipitation was performed. Lysates from HHV-6-infected cells were incubated for 4 h with an anti-gH MAb (1D3 or 2E4 [generously provided by Gabriella Campadelli-Fiume, University of Bologna, Bologna, Italy]), an anti-gQ MAb (AU100-119), or an anti-gB MAb (OHV-1) and were immunoprecipitated with protein G-Sepharose beads as described previously (24). The beads with each immune complex were then incubated with 400 μl of 10-fold-concentrated culture supernatants containing CD46(309his) or CD4(387his) for 4 h. After being washed, the immunoprecipitates were solubilized with sample buffer (0.1 M Tris-HCl [pH 6.8], 20% glycerol, 4% SDS, 5% 2-mercaptoethanol, and 0.2% bromophenol blue), separated by SDS-PAGE, and transferred to polyvinylidene difluoride membranes. The membranes were then incubated with the His probe, anti-gH serum, or MAbs against gQ or gB. As shown in Fig. 4, although MAbs 1D3 (Fig. 4B, panel b, lane 2), 2E4 (Fig. 4B, panel b, lane 4) and A100-119 (Fig. 4A, panel b, lane 2) coprecipitated with CD46(309his), MAb OHV-1 (Fig. 4C, panel b, lane 2) did not, and none of the MAbs coprecipitated with CD4(387his) (Fig. 4A, panel b, lane 1; Fig. 4B, panel b, lanes 1 and 3; and Fig. 4C, panel b, lane 1).
FIG. 4.
Immunoprecipitation with MAbs, AU100-119, 1D3, 2E4, and OHV-1 followed by immunoblot analyses of the proteins precipitated with the MAbs. The lysates from HHV-6A-infected cells were immunoprecipitated with one of the following MAbs: AU100-119, anti-gQ (A); 1D3 or 2E4, anti-gH (B); and OHV-1, anti-gB (C). The immune complexes were bound to protein G beads and further incubated with CD46(309his) (panel A, lanes 2; panel B, lanes 2; panel B, lanes 4; and panel C, lanes 2) or CD4(387his) (panel A, lanes 1; panel B, lanes 1; panel B, lanes 3; and panel C, lanes 1). The immunoprecipitated proteins were analyzed by 8% or 10% SDS-PAGE followed by immunoblotting with one of the following: AU100-119 MAb (panel A, panel a), anti-gH serum (panel B, panel a) or OHV-1 MAb (panel C, panel a), or His probe (panels A, B, and C, panels b). The MAbs precipitated each protein (panels A, B, and C, panels a). The MAbs AU100-119 (panel A, panel b, lane 2), ID3 (panel B, panel b, lane 2) and 2E4 (panel B, panel b, lane 4) coprecipitated CD46, but OHV-1 did not (panel C, lanes 2). None of the MAbs coprecipitated CD4 (panels A and B, panels b, lane 1; panel B, panel b, lane 3; and panel C, panel b, lane 1). Input of CD46 (panel A, pane l b, lane 4, and panel C, panel b, lane 3) and CD4 (panel A, panel b, lane 3, and panel C, panel b, lane 4). Numbers beside panels show kilodaltons.
These data indicated that gH and gQ were coprecipitated with CD46(309his) by each MAb but gB was not. Moreover, both MAbs against gH, 2E4 and 1D3, can neutralize virus infectivity and prevent polykaryocyte formation, with 2E4 acting more efficiently (2, 7). Therefore, 2E4 and 1D3 can be described as anti-fusion MAbs (2). Both of these MAbs recognize epitopes in the external domain of gH (2). Our results indicate that the epitopes that are recognized by both MAbs may not be in a CD46-binding domain and that the neutralizing activity of the MAbs may not be due to its interference with this interaction. These results show that CD46 associates with gH-gL-gQ-80K as a complex.
To identify the viral ligand(s) of CD46 in more detail, each protein (gQ, gH, or gL) was transiently expressed in mammalian cells. However, when mammalian cells were transfected with a plasmid containing the gQ gene alone (27), the expressed gQ protein was completely sensitive to endo H (unpublished data) and no gQ-80K with complex N-linked oligosaccharides was detected in the transfected cells, indicating that the gQ-80K that associates with gH-gL may be processed a different way in HHV-6-infected cells. Therefore, using recombinant baculovirus technology as described above to induce further glycosylation, the gQ protein was expressed and secreted with the honeybee melittin signal peptide as an Fc fusion protein in insect cells. A 1,872-bp cDNA fragment of the gQ gene previously described (27), corresponding to amino acids 33 to 656 (in which amino acid 33 is the first amino acid after the predicted signal peptide sequence), was amplified by PCR. The primer sequences were as follows: 5′-ggatccAACGGCCCACCGACGTGCGGGAA-3′ for the amino-terminal primer and 5′-ggatccCTGTAATTTGTGTAATTTAATAAGAG-3′ for the carboxy-terminal primer (the BamHI site is indicated by lowercase letters). The PCR product was inserted into pFastBac1-Msp-Fc to express the protein fused with the NH2-terminal honeybee melittin signal peptide and the COOH-terminal immunoglobulin G Fc (AgQ-Fc). A baculovirus bearing AgQ-Fc was prepared as described above. After 3 days of infection, the culture supernatants were collected and concentrated 10-fold. Samples of concentrated supernatant (300 μl) were incubated with protein G-Sepharose beads. After being washed, the AgQ-Fc-bound beads were incubated with 400 μl of the concentrated culture supernatant containing either CD46(309his) or CD4(387his). After being washed, the precipitated proteins were immunoblotted with anti-gQ MAb or His probe. As shown in Fig. 5A, AgQ-Fc was unable to bind either CD46(309his) (Fig. 5A, panel b, lane 2) or CD4(387his) (Fig. 5A, panel b, lane 1), indicating that gQ modified in insect cells alone was unable to bind to CD46.
FIG. 5.
Neither soluble gQ-Fc alone nor gH-gL complex alone binds to CD46. (A) Soluble HHV-6A gQ-Fc was incubated with protein G-Sepharose beads. The AgQ-Fc-bound beads were incubated with CD46(309his) (lanes 2) or CD4(387his) (lanes 1), and then the precipitated proteins were immunoblotted with anti-gQ MAb (panel a) or His probe (panel b). AgQ-Fc was unable to bind either CD46 (panel b, lane 2) or CD4 (panel b, lane 1). Results for input of CD46 (panel b, lane 4) and CD4 (panel b, lane 3) are shown. (B) COS-7 cells were infected with the recombinant vaccinia virus vTF7 and subsequently transfected with plasmids pcDNA-UgH and pcDNA-UgL. Lysates were prepared and immunoprecipitated with 1D3, an anti-gH MAb. The immune complexes were incubated with protein G-Sepharose beads, and the complex-bound beads were incubated with CD46(309his) (panels a and b, lanes 2) or CD4(387his) (panels a and b, lanes 1). The precipitated proteins were immunoblotted with anti-gL antiserum (panel a) or His probe (panel b). The MAb 1D3 coprecipitated gL (panel a, lanes 1 and 2) with gH but did not coprecipitate either CD46 (panel b, lane 2) or CD4 (panel b, lane 1). Results for input of CD46 (panel b, lane 4) and CD4 (panel b, lane 3) are shown. Numbers beside panels indicate kilodaltons.
A DNA fragment coding for gL from U1102 (HHV-6A) was amplified by PCR with the following primers: 5′-gctagcAACCATGGAACTTTTACTATTTG-3′ for the amino-terminal primer and 5′-gatatcTTATGTGTTTCTAATCAGAAT-3′ for the carboxy-terminal primer (restriction enzyme sites are indicated by lowercase letters). The PCR product was inserted into pcDNA 3.1/myc-His (Invitrogen) via NheI and EcoRV restriction sites. The resulting expression plasmid, pcDNA-UgL, encoded HHV-6A gL. pcDNA-UgH, an expression plasmid encoding HHV-6A gH, was constructed as follows. The plasmid PTMUGH (U1102 gH) (36) was digested with EcoRI and SalI, and the fragment encoding gH was inserted into pcDNA3.1/zeo (Invitrogen) via EcoRI and XhoI sites. Using Lipofectamine (Invitrogen) according to the manufacturer's protocol, the pcDNA-UgH and pcDNA-UgL plasmids under the control of the T7 promoter were then introduced into COS-7 cells already infected with the recombinant vaccinia virus vTF7 (expressing T7 RNA polymerase). vTF7 was a kind gift from Bernard Moss (National Institutes of Health, Bethesda, Md.). After 18 h of cultivation, the infected-transfected cells were collected and lysed. The lysates were incubated with the anti-gH MAb 1D3, and the immune complex was incubated with protein G-Sepharose beads. After being washed, the complex-bound beads were incubated with CD46(309his) or CD4(387his). After being washed, the precipitated proteins were immunoblotted with anti-gH or anti-gL antiserum or His probe. The MAb 1D3 coprecipitated gL (Fig. 5B, panel a) with gH but did not coprecipitate CD46 (Fig. 5B, panel b, lane 2), indicating that the gH-gL complex alone was unable to bind to CD46 and that the interaction with CD46 might require the additional association with gQ-80K modified in HHV-6-infected cells.
The entry of viruses into cells is a complex process that is still incompletely understood. In several cases, it appears to require not only a cellular receptor to interact with the virus attachment protein but also at least one additional molecule to interact with the virus and facilitate penetration. Studies on other herpesviruses have provided indirect evidence of a role for homologs of the gH-gL molecules in membrane fusion. In HHV-6, a gH-specific MAb inhibits virus-induced fusion in cultured cells (15). Likewise, in Epstein-Barr virus (EBV), a MAb specific for gp85 inhibits membrane fusion but not virus attachment in cultured B-cell lines (18). A mutant pseudorabies virus that lacks gH attaches to cultured cells but does not penetrate them unless fusion between the virus and cell is induced artificially by treatment with polyethylene glycol (4). Affinity-purified gH-gL from bovine herpesvirus 1 blocks virus penetration into MDBK cells but not the initial attachment (37).
In herpesvirus virions, gB and gH-gL function after virus attachment and during virus entry by promoting membrane fusion (11, 29). Studies by Mori et al. and others showed that fusion from without induced by HHV-6 or human cytomegalovirus was specifically inhibited by antibodies against gH and gB, confirming that both these glycoproteins are involved in the process (19, 20). In varicella-zoster virus, gH-gL is intrinsically fusogenic, in that transfected HeLa cells expressing both genes have been shown to undergo extensive polykaryocytosis (6). Polyclonal antibodies and MAbs against herpes simplex virus 1 gL can efficiently block cell fusion by syncytial mutants, indicating a role for gL in this fusion process (22). Pseudorabies virus gL is necessary for virus penetration and cell-cell spread in cell culture (12). These previous studies indicate that the gH-gL complex is essential for fusion between the virion envelope and the cellular cytoplasmic membrane during penetration. In the case of EBV, viral envelope fusion with the host cell membrane requires the additional interaction of the ternary EBV glycoprotein gp82-gp25-gp42 (gH-gL-gp42) complex with its cellular ligand (14, 38). A MAb that inhibits the fusion of EBV and that had previously been assumed to be specific for gH is actually directed against the 42-kDa component of the complex (14, 35). The HLA class II protein HLA-DR binds to gp42 and can serve as a coreceptor for EBV entry (13, 33). Here, we showed that CD46 associates with the gH-gL-gQ-80K complex of HHV-6A. In agreement with our study, CD46 has been found to function as a fusion receptor by interacting with the complex in the process of virus entry. The steric conformation of the gH-gL-gQ-80K complex might be important for the virus entry process. However, whether one glycoprotein of the complex binds to CD46 directly, analogous to the interaction between EBV gp42 and HLA class II, or whether the steric conformation of the gH-gL-gQ-80K complex itself is required for the interaction with CD46 needs to be investigated.
Acknowledgments
This study was supported in part by a grant-in-aid for COE Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
We thank Gabriella Campadelli-Fiume (University of Bologna, Italy) for providing MAbs, Yoshimi Takai (Osaka University, Japan) and Tsukasa Seya (Osaka Medical Center for Cancer and Cardiovascular Disease, Japan) for providing plasmids, and Yun Bao Jiang and Sayoko Yonemoto (Osaka University) for technical assistance.
REFERENCES
- 1.Ablashi, D. V., N. Balachandran, S. F. Josephs, C. L. Hung, G. R. Krueger, B. Kramarsky, S. Z. Salahuddin, and R. C. Gallo. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184:545-552. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. A., and U. A. Gompels. 1999. N- and C-terminal external domains of human herpesvirus-6 glycoprotein H affect a fusion-associated conformation mediated by glycoprotein L binding the N terminus. J. Gen. Virol. 80:1485-1494. [DOI] [PubMed] [Google Scholar]
- 3.Aubin, J. T., H. Collandre, D. Candotti, D. Ingrand, C. Rouzioux, M. Burgard, S. Richard, J. M. Huraux, and H. Agut. 1991. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J. Clin. Microbiol. 29:367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babic, N., B. G. Klupp, B. Makoschey, A. Karger, A. Flamand, and T. C. Mettenleiter. 1996. Glycoprotein gH of pseudorabies virus is essential for penetration and propagation in cell culture and in the nervous system of mice. J. Gen. Virol. 77:2277-2285. [DOI] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume, G., S. Guerrini, X. Liu, and L. Foa-Tomasi. 1993. Monoclonal antibodies to glycoprotein B differentiate human herpesvirus 6 into two clusters, variants A and B. J. Gen. Virol. 74:2257-2262. [DOI] [PubMed] [Google Scholar]
- 6.Duus, K. M., C. Hatfield, and C. Grose. 1995. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology 210:429-440. [DOI] [PubMed] [Google Scholar]
- 7.Foa-Tomasi, L., A. Boscaro, S. di Gaeta, and G. Campadelli-Fiume. 1991. Monoclonal antibodies to gp100 inhibit penetration of human herpesvirus 6 and polykaryocyte formation in susceptible cells. J. Virol. 65:4124-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 9.Greenstone, H. L., F. Santoro, P. Lusso, and E. A. Berger. 2002. Human herpesvirus 6 and measles virus employ distinct CD46 domains for receptor function. J. Biol. Chem. 277:39112-39118. [DOI] [PubMed] [Google Scholar]
- 10.Iwata, K., T. Seya, Y. Yanagi, J. M. Pesando, P. M. Johnson, M. Okabe, S. Ueda, H. Ariga, and S. Nagasawa. 1995. Diversity of sites for measles virus binding and for inactivation of complement C3b and C4b on membrane cofactor protein CD46. J. Biol. Chem. 270:15148-15152. [DOI] [PubMed] [Google Scholar]
- 11.Keay, S., and B. Baldwin. 1991. Anti-idiotype antibodies that mimic gp86 of human cytomegalovirus inhibit viral fusion but not attachment. J. Virol. 65:5124-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klupp, B. G., W. Fuchs, E. Weiland, and T. C. Mettenleiter. 1997. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J. Virol. 71:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, D. X., U. A. Gompels, L. Foa-Tomasi, and G. Campadelli-Fiume. 1993. Human herpesvirus-6 glycoprotein H and L homologs are components of the gp100 complex and the gH external domain is the target for neutralizing monoclonal antibodies. Virology 197:12-22. [DOI] [PubMed] [Google Scholar]
- 16.Liu, D. X., U. A. Gompels, J. Nicholas, and C. Lelliott. 1993. Identification and expression of the human herpesvirus 6 glycoprotein H and interaction with an accessory 40K glycoprotein. J. Gen. Virol. 74:1847-1857. [DOI] [PubMed] [Google Scholar]
- 17.Lublin, D. M., M. K. Liszewski, T. W. Post, M. A. Arce, M. M. Le Beau, M. B. Rebentisch, L. S. Lemons, T. Seya, and J. P. Atkinson. 1988. Molecular cloning and chromosomal localization of human membrane cofactor protein (MCP). Evidence for inclusion in the multigene family of complement-regulatory proteins. J. Exp. Med. 168:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, N., and L. M. Hutt-Fletcher. 1988. A monoclonal antibody to glycoprotein gp85 inhibits fusion but not attachment of Epstein-Barr virus. J. Virol. 62:2366-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne, R. S., D. A. Paterson, and J. C. Booth. 1998. Human cytomegalovirus glycoprotein H/glycoprotein L complex modulates fusion-from-without. J. Gen. Virol. 79:855-865. [DOI] [PubMed] [Google Scholar]
- 20.Mori, Y., T. Seya, H. L. Huang, P. Akkapaiboon, P. Dhepakson, and K. Yamanishi. 2002. Human herpesvirus 6 variant A but not variant B induces fusion from without in a variety of human cells through a human herpesvirus 6 entry receptor, CD46. J. Virol. 76:6750-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori, Y., P. Akkapaiboon, X. Yang, and K. Yamanishi. 2003. The human herpesvirus 6 U100 gene product is the third component of the gH-gL glycoprotein complex on the viral envelope. J. Virol. 77:2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novotny, M. J., M. L. Parish, and P. G. Spear. 1996. Variability of herpes simplex virus 1 gL and anti-gL antibodies that inhibit cell fusion but not viral infectivity. Virology 221:1-13. [DOI] [PubMed] [Google Scholar]
- 23.Okuno, T., H. Sao, H. Asada, K. Shiraki, M. Takahashi, and K. Yamanishi. 1990. Analysis of a glycoprotein of human herpesvirus 6 (HHV-6) using monoclonal antibodies. Virology 176:625-628. [DOI] [PubMed] [Google Scholar]
- 24.Okuno, T., H. Shao, H. Asada, K. Shiraki, M. Takahashi, and K. Yamanishi. 1992. Analysis of human herpesvirus 6 glycoproteins recognized by monoclonal antibody OHV1. J. Gen. Virol. 73:443-447. [DOI] [PubMed] [Google Scholar]
- 25.Papanikolaou, E., V. Kouvatsis, G. Dimitriadis, N. Inoue, and M. Arsenakis. 2002. Identification and characterization of the gene products of open reading frame U86/87 of human herpesvirus 6. Virus Res. 89:89-101. [DOI] [PubMed] [Google Scholar]
- 26.Pfeiffer, B., Z. N. Berneman, F. Neipel, C. K. Chang, S. Tirwatnapong, and B. Chandran. 1993. Identification and mapping of the gene encoding the glycoprotein complex gp82-gp105 of human herpesvirus 6 and mapping of the neutralizing epitope recognized by monoclonal antibodies. J. Virol. 67:4611-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeiffer, B., B. Thomson, and B. Chandran. 1995. Identification and characterization of a cDNA derived from multiple splicing that encodes envelope glycoprotein gp105 of human herpesvirus 6. J. Virol. 69:3490-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian, G., C. Wood, and B. Chandran. 1993. Identification and characterization of glycoprotein gH of human herpesvirus-6. Virology 194:380-386. [DOI] [PubMed] [Google Scholar]
- 29.Rasmussen, L., S. Resta, and T. Merigan. 1991. Human cytomegalovirus glycoprotein-receptor interactions. Transplant. Proc. 23:60-63. [PubMed] [Google Scholar]
- 30.Sakisaka, T., T. Taniguchi, H. Nakanishi, K. Takahashi, M. Miyahara, W. Ikeda, S. Yokoyama, Y.-F. Peng, K. Yamanishi, and Y. Takai. 2001. Requirement of interaction of nectin-1α/HveC with afadin for efficient cell-cell spread of herpes simplex virus type 1. J. Virol. 75:4734-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salahuddin, S. Z., D. V. Ablashi, P. D. Markham, S. F. Josephs, S. Sturzenegger, M. Kaplan, G. Halligan, P. Biberfeld, F. Wong-Staal, B. Kramarsky, et al. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596-601. [DOI] [PubMed] [Google Scholar]
- 32.Santoro, F., P. E. Kennedy, G. Locatelli, M. S. Malnati, E. A. Berger, and P. Lusso. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817-827. [DOI] [PubMed] [Google Scholar]
- 33.Spriggs, M. K., R. J. Armitage, M. R. Comeau, L. Strockbine, T. Farrah, B. Macduff, D. Ulrich, M. R. Alderson, J. Müllberg, and J. I. Cohen. 1996. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR β chain and inhibits antigen presentation. J. Virol. 70:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanton, R., J. D. Fox, R. Caswell, E. Sherratt, and G. W. Wilkinson. 2002. Analysis of the human herpesvirus-6 immediate-early 1 protein. J. Gen. Virol. 83:2811-2820. [DOI] [PubMed] [Google Scholar]
- 35.Strnad, B. C., T. Schuster, R. Klein, R. F. Hopkins III, T. Witmer, R. H. Neubauer, and H. Rabin. 1982. Production and characterization of monoclonal antibodies against the Epstein-Barr virus membrane antigen. J. Virol. 41:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda, K., M. Haque, T. Sunagawa, T. Okuno, Y. Isegawa, and K. Yamanishi. 1997. Identification of a variant B-specific neutralizing epitope on glycoprotein H of human herpesvirus-6. J. Gen. Virol. 78:2171-2178. [DOI] [PubMed] [Google Scholar]
- 37.van Drunen Littel-van den Hurk, S., S. Khattar, S. K. Tikoo, L. A. Babiuk, E. Baranowski, D. Plainchamp, and E. Thiry. 1996. Glycoprotein H (gII/gp108) and glycoprotein L form a functional complex which plays a role in penetration, but not in attachment, of bovine herpesvirus 1. J. Gen. Virol. 77:1515-1520. [DOI] [PubMed] [Google Scholar]
- 38.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyatt, L. S., N. Balachandran, and N. Frenkel. 1990. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J. Infect. Dis. 162:852-857. [DOI] [PubMed] [Google Scholar]
- 40.Yamanishi, K., T. Okuno, K. Shiraki, M. Takahashi, T. Kondo, Y. Asano, and T. Kurata. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065-1067. [DOI] [PubMed]