Abstract
Infection with a viral pathogen triggers several pathways in the host cell that are crucial to eliminating infection, as well as those that are used by the virus to enhance its replication and virulence. We have here used suppression subtractive hybridization and cDNA microarray analyses to characterize the host transcriptional response in an avian pneumovirus model of infection. The results of our investigations reveal a dynamic host response that includes the regulation of genes with roles in a vast array of cellular functions as well as those that have not been described previously. The results show a considerable upregulation in transcripts representing the interferon-activated family of genes, predicted to play a role in virus replication arrest. The analysis also identified transcripts for proinflammatory leukocyte chemoattractants, adhesion molecules, and complement that were upregulated and may account for the inflammatory pathology that is the hallmark of viral respiratory infection. Interestingly, alterations in the transcription of several genes in the ubiquitin and endosomal protein trafficking pathways were observed, suggesting a role for these pathways in virus maturation and budding. Taken together, the results of our investigations provide key insights into individual genes and pathways that constitute the host cell's response to avian pneumovirus infection, and they have enabled the development of resources and a model of host-pathogen interaction for an important avian respiratory tract pathogen.
Host-virus interactions include a complex interplay of molecular pathways directed by the host to prevent viral replication and countermeasures by the virus to favor its propagation. Elucidation of these host- and virus-directed responses provides new insights into the biology of mechanisms governing host antiviral strategies as well as the pathogenesis of viral infections. It is being increasingly recognized that an efficient means of characterizing the molecular basis of the host-pathogen relationship is profiling of genes whose expression is altered during the course of infection. This provides not only a molecular description of factors that engender host resistance or viral pathogenicity but also a framework for understanding the molecular basis of the pathology associated with the disease.
Avian species are highly susceptible to respiratory tract infections due to the unusual anatomical and immunological features of their respiratory tracts (45). Despite the great morbidity and mortality associated with these infections, very little is known about the molecular basis of disease pathogenesis and the host's response to infection. Among the major pathogens associated with respiratory disease in birds is the avian pneumovirus (APV), a member of the Paramyxoviridae family of enveloped viruses and the causal agent of acute and highly contagious respiratory tract infections in poultry species and wild bird populations worldwide (2). Infected birds typically exhibit severe inflammatory changes such as swollen sinuses, nasal and ocular discharge, and respiratory distress. The disease is characterized histopathologically by acute respiratory tract inflammatory changes including diffuse cellular infiltration, hemorrhage, and dense submucosal infiltration with heterophils (29).
We describe here the application of suppression subtractive hybridization (SSH) in conjunction with cDNA microarray analysis to explore the alterations in host cell transcriptional program in an APV model of infection. The present study provides intriguing information on the changes in host gene expression upon infection with a pneumovirus, which could serve as a foundation for future investigations on the roles of various genes and pathways in the pathogenesis of this important disease in avian species.
MATERIALS AND METHODS
Tissue culture and virus infection.
Primary chicken embryo cells were prepared from 10-day-old embryonated specific-pathogen-free chicken eggs (SPAFAS, North Franklin, Conn.) and grown in a medium containing equal quantities of Leibovitz and McCoy's media (Sigma, St. Louis, Mo.) with 5% fetal bovine serum, 100 IU of penicillin G/ml, and 100 mg of dihydrostreptomycin/ml in 150-mm-diameter tissue culture dishes. When 100% confluent, cells were passed once and plated into 150-mm-diameter dishes. After 24 h of incubation, cells were ∼95% confluent; at this time they were infected with the Colorado strain of APV subtype C (5) at a multiplicity of infection of 0.25 PFU per cell in a volume of 10 ml of medium and were allowed to adsorb at 37°C for 1 h. Following adsorption, 25 ml of Leibovitz McCoy's medium was added, and flasks were incubated at 37°C. Cells used as uninfected controls were mock treated (with sterile media) and processed in a fashion similar to that of the infected cells. Cells were harvested at 2.5, 24, 48, and 96 h postinfection (p.i.) from APV- and mock-infected cultures, respectively.
Purification of poly(A)+ mRNA.
Adherent APV-infected and control cells were washed once with phosphate-buffered saline, followed by lysis and homogenization in Trizol reagent (Invitrogen Life Technologies, Carlsbad, Calif.). Total RNA was purified according to the manufacturer's instructions. Poly(A)+ mRNA was extracted from the total RNA by using Oligotex mRNA spin columns (Qiagen, Valencia, Calif.) per the manufacturer's instructions.
Construction of normalized subtracted cDNA libraries.
A forward subtracted library was constructed in which poly(A)+ mRNA from APV-infected cells served as the “tester,” or the population whose upregulated transcripts were to be identified, and poly(A)+ mRNA from mock-infected control cells was designated the “driver,” or the population whose transcripts served as a reference for cDNA subtraction. APV-infected and mock-treated cell transcripts were also used conversely as driver and tester samples, respectively, to construct a reverse subtracted library. The differentially expressed cDNAs (targets) are present in the tester cDNA but are absent (or present at lower levels) in driver cDNAs. The SSH procedure (9) was followed to construct forward and reverse subtracted libraries by using the PCR-Select cDNA subtraction kit (Clontech, Palo Alto, Calif.). Briefly, 2 μg of the poly(A)+ mRNA from each of the tester and driver, collected at 96 h p.i., was converted to double-stranded cDNA by reverse transcription (RT). Both tester and driver cDNAs were digested with RsaI to produce shorter blunt-ended fragments. Only the digested tester cDNA was subdivided into two portions, each of which was ligated with a different adapter sequence, resulting in two populations of tester. After ligation, a series of two hybridization steps was performed. During the first hybridization, an excess of driver was added to each tester, denatured, and allowed to anneal. Due to the second-order kinetics of hybridization and the suppression PCR effect, the concentrations of high- and low-abundance target single-stranded molecules in the tester are normalized (43) and the target sequences in the tester become significantly enriched for differentially expressed genes. In the second hybridization, the two first-hybridization reaction products were mixed with each other and with fresh denatured driver cDNA. The populations of normalized and subtracted single-stranded target cDNAs anneal with each other, forming double-stranded hybrids with different adapter sequences at their 5′ ends. The adapter ends were filled in with DNA polymerase, and the subtracted hybrid molecules were specifically amplified by nested PCR using adapter-specific primer pairs.
Cloning of the SSH libraries into the TA vector.
Amplified products from the secondary nested PCR, constituting the subtracted target cDNAs, were ligated with the pGEM-T plasmid vector (Promega, Madison, Wis.) and transformed into maximum-efficiency Escherichia coli DH5α cells (Life Technologies). Subsequently, transformed bacteria were plated onto Luria-Bertani agar plates containing ampicillin (100 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal; 50 μg/ml), and isopropyl-β-d-thiogalactoside (IPTG; 100 μM), followed by overnight incubation at 37°C. Random recombinant white colonies were selected and cultured in Luria-Bertani broth containing ampicillin (100 μg/ml), and plasmid extraction was performed with the QIAwell 96-well plasmid purification system (Qiagen). A total of 960 forward and reverse subtracted clones were sequenced, and the sequences were identified based on homology searches with public genetic databases (Swiss-Prot, TrEMBL, PIR, NRL-3D, and GenPept). Sequence data were analyzed and edited for quality and vector sequences by using Phred-Phrap/Consed analysis software.
cDNA microarray construction.
A 2,950-element cDNA microarray chip containing 960 SSH clones was constructed as described elsewhere (34, 35). In brief, 576 forward SSH and 384 reverse SSH clone inserts were amplified by PCR using a primer pair corresponding to the flanking adapter sequences (Clontech). All PCR products were visualized on 1% agarose gels to ensure quality and adequate amplification, followed by purification with MultiScreen PCR plates (Millipore, Bedford, Mass.). PCR products were printed in triplicate onto poly-l-lysine-coated glass slides by employing a Microgrid II robot (BioRobotics, Boston, Mass.). Three bacterial genes (from Pasteurella multocida) and one plant gene (the Arabidopsis thaliana gene encoding the chlorophyll a/b-binding protein [Cab]) were included as negative controls, and total cellular cDNA was included as a positive control. All control elements were spotted 14 times each on the array.
Fluorescent probe labeling and hybridization.
Poly(A)+ RNA (300 ng) purified from APV-infected and mock-infected cells at 2.5, 24, 48, and 96 h p.i. was reverse transcribed by using an oligo(dT)12-18 primer, deoxynucleoside triphosphates, aminoallyl dUTP, and Superscript II reverse transcriptase (Invitrogen Life Technologies). cDNAs from infected and control samples were labeled with the monofunctional dyes Cy5 and Cy3 (Amersham, Piscataway, N.J.), respectively, followed by hybridization with the spotted array at 67°C for 5 h. For all hybridizations, A. thaliana Cab gene mRNA (Stratagene, La Jolla, Calif.) was spiked into the cDNA synthesis reactions of both samples, and information from this mRNA was later used for data normalization. Prior to final hybridizations, hybridization conditions, such as the optimum amount of labeled probe, the hybridization buffer composition, temperature, and washing stringencies, were standardized to ensure maximum specificity and sensitivity, and all hybridizations were performed with the optimized conditions, at which there was no appreciable cross-hybridization with the control spots. The complete experiment, including primary culture of cells, virus infection, and RNA isolation, was performed twice, with probe synthesis and hybridizations performed at least twice per independent experiment. This plan resulted in 12 independent Cy5/Cy3 intensity ratio data points for each spotted cDNA at each time point. Images of the hybridized arrays were acquired by laser confocal scanning (Scanarray 5000; GSI Lumonics, Watertown, Mass.) and analyzed with Quantarray, version 3.0 (GSI Lumonics), and Spotfire Decision Site, version 6.5 (http://www.spotfire.com), software.
Data analysis.
Prior to the import of gene identifiers and descriptions into the data set, the raw data were analyzed as follows for quality control: (i) individual spots were flagged for quality and discarded during analysis and quantification of spot intensities, (ii) local background fluorescence was subtracted from the fluorescence intensity of each spot for both the Cy5 and Cy3 channels, (iii) the entire data set was normalized for both channels based on the A. thaliana (Cab gene) spike control, (iv) spots with high background intensity for either dye were eliminated, (v) replicate spots that had Cy5/Cy3 intensity ratios 2 or more standard deviations higher than the mean intensity ratio were discarded, and (vi) replicate spot ratios were averaged, and differential expression ratios were determined. Further data analysis and visualizations of expression profiles were performed using Spotfire Decision Site software, version 6.5.
Differential expression analysis by real-time RT-PCR.
The differential expression of a group of 20 selected genes was further validated by real-time quantitative RT-PCR using SYBR green-based detection (ABI) that was run on an ABI 7700 fluorescent sequence detection system (Perkin-Elmer, Foster City, Calif.). Selection of genes for further validation was based on (i) previously known critical functions of the encoded proteins in the context of the host-virus relationship and (ii) their representation of the whole spectrum of expression patterns from very strong induction to weak induction and significant repression by APV infection. The quality and specificity of amplified products were confirmed by visualization on a 2% agarose gel.
RESULTS AND DISCUSSION
We have utilized a combined approach of SSH in conjunction with cDNA microarray analysis to identify genes that are differentially regulated during infection with APV. Of the 960 SSH clones characterized, a total of 507 (53%) were unique genes, with the remainder representing redundant sequences or different fragments of the same transcript. Of these, a slight majority (296 expressed sequence tags, or 58%) had no homologues in the public sequence databases and 48 (9%) were similar to genes with unknown functions in other eukaryotes. The remaining (163) expressed sequence tags had homology to sequences in chickens, mice, and/or humans that had been identified previously.
cDNA microarray analysis revealed that the expression levels of 352 genes were significantly altered over the 96-h period. Of these genes, 268 transcripts showed an increase and 84 showed a decrease in expression levels; a partial list is given in Table 1. Transcripts representing several functional classes were perturbed by APV infection; these included many genes with known roles in blocking virus replication and others with a likely role in facilitating virus propagation. The major functional classes of genes that were transcriptionally altered during infection are summarized below.
TABLE 1.
Host genes regulated in expression by APV infectiona
| Accession no.b | Gene or gene product | Fold change in transcription (SD)c at the following time P.i.:
|
|||
|---|---|---|---|---|---|
| 2.5 h | 24 h | 48 h | 96 h | ||
| Avian pneumovirus gene transcripts | |||||
| EMB, Q9QF48 | Nucleocapsid protein | NC | 19.57 (2.50) | 50.40 (1.87) | 67.56 (5.18) |
| EMB, Q9QF47 | Phosphoprotein | NC | 28.33 (3.16) | 25.99 (2.07) | 58.73 (4.07) |
| EMB, Q90244 | Matrix protein | NC | 14.54 (2.22) | 21.14 (2.66) | 44.93 (2.68) |
| EMB, Q9QD13 | Fusion glycoprotein | NC | 23.72 (2.09) | 35.42 (2.87) | 65.62 (7.10) |
| EMB, Q9QF45 | Matrix glycoprotein M2 ORF2 | 3.04 (0.28) | 24.87 (0.92) | 21.44 (1.55) | 70.08 (6.94) |
| EMB, Q9QF46 | Matrix glycoprotein M2 ORF1 | NC | 20.62 (1.57) | 30.83 (2.92) | 65.66 (3.89) |
| EMB, P87509 | RNA-dependent RNA polymerase | NC | 14.20 (1.08) | 22.73 (1.68) | 37.29 (2.73) |
| IFN-induced transcripts | |||||
| SWP, MX_CHICKd | IFN-induced GTP-binding protein Mx | NC | 14.20 (2.41) | 38.63 (2.09) | 61.38 (6.06) |
| SWP, INI2_PANTRd | IFN-induced protein 6-16 precursor | NC | 17.39 (1.95) | 13.87 (1.84) | 30.64 (2.40) |
| SWP, IFT1_HUMANd | IFN-induced 56-kDa protein | 6.89 (1.87) | 32.28 (3.58) | 19.70 (2.90) | 20.90 (4.13) |
| EMB, AAH04977d | IFN-induced protein with TPR 4 | 4.38 (1.28) | 12.60 (3.12) | 18.67 (3.21) | 6.63 (0.64) |
| SWP, STA1_HUMANd | Signal transducer and activator of transcription 1-α/β | NC | 16.36 (1.98) | 33.77 (3.41) | 54.86 (2.13) |
| EMB, AAH02704 | Similar to signal transducer and activator of transcription 1 | NC | 11.32 (1.37) | 7.89 (1.10) | 9.54 (0.80) |
| SWP, IFT5_HUMAN | Retinoic acid- and IFN-inducible 58-kDa protein (RI58) | 2.80 (0.72) | 31.19 (3.80) | 28.30 (3.12) | 22.23 (3.07) |
| EMB, Q9QXH3d | IFNα-inducible protein P27-H | NC | 15.28 (1.96) | 12.19 (1.26) | 23.67 (4.16) |
| Proinflammatory chemoattractants and adhesion molecules | |||||
| SWP, EMF1_CHICKd | CXC chemokine 9E3 (human IL-8 homologue) | 2.39 (0.57) | 7.30 (1.39) | 39.32 (5.46) | 53.84 (5.30) |
| EMB, O73912 | CXC chemokine K60 | 2.01 (0.57) | 2.43 (0.61) | 18.64 (2.94) | 18.33 (2.84) |
| EMB, O35131d | Complement C3F | NC | 2.24 (0.15) | 10.03 (0.77) | 12.91 (1.20) |
| EMB, O42402d | Ornithokinin receptor | NC | NC | 2.32 (0.36) | 2.98 (0.41) |
| Ubiquitin-proteasome system | |||||
| SWP, PRSX_SPETR | 26S protease regulatory subunit S10B | NC | NC | 2.37 (0.21) | 2.21 (0.35) |
| SWP, UBPI_HUMANd | Ubiquitin carboxyl-terminal hydrolase 18 (Ubp) | 2.13 (0.41) | 11.94 (1.31) | 6.23 (1.32) | 11.83 (2.51) |
| EMB, AAH01874d | Similar to sequestosome 1 | 2.01 (0.24) | NC | 2.29 (0.26) | 7.78 (1.04) |
| SWP, CUL1_MOUSEd | Cullin homologue 1 (CUL-1) | NC | NC | NC | 2.13 (0.56) |
| EMB, O88544 | COP9 complex subunit 4 | NC | NC | 2.00 (0.21) | 2.20 (0.20) |
| Vesicular protein trafficking and fusion | |||||
| EMB, Q9Y4Z6 | Vacuolar protein sorting (Vps45) | NC | NC | 5.18 (1.11) | 4.44 (1.57) |
| EMB, Q9D872 | Signal recognition particle receptor B subunit | NC | NC | NC | 2.30 (0.50) |
| EMB, Q98932 | Rab5C-like protein | NC | 9.45 (0.88) | 30.34 (1.62) | 50.68 (2.39) |
| EMB, Q9JMJ6 | Syntaxin 7 | NC | NC | 2.04 (0.37) | 3.42 (0.74) |
| EMB, Q9Y5P9 | Endocytic receptor Endo180 | NC | 2.38 (0.14) | 3.06 (0.34) | 4.33 (0.15) |
| SWP, CLH1_HUMAN | Clathrin heavy chain 1 (CLH-17) | NC | NC | NC | 2.82 (0.85) |
| Antigen presentation | |||||
| EMB, O42404d | CD80-like protein precursor | NC | NC | 2.68 (0.32) | 4.67 (0.56) |
| Transcription regulation | |||||
| SWP, TMF1_HUMANd | TATA element modulatory factor (TMF) | NC | NC | 3.24 (0.28) | 4.83 (0.53) |
| SWP, PTB_PIG | Polypyrimidine tract-binding protein (PTB) | NC | NC | NC | 2.48 (0.38) |
| EMB, O95320 | U5 snRNP-specific 40-kDa protein | NC | NC | 4.58 (0.53) | 7.16 (1.88) |
| EMB, Q9WVG7 | Odd-skipped related 1 protein | NC | −1.89 (0.06) | −2.15 (0.08) | −2.02 (0.11) |
| Translation | |||||
| SWP, IF37_MOUSE | Translation initiation factor 3 subunit (eIF-3) | NC | 2.03 (0.29) | 5.18 (0.65) | 6.45 (0.78) |
| SWP, RL3_BOVIN | 60S ribosomal protein L3 | NC | NC | 9.22 (0.60) | 11.15 (0.98) |
| SWP, PDI_CHICK | Protein disulfide isomerase (PDI) | NC | NC | −2.32 (0.03) | −2.11 (0.01) |
| SWP, IF2P_HUMAN | Translation initiation factor 2 (IF-2) | NC | −2.20 (0.06) | −3.20 (0.03) | NC |
| EMB, Q9PTD6 | Ribosomal protein S6 | NC | NC | −3.00 (0.01) | −1.87 (0.04) |
| SWP, RL5B_XENLA | 60S ribosomal protein L5B | NC | NC | −2.67 (0.02) | −2.54 (0.05) |
| SWP, EF1A_CHICKd | Elongation factor 1-alpha 1 | NC | NC | −2.46 (0.03) | NC |
| SWP, NPM_CHICK | Nucleophosmin (NPM) (nucleolar phosphoprotein B23) | NC | NC | −2.18 (0.06) | NC |
| Cell growth and proliferation | |||||
| SWP, QSP_CHICK | Quiescence-specific protein precursor (P20K) | NC | 3.97 (0.81) | 5.02 (0.33) | 7.92 (0.64) |
| SWP, DIA_DROME | Diaphanous protein | NC | NC | −2.06 (0.03) | NC |
| Signal transduction | |||||
| EMB, Q9U9S7 | Adenylyl cyclase | 2.18 (0.16) | 8.91 (0.61) | 9.47 (0.23) | 16.52 (2.03) |
| EMB, Q08623 | PKC-zeta-interacting protein (ZIP) | NC | NC | 2.41 (0.18) | 6.76 (0.89) |
| RNA regulation | |||||
| SWP, RSFR_CHICK | RNase homologue precursor (RSFR) | NC | NC | NC | 2.54 (0.59) |
| Plasma/nuclear membrane proteins | |||||
| SWP, SSGP_VOLCA | Sulfated surface glycoprotein 185 (SSG 185) | 2.22 (0.28) | 20.78 (1.73) | 16.18 (0.89) | 40.35 (4.27) |
| SWP, N107_HUMAN | Nuclear pore complex protein NUP107 | NC | NC | 2.23 (0.28) | 2.08 (0.33) |
| Heat shock and stress response | |||||
| EMB, Q9JII7 | Arsenite-inducible RNA-associated protein | NC | NC | 2.95 (0.27) | 4.77 (0.70) |
| SWP, HS47_CHICK | 47-kDa heat shock protein precursor | NC | NC | −2.33 (0.03) | −2.05 (0.02) |
| SWP, ENPL_CHICK | Heat shock 108-kDa protein | NC | NC | −2.31 (0.04) | NC |
| EMB, O60884 | DNAJ protein | NC | NC | −1.98 (0.03) | NC |
| Cell cycle | |||||
| SWP, CGA2_CHICK | Cyclin A2 | NC | NC | 2.91 (0.56) | 3.95 (1.22) |
| SWP, CCT2_HUMAN | Cyclin T2 | NC | NC | 3.18 (0.54) | 4.82 (0.82) |
| Transporter proteins | |||||
| SWP, ABF2_HUMAN | Iron-inhibited ABC transporter 2 (ABCF2) | NC | NC | NC | 3.29 (0.59) |
| SWP, ATCP_HUMAN | Calcium-transporting ATPase 1 plasma membrane | NC | NC | NC | 2.22 (0.44) |
| Tumor-associated proteins | |||||
| EMB, Q9H3G6 | Melanoma differentiation associated protein-5 | NC | NC | 2.15 (0.20) | 2.56 (0.32) |
| EMB, O95140 | CPRP1 | NC | NC | 2.41 (0.57) | 3.37 (0.90) |
| SWP, CSR2_CHICK | Cysteine-rich protein 2 (CRP2) | NC | NC | −1.71 (0.04) | −3.22 (0.03) |
| EMB, O00199 | Integral membrane serine protease seprase | NC | −1.75 (0.06) | −3.32 (0.02) | −4.05 (0.04) |
| EMB, Q9JJK2 | Testis-specific adriamycin sensitivity protein | NC | NC | NC | −2.00 (0.10) |
| Cytoskeleton and motor proteins | |||||
| SWP, ACT2_XENTR | Actin alpha 2 | NC | NC | −2.79 (0.03) | −4.12 (0.01) |
| EMB, Q61852 | Actin gamma 2 | NC | NC | −2.98 (0.02) | −3.84 (0.02) |
| SWP, ACT2_HALRO | Actin 2/4/4A | NC | NC | −2.56 (0.04) | −3.68 (0.02) |
| SWP, ACT1_ORYLA | Actin (MA1) | NC | NC | −2.64 (0.03) | −4.08 (0.02) |
| EMB, Q9QZ83 | Gamma actin-like protein | NC | NC | −2.13 (0.02) | NC |
| PIR, S06117 | Myosin heavy chain | NC | NC | −3.64 (0.03) | −2.97 (0.05) |
| EMB, Q9NU67 | Retinitis pigmentosa 2 (RP2) | NC | NC | 5.97 (0.59) | 8.67 (1.61) |
| SWP, KIF2_HUMAN | Kinesin-like protein KIF2 | NC | NC | 3.75 (0.56) | 6.63 (0.87) |
| Protein degradation/lysosomal | |||||
| SWP, FUCO_HUMAN | Tissue alpha-l-fucosidase precursor | NC | NC | −2.44 (0.03) | −3.62 (0.02) |
| Mitochondrial protein | |||||
| SWP, COX1_CHICK | Cytochrome c oxidase polypeptide 1 | NC | NC | −1.69 (0.04) | −1.73 (0.03) |
| Extracellular matrix and cell adhesion | |||||
| SWP, HAS2_CHICK | Hyaluronan synthase 2 | NC | NC | 2.00 (0.25) | 4.60 (0.95) |
| EMB, O35103 | Osteomodulin | NC | NC | 3.13 (0.43) | 4.08 (0.96) |
| SWP, ITB1_CHICK | Integrin beta-1 precursor (CSAT antigen) | NC | NC | −1.83 (0.08) | −3.42 (0.05) |
| SWP, TSP2_CHICKd | Thrombospondin 2 precursor | NC | NC | −3.39 (0.04) | −8.24 (0.01) |
| EMB, Q91002 | Thrombospondin-4 | NC | NC | NC | −3.35 (0.10) |
| SWP, CA11_CHICK | Collagen alpha 1 (I) chain precursor | NC | NC | −2.28 (0.05) | −2.74 (0.03) |
| SWP, CA13_CHICK | Collagen alpha 1 (III) chain | NC | NC | −1.77 (0.03) | −2.65 (0.02) |
| SWP, CA18_HUMAN | Collagen alpha 1(VIII) chain precursor | NC | NC | NC | −1.82 (0.19) |
| SWP, CA21_CHICKd | Collagen alpha 2 (I) chain precursor | NC | NC | −2.34 (0.02) | −4.32 (0.01) |
| SWP, CA25_HUMAN | Collagen alpha 2(V) chain precursor | NC | NC | −2.12 (0.02) | −3.41 (0.02) |
| EMB, Q9DE68 | Dermatan sulfate proteoglycan decorin | NC | NC | NC | −2.15 (0.03) |
| EMB, O94769 | Extracellular matrix protein | NC | NC | −2.68 (0.02) | −2.26 (0.03) |
| SWP, LUM_CHICKd | Lumican precursor (keratan sulfate proteoglycan) | NC | NC | −2.09 (0.02) | −4.93 (0.02) |
| SWP, PEDF_MOUSE | Pigment epithelium-derived factor precursor | NC | NC | −2.09 (0.03) | −2.67 (0.05) |
| SWP, LYOX_CHICK | Protein-lysine 6-oxidase precursor | NC | NC | −1.71 (0.07) | NC |
| Metabolism | |||||
| SWP, RPIA_MOUSE | Ribose 5-phosphate isomerase | NC | NC | 4.55 (0.50) | 8.80 (0.66) |
| EMB, AAH02559 | High-glucose-regulated protein 8 | NC | 3.27 (0.24) | 13.94 (1.50) | 19.51 (2.20) |
| SWP, PKBS_BOVIN | Peripheral-type benzodiazepine receptor (PBR) | NC | 3.04 (0.20) | 9.07 (0.86) | 14.11 (0.86) |
| SWP, PWP2_HUMAN | Periodic tryptophan protein 2 homologue (PWP2) | NC | NC | 3.05 (0.64) | 4.02 (0.40) |
| SWP, P5CS_HUMAN | Delta 1-pyrroline-5-carboxylate synthetase (P5CS) | NC | NC | −3.24 (0.02) | −2.55 (0.03) |
| SWP, GNA1_DROME | Probable glucosamine-phosphate N-acetyltransferase | NC | NC | 2.35 (0.30) | 3.17 (0.58) |
| EMB, O15460 | Prolyl 4-hydroxylase alpha (ii) subunit | NC | NC | NC | 2.12 (0.49) |
| SWP, PNAD_PIG | Protein N-terminal asparagine amidohydrolase | NC | NC | 3.39 (0.41) | 2.92 (0.36) |
| EMB, AAB34334 | Protein phosphatase 1 gamma 1 | NC | NC | NC | 2.04 (0.44) |
| SWP, ADRO_MOUSE | NADPH: adrenodoxin oxidoreductase | NC | NC | 2.12 (0.31) | 2.73 (0.27) |
| SWP, AT5C_HUMAN | Potential phospholipid-transporting ATPase VC | NC | NC | −2.04 (0.01) | −2.63 (0.06) |
| EMB, AAH01741 | Similar to glyoxalase I | NC | NC | −2.91 (0.06) | −2.67 (0.04) |
| EMB, AAD02474 | Glyceraldehyde-3-phosphate dehydrogenase | NC | NC | −2.23 (0.01) | NC |
| SWP, ENOA_CHICK | Alpha enolase (2-phospho-D-glycerate hydrolyase) | NC | NC | −2.11 (0.03) | NC |
| SWP, APA1_CHICK | Apolipoprotein A-1 precursor | NC | NC | NC | −1.93(0.02) |
| SWP, FTDH_RAT | 10-Formyltetrahydrofolate dehydrogenase | NC | −1.77 (0.07) | NC | NC |
| SWP, ATP6_CHICK | ATP synthase A chain | NC | NC | −1.93 (0.03) | NC |
| Cell physiology and development | |||||
| SWP, FKBB_HUMAN | 12.6-kDa FK506-binding protein | NC | NC | −1.99 (0.05) | NC |
| SWP, FM14_MOUSE | Formin 1 isoform IV | NC | NC | NC | 2.69 (0.69) |
| SWP, GRP1_PETHY | Glycine-rich cell wall structural protein 1 | NC | NC | −3.44 (0.02) | −3.83 (0.04) |
| EMB, P97487 | Hippocampal amyloid protein | NC | NC | −1.82 (0.04) | −1.86 (0.06) |
| Unknown function | |||||
| EMB, Q9D0V1 | 1110067L12RIK protein | NC | NC | NC | 2.11 (0.37) |
| EMB, Q9D9V7 | 1700027J05RIK protein | NC | NC | NC | 2.28 (0.31) |
| EMB, Q9D9D0 | 1700101G24RIK protein | NC | NC | 2.44 (0.38) | 3.45 (0.69) |
| EMB, Q9CY20 | 2510027N19RIK protein | NC | NC | 2.67 (0.38) | 5.26 (1.03) |
| EMB, Q9D0E4 | 2610021K23RIK protein | NC | NC | NC | 2.82 (0.38) |
| EMB, Q9D2N6 | 4631426J05RIK protein | NC | NC | 2.92 (0.41) | 7.45 (1.23) |
| EMB, Q9D5P8 | 4930402H24RIK protein | NC | NC | 2.43 (0.36) | 4.78 (0.95) |
| EMB, Q9NUM1 | CDNA FLJ11277 fis, clone PLACE1009404 | 2.19 (0.32) | 3.67 (0.44) | 2.37 (0.24) | 5.29 (0.61) |
| EMB, Q9H8V6 | CDNA FLJ13201 fis, clone NT2RP3004498 | NC | NC | NC | 3.15 (0.39) |
| EMB, Q9H812 | CDNA FLJ14005 fis, clone Y79AA1002361 | NC | NC | 2.61 (0.49) | 3.08 (0.47) |
| EMB, Q9H7T5 | CDNA FLJ14273 fis, clone PLACE1004913 | NC | NC | −2.16 (0.05) | NC |
| EMB, Q9NXP3 | CDNA FLJ20129 fis, clone COL06190 | NC | NC | NC | −3.59 (0.10) |
| EMB, Q9NWV4 | CDNA FLJ20580 fis, clone REC00516 | NC | NC | −2.07 (0.07) | −3.18 (0.03) |
| EMB, Q9VKV8 | CG5343 protein | NC | NC | 2.21 (0.36) | 2.70 (0.52) |
| EMB, Q9VLT1 | CG8683 protein | NC | NC | NC | −3.31 (0.10) |
| IGI, RRN01291 | KIAA0613 protein | NC | NC | NC | 2.31 (0.45) |
| EMB, AAH02380 | Unknown (protein for IMAGE:2961244) | NC | NC | −2.05 (0.02) | −2.28 (0.03) |
| GEN, CAC35438 | Unnamed protein product | NC | 2.98 (0.39) | 2.94 (0.28) | 7.72 (1.05) |
| EMB, AAH05857 | Unknown (protein for MGC:2910). | NC | 2.89 (0.48) | 3.64 (0.51) | 8.65 (0.94) |
| EMB, AAH02575 | Unknown (protein for IMAGE:3161568) | NC | NC | 3.58 (0.60) | 6.57 (0.92) |
| GEN, CAC22485 | Unnamed protein product | NC | 2.28 (0.49) | 3.56 (0.60) | 5.59 (1.38) |
| EMB, CAB66533 | Hypothetical 22.5-kDa protein | NC | NC | 2.69 (0.65) | 4.74 (0.83) |
| EMB, Q9Y2J9 | KIAA0995 protein | NC | NC | 3.17 (0.52) | 4.31 (0.39) |
| EMB, Q9ULD7 | KIAA1283 protein | NC | NC | −2.15 (0.06) | −2.38 (0.06) |
| EMB, Q9P2H7 | KIAA1370 protein | NC | NC | 2.38 (0.32) | 3.31 (0.51) |
| EMB, Q9H3F0 | MSTP034 | NC | NC | NC | 2.41 (0.51) |
| EMB, Q9JM62 | Polyposis locus protein 1-like 1 | NC | NC | −1.74 (0.04) | −1.76 (0.05) |
| EMB, Q9D0Z1 | Q9D0Z1 1110039P19RIK protein | NC | NC | −2.64 (0.03) | −3.10 (0.05) |
| EMB, Q9H8C3 | Q9H8C3 CDNA FLJ13770 FIS, CLONE PLACE4000269 | NC | NC | 2.42 (0.30) | 4.10 (0.55) |
| IGI, RCHU01422 | RCHU01422 hypothetical protein | NC | NC | 2.45 (0.17) | 3.01 (0.40) |
| SWP, RNF6_HUMANd | Ring finger protein 6 | NC | NC | NC | 4.38 (1.38) |
| EMB, AAH06414 | Similar to KIAA0952 protein | NC | NC | 3.78 (0.48) | 6.86 (0.99) |
| EMB, Q9LGZ9 | Genomic DNA, chromosome 3, BAC clone:F1D9 | NC | 17.78 (1.60) | 30.55 (1.67) | 65.98 (3.70) |
| Hypothetical proteins | |||||
| EMB, Q9Y520 | Hypothetical 295.8-kDa protein | NC | NC | NC | 3.74 (0.55) |
| EMB, Q9H0P6 | Hypothetical 38.9-kDa protein | NC | NC | NC | 2.73 (0.33) |
| SWP, YG1W_YEAST | Hypothetical 44.2-kDa protein in RME1-TFC4 intergenic region | NC | NC | 4.83 (0.61) | 12.00 (1.17) |
| EMB, Q9Y3V7 | Hypothetical 63.3-kDa protein | NC | NC | NC | −2.04 (0.09) |
| SWP, Y052_HUMAN | Hypothetical protein KIAA0052 | NC | 2.57 (0.47) | 2.57 (0.15) | 11.54 (2.65) |
| EMB, AAH02877 | Similar to hypothetical protein FLJ1158 | NC | NC | NC | 4.33 (1.47) |
| EMB, AAH06213 | Similar to RIKEN cDNA 1200013A08 gene | NC | NC | 2.20 (0.50) | 2.04 (0.36) |
The APV infection time course experiment was repeated twice, and hybridization of SSH cDNA chips with fluorescent labeled probes was repeated twice per experiment. The table lists clones isolated by SSH and validated to be differentially expressed by microarray analysis. Only genes with twofold or greater changes in transcription in APV-infected cells relative to that in uninoculated controls are shown. Differentially expressed genes with no homologues in databases, which are designated “unknown,” are not shown.
From the Swiss Prot (SWP) and EMBL (EMB) databases. Regulated transcripts are classified according to functional groups.
Values are average Cy5-to-Cy3 ratios from four replicate experiments, along with their standard deviations which are given (in parentheses) only for transcripts showing twofold or greater changes in expression. NC, no change. SD, standard deviation.
Results were confirmed by real-time RT-PCR.
IFN response.
Among the genes and pathways altered, transcriptional change in the interferon (IFN)-regulated class of antiviral genes was the most striking. Several IFN-stimulated genes (ISGs) were strongly induced (ranging from 2- to 61-fold induction); their expression showed increases as early as 2.5 h p.i. and continued to increase over the entire 96-h time course. Alpha/beta IFNs are a family of cytokines that are produced in response to viral infection (33) and constitute a first line of innate defense against viral infection by inducing the expression of several ISGs, of which the antiviral protein Mx, the double-stranded-RNA-activated protein kinase R (PKR), the 2′-5′ oligoadenylate synthetase (2-5A), and the IFN-induced 56-kDa protein (IFI-56K) are well characterized (14, 33).
The gene encoding the Mx protein was one of the most strongly induced (more than 60-fold) during infection. Mx proteins constitute a group of large antiviral GTPases that are key components of the IFN-induced antiviral defenses (39) and mediate their effect by sequestering viral nucleocapsids, thus rendering them inaccessible for replication (21). Owing to their significance, Mx proteins are evolutionarily conserved in all vertebrates analyzed so far, including chickens, mammals, and fish. Activity against orthomyxoviruses, paramyxoviruses, rhabdoviruses, bunyaviruses, and togaviruses has previously been demonstrated for human MxA, and activity against orthomyxoviruses has been demonstrated for mouse Mx1 (33). Although up-regulation of Mx expression has been observed during infection with a wide range of viruses such as hepatitis C virus, herpes simplex virus, and human cytomegalovirus (1, 26, 53), its antiviral potency in these cases remains to be determined. Similarly, it is not known if the avian Mx protein has an antiviral effect toward APV or other avian respiratory viruses. Sequence analysis suggests that the avian Mx gene contains an IFN-stimulated response element (ISRE), common to all ISG promoters, that is strongly activated upon IFN stimulation (39). The massive induction of Mx gene expression during the course of APV infection suggests its possible activity as an antiviral ISG, and thereby it could have an important role in APV pathogenesis.
The gene encoding the IFI-56K protein was up-regulated early (2.5 h) in infection and continued to accumulate over time. It is known that IFN creates an antiviral state, in part, by halting mRNA translation. The IFI-56K protein contains eight tetratricopeptide repeat (TPR) motifs that mediate protein-protein interaction by binding the P48 subunit of translation initiation factor eIF-3, thus inhibiting protein synthesis (14). By using the gene array approach, enhanced expression of IFI-56K was also found during infections with hepatitis C virus, herpes simplex virus, and cytomegalovirus (1, 26, 53). The considerable (6- to 32-fold) induction of the IFI-56K gene suggests a role for this protein in inhibiting the translation complex during APV infection. Interestingly, a gene encoding one of the eIF-3 subunits in the translational machinery predicted to be inhibited by IFI-56K showed increased (two- to sixfold) expression after the onset of the increase in IFI-56K transcription; this gene may represent a virus-induced countermeasure to overcome the IFI-56K-mediated block of eIF-3, an intriguing hypothesis that requires further investigation.
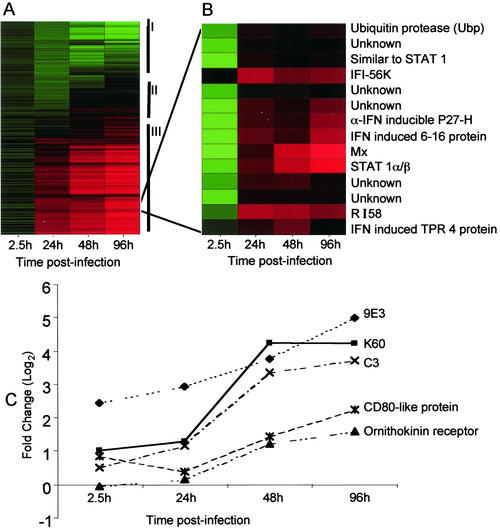
The analysis also showed that the genes encoding RI58 and IFN-induced protein with TPR 4 were up-regulated. Both of these are ISGs whose roles in the IFN pathway remain to be elucidated. Interestingly, however, both possess TPR motifs, have high homology with IFI-56K (50), and in our studies were found to be up-regulated with profiles similar to that of IFI-56K (Fig. 1B). It is therefore tempting to speculate that they might have a collaborative regulatory function similar to that of IFI-56K during infection.
FIG. 1.
Alteration in host transcriptional program by APV infection. (A) The expression pattern of the 960 cDNAs analyzed by microarray is represented as a hierarchical cluster. Each row represents an individual cDNA element spotted on the array, and each column represents the expression states of cDNAs at a particular time point p.i. Each expression data point represents the ratio of the fluorescence intensity of the cDNA from APV-infected cells to the fluorescence intensity of the cDNA from mock-treated reference cells and is the average value of 12 data points. The cluster is subdivided into three groups indicated by roman numerals at the right, consisting of genes that were repressed (I) (green), genes that were induced (III) (red), and genes whose expression did not change (II) (black). (B) Upon principal-component analysis, all IFN-α/β-inducible genes and five unknown genes clustered together, exhibiting very similar temporal profiles. Red represents up-regulation of expression. (C) The chemokine (9E3, K60), complement (C3), and adhesion molecule (CD80-like protein, ornithokinin receptor) genes whose transcription was perturbed by APV infection also follow similar expression dynamics and cluster together.
The transcript encoding STAT1, a latent cytoplasmic transcription factor that plays a central role in the IFN-α/β signaling pathway (6), was strongly induced by APV (>50-fold). STAT1 not only serves as an IFN signaling molecule, but its own expression is also induced by IFN (7). Although the IFN transcript was not isolated by SSH, up-regulation of STAT1, a hallmark of the IFN signaling cascade, suggests that the induction of ISGs was perhaps due to IFN stimulation. However, this does not exclude the possibility that induction of a few or all ISGs identified here may be partially attributed to APV infection itself and its double-stranded RNA replication intermediates by their own IFN-independent signaling pathways. Overall, the data suggest that the increased expression of STAT1 may account for the early and strong up-regulation of the ISGs observed soon after APV infection.
It is interesting that PKR and 2-5A were not isolated here by SSH as differentially expressed genes. Either they were not induced during infection, were activated to a level below the assay detection limit, or were indeed isolated but are among the unknown differentially expressed cDNAs that have no homologue in the database, since the chicken sequences for these genes are not known.
Together, the data reveal massive induction of ISGs during APV infection. While the exact role of this enhanced ISG expression during APV infection is unclear, it is noteworthy that paramyxoviruses in general are known to block the IFN signaling cascade so as to circumvent IFN antiviral responses in a virus-specific fashion (10). Similarly, the human and bovine respiratory syncytial viruses (HRSV and BRSV, respectively), both close relatives of APV, although they do not inhibit IFN-α/β or IFN-γ production or signaling, do antagonize IFN-induced antiviral proteins such as Mx through the action of the viral NS1 and NS2 proteins and thus are resistant to the effects of IFN (37). Our analyses suggest that, like HRSV and BRSV, APV does not block IFN production or signaling, as indicated by up-regulation of several ISGs. However, unlike these other viruses, APV lacks the genes encoding NS1 and NS2 proteins and thus may be sensitive to the antiviral effects of Mx and other ISGs. Alternatively, if APV does possess any IFN resistance mechanism, it could conceivably operate as an event downstream of the IFN signaling cascade and at the level of action of ISG-encoded proteins. Future investigations to elucidate the effects of IFN-triggered proteins identified in this study on APV replication will be of immense significance for understanding their role in APV biology.
Proinflammatory chemokines, complement, and adhesion molecules.
Chemokines are a superfamily of chemoattractant cytokines that recruit immune cells to the site of infection with viruses, bacteria, or fungi. Based on the number and relative positions of the conserved cysteine residues, they are classified into the CXC, CC, C, and CX3C subfamilies (32). Although well known for their immunomodulatory and chemoattractive activities, chemokines have been implicated in pathological injury caused by severe inflammation. Here we observed significant induction by APV of genes that are well-known mediators of proinflammatory responses. The genes encoding the proinflammatory CXC chemokine mRNAs, including 9E3 (chicken homologue of human interleukin 8 [IL-8]) (19) and K60 (42), two of the four chemokines known in chickens, were considerably up-regulated (53- and 18-fold, respectively). While these proteins have not been previously implicated in APV pathogenesis, their chemoattractant properties are in agreement with the type of cellular infiltrate present in the avian respiratory tract during infection. 9E3 and K60 are both CXC chemokines and strong chemoattractants for heterophils (the avian equivalents of neutrophils) (42), which are the predominant infiltrating leukocytes in the APV-infected respiratory tract. This observation is consistent with the HRSV-induced up-regulation of IL-8 expression (22, 52) and neutrophil infiltration seen during human infections; along with other chemokines, IL-8 is believed to be the cause of airway inflammation and immunopathogenic injury caused by neutrophils (47, 52). While the exact mechanisms governing the inflammation and recruitment of circulating heterophils into the respiratory tracts of APV-infected birds remain unknown, induction of 9E3 and K60 provides a plausible explanation for the extensive inflammation and swelling of the face, inflammatory heterophil infiltration, exudation, and edema of sinuses of birds during APV infection.
There was strong induction of the gene encoding the complement pathway protein C3 (2- to 12-fold induction over the time course). The complement system is a critical component of the host innate defense mechanisms against a variety of pathogens. It is composed of multiple factors that act in concert to kill virus-infected cells, bacteria, and parasites (reviewed in reference 11). The C3 complement is the most abundant protein, has leukocyte chemoattractant properties, and plays a pivotal role in initiating the complement cascade. The transcriptional activation and role of C3 complement in APV infection have not been reported previously. In human infections with HRSV, it has been shown that HRSV stimulates C3 production in infected cells, which mediates cell lysis through neutrophil-dependent cytotoxicity (20); this, in conjunction with chemokines, is considered to be the basis of infection-related pathology.
The results also show elevated expression (>2-fold) of the gene encoding the ornithokinin receptor, the only known chicken receptor for proinflammatory peptides known as kinins, which are responsible for changes associated with inflammation such as vasodilation, increased vascular permeability, and pain (38). Although further investigation is required to confirm the role of activated 9E3, K60, C3, and ornithokinin receptor genes in APV pathogenesis, the results strongly suggest that the inflammation and migration of heterophils into the respiratory tract triggered by these factors are likely to play an important role in APV-related pathology.
Ub-proteasome pathway.
Several genes belonging to the ubiquitin (Ub)-proteasome pathway, including those encoding the 26S proteasome subunit p42, COP9 complex subunit 4, and E3 ubiquitin ligase CUL-1, the gene similar to sequestosome 1, and the gene encoding deubiquitinating enzyme 18, were induced during APV infection. The functions of many of these genes have been well characterized in simple and higher eukaryotes. The Ub-proteasome pathway comprises a major proteolytic system that regulates the dynamics of proteins (17). Multi-Ub chains are added to proteins to tag them for degradation by the 26S proteasome, a multiproteolytic enzyme complex. Ubiquitination is accomplished by a set of enzymes including E1 (Ub activating), E2 (Ub conjugating), and E3 (Ub-substrate ligase). A gene similar to sequestosome 1 encodes a nonproteasomal Ub binding protein, p62, that binds preferentially with multi-Ub and forms a novel cytoplasmic structure called a sequestosome, which contains Ub-protein complexes and is implicated in Ub-mediated signaling and/or protein degradation by virtue of its role as a temporary storage place for Ub-protein complexes (41, 46). Induction of several Ub-proteasome pathway proteins by APV indicates enhanced activity of this proteolytic machinery during infection. In conjunction with the IFI-56K inhibition of protein translation described above, this may be a parallel or alternative host antiviral response to enhance proteolysis. This hypothesis is supported by the fact that several of the proteasome system genes are induced by IFN-α/β stimulation (8). Alternatively, this response may serve to enhance the production of viral antigenic peptides for display in the context of major histocompatibility complex class I molecules to virus-specific T lymphocytes for triggering of an antiviral immune response.
Given the induction of the Ub system genes that mediate proteolysis, an intriguing observation is the up-regulation of the Ub protease (Ubp) or deubiquitinating enzyme during APV infection. Ubp is known to cleave Ub tags from proteins and render them resistant to proteasome degradation (17). Induction of this enzyme together with the gene similar to sequestosome 1 occurred by 2.5 h p.i., and expression remained elevated (maximum, 11.9-fold) throughout the experiment. While Ubps are substrate specific and the functional relevance of the observed up-regulation remains to be determined, the enhanced expression of Ubp can be interpreted in several ways. (i) It may be a virus-directed virulence mechanism to prevent Ub-mediated degradation of nascent viral proteins in order to ensure successful production of virus progeny. In this context, it is noteworthy that Ubp and IFI-56K have similar induction profiles (Fig. 1B). Both genes are up-regulated very soon after infection, at 2.5 h p.i., and remain activated throughout the experiment, suggesting that activation of Ubp may be a virus-induced response to stabilize proteins in the presence of an IFI-56K-mediated translation block. (ii) Since proteasome-generated peptides are presented in the context of major histocompatibility complex class I molecules on the surfaces of infected cells to T lymphocytes, Ubp activation may be an exquisite virulence mechanism to prevent antigen presentation and escape host immune recognition right from the beginning of host invasion. This could possibly be the basis of the poor adaptive immunity of poultry flocks, which repeatedly experience APV outbreaks with the same virus strain even during their short life spans and are more susceptible to secondary infections when exposed to APV. (iii) The increased activity of Ubp leads to higher levels of free intracellular Ub, which along with other vesicular-system proteins, plays a key role in the maturation and budding of a variety of viruses and could have similar functions in APV replication.
Interestingly, a variety of other virus types exploit the Ub-proteasome pathway to facilitate their replication and evade host immunity. For instance, porcine reproductive and respiratory syndrome (PRRS) virus (51) and human cytomegalovirus (53) infections cause up-regulation of Ubp expression, which in the case of the PRRS virus is thought to be a mechanism for escaping host immunity (51).
Vesicular protein trafficking and fusion.
Enveloped viruses are packaged, mature, and exit from the host cell by budding (pinching off) from the cell membrane. Viral proteins and nucleocapsids utilize the host cell machinery for their transport to the plasma membrane. An interesting group of vacuolar protein sorting (Vps) genes showed altered expression upon APV infection. These include genes involved in protein sorting, trafficking, vesicle formation, membrane tethering, and fusion such as those encoding Vps45, Rab5c-like protein, syntaxin 7, clathrin heavy chain, signal recognition particle receptor, and the endocytic receptor Endo180. The genes encoding the Vps45 and signal recognition particle receptor were induced >5- and >2-fold, respectively, and are thought to play critical roles in vacuolar protein sorting from the Golgi apparatus through the endosomal pathway to the lysosome (vacuole in yeast) (4). The gene encoding the Rab5c-like protein was also substantially induced during APV infection. It functions in the transport of cargo in clathrin-coated vesicles to the early endosomes (36). Rab5c-like proteins are small GTPases that complex with other proteins, perform functions such as transport vesicle formation and tethering of vesicles at their target membranes, and are also known to recruit SNARE proteins (such as syntaxins) that are responsible for fusion of membranes at the Rab5-tethered sites (24). Endo180 functions as a recycling molecule and mediates the transport of glycosylated proteins to the endosomal compartment (40). The membrane-bound proteins are sorted by the Vps pathway for ultimate degradation in the lysosome, which serves as a mechanism for regulating cellular proteins. Syntaxin 7, in addition to fusing membranes, is involved along with Vps45 in the formation of multivesicular bodies, which are endosomal vesicles containing cargos brought in by the Vps machine for lysosomal degradation (27). It appears, therefore, that upon infection, several genes that are all known to serve various functions in the Vps pathway are up-regulated.
The role of the Vps and Ub pathways in the replicative biology of viruses is being increasingly recognized. Over recent years, fairly diverse viruses such as those in the retrovirus, filovirus, and rhabdovirus families have been witnessed to hijack and exploit the host Vps and Ub pathways for their packaging and egress from the cell (31). It is believed that the virus budding phenomenon is topologically similar to multivesicular-body formation in the Vps pathway (in both mechanisms, the membrane invaginates away from the cytoplasm) and that viruses therefore can usurp this machinery in an analogous fashion for viral bud formation and exocytosis (12). Though far from being completely understood, most of the information in this arena comes from studies on human immunodeficiency virus type 1 (HIV-1) and other quite divergent viruses (reviewed in reference 31). Exploitation of the Vps pathway for budding, however, requires a conserved functional consensus sequence known as the late assembly (L) domain. First identified in Rous sarcoma virus (48), L domains have been identified in many RNA viruses, and thus use of the Vps pathway proteins is believed to be a broadly applicable mechanism of virus budding. Although all L domains have the same budding function, the motif sequence is variable across viruses. The sequence motif PPxY in the Rous sarcoma virus Gag and vesicular stomatitis virus matrix (M) proteins and the sequence motifs PTAPP and YxxL in Gag proteins of HIV-1 and equine infectious anemia virus, respectively, have been identified as L domain consensus sequences (16, 31). The current model suggests that the L domain mediates ubiquitination of viral Gag or M protein by Ub ligases. The Ub-tagged protein then recruits Vps pathway proteins (such as Vps4 and Tsg101 in HIV-1) that ultimately mediate the budding process (31). The budding machinery is also known to require active proteasomes, free Ub (15, 16, 30), and the recruitment of the Vps-endosomal protein machinery to target viral proteins for final budding (12, 31). Given the use of an analogous mechanism for budding by fairly divergent virus families, it could conceivably be a pathway for virus egress from the host cell for other viruses as well that have not yet been studied from this perspective.
Importantly, in addition to Vps pathway protein transcripts, many genes with functions in the Ub pathway were also transcriptionally induced upon infection with APV. The molecular mechanism of budding in paramyxoviruses in general is virtually unknown, and the existence of the L domain also has not been reported for any of the family members. Aside from the knowledge that the paramyxovirus matrix (M) protein has a role in virus assembly, the molecular mechanisms that drive M protein-directed virus assembly at the limiting membrane remain undescribed. However, the M proteins of Sendai virus (a paramyxovirus) and human parainfluenza virus (a close relative of APV) (both classified under the same order, Mononegavirales) and that of influenza virus (an orthomyxovirus) have been shown to be capable of budding out of cells in the form of virus-like particles when expressed alone in the absence of other viral proteins (3, 13, 44), a feature reminiscent of the Gag and M proteins of retroviruses, rhabdoviruses, and filoviruses that is attributed to the functional L domain in their sequences (reference 15 and references therein). Significant evidence, therefore, suggests that M proteins of paramyxoviruses (3, 44) complete the budding process perhaps in a fashion similar to that of M and Gag proteins of rhabdoviruses, filoviruses, and retroviruses, which employ their L domains (16, 31) in recruiting host machinery for late stages of budding. Hence, paramyxovirus M proteins might also have a functional domain(s) that could be utilized in a similar manner.
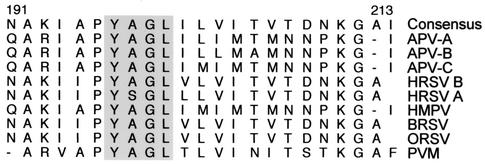
In order to investigate if APV and other pneumoviruses possess any of the known L domain consensus sequences, we aligned the M protein sequences of all known pneumoviruses that infect a variety of host species. Our analysis shows that the M proteins of all pneumoviruses possess the L domain consensus sequence YxxL, located in the C-terminal region (Fig. 2). The putative L domain was found to span residues 197 to 200 with a consensus sequence of YAGL for APV-A, -B, and -C, HRSV B, human metapneumovirus, BRSV, ovine RSV, and pneumonia virus of mice. The sequence YSGL was present instead at the same location in HRSV A and had a single residue substitution of A to S at position 198 but still conformed to the YxxL L domain consensus. Therefore, based on (i) the fact that the M protein is sufficient for mediating budding in a variety of paramyxoviruses, (ii) the presence of a highly conserved putative L domain consensus in M protein sequences of all known pneumoviruses, and (iii) the enhanced expression of several Vps and Ub pathway proteins during APV infection, it is reasonable to hypothesize that APV employs the M protein to usurp the Vps and Ub pathways during late assembly and egress from the host cell. Based on this, we have developed a model for the recruitment of Vps proteins by the APV M protein (Fig. 3) for virus budding. Further studies testing this model and examining the role of the putative L domain of the APV M protein in recruiting Vps proteins are likely to provide important insights into pneumovirus biology and also identify the mechanisms involved in the final stages of paramyxovirus budding and maturation.
FIG. 2.
Amino acid sequence comparison of the M protein fragments (residues 191 to 213) of pneumoviruses. M protein sequences of APV-A (GenBank accession no. X58639), APV-B (U37586), APV-C (AF262571), HRSV B (AAB82433), human metapneumovirus (HMPV) (AF371337), BRSV (NC_001989), ovine RSV (ORSV) (U02470), and pneumonia virus of mice (PVM) (U66893) were aligned by the ClustalW method (DNAStar, Madison, Wis.). The putative L domain (YxGL) spanning residues 197 to 200 is shaded.
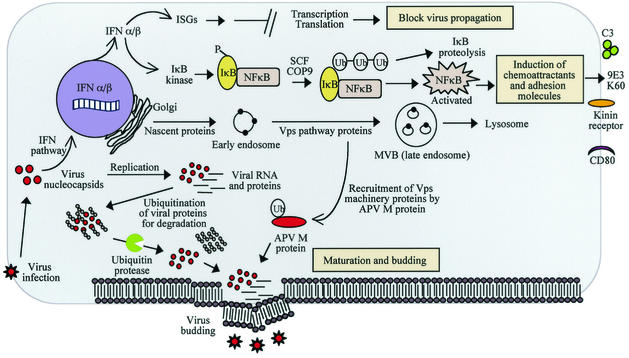
FIG. 3.
Proposed “synthesis” model for APV molecular pathogenesis in an infected host cell. Infection of the host cell by APV triggers host antiviral defenses comprising the IFN pathway, leading to the activation of several ISGs known to impact virus propagation. The data suggest that IFN may be important for the induction of chemokine and adhesion molecule genes by mediating IκB degradation and NF-κB activation facilitated by IκB ubiquitin ligase (SCF). The proposed mechanism of recruitment of host Vps pathway proteins (Vps45, Rab5c, clathrin, Endo180, and syntaxin 7) by APV for completion of its replicative cycle and budding is shown. The model predicts that the putative L domain in the APV M protein undergoes ubiquitination and thereby interacts with one or more of the Vps proteins. The complex thus formed directs more of the Vps pathway factors to the cell membrane to form an active budding complex for the egress of virus progeny from the host cell. The possible role of Ubp in aiding viral proteins to escape proteasomal degradation is also presented.
Antigen presentation.
There was enhanced expression of the gene encoding the CD80-like surface protein, which is the chicken homologue of mammalian CD80 (B7-1) and a ligand for the T-cell proteins CD28 and CTLA4 and which acts as a costimulatory molecule for T-cell activation (28). B7-1 is known to be expressed only on “professional” antigen-presenting cells (macrophages, dendritic cells, and B lymphocytes). Induction of CD80 in response to APV infection in non-antigen-presenting cells was therefore unexpected. Elevated transcription of the CD80 gene was found by SSH and microarray analyses and was also confirmed by real-time RT-PCR (Tables 1 and 2). Interestingly, however, recent studies have identified B7 expression in a variety of cell types, for instance, epidermal, thyroid, and tumor cells (18, 23, 49). Further, the regulation of B7-1 expression in epidermal cells and hepatocytes infected with Leishmania major and hepatitis C virus, respectively, is thought to be important for immune stimulation (25). Moreover, enhancement of B7-1 expression has also been shown to occur in response to IFN-α stimulation (49). Hence, up-regulation of B7-1 transcription in APV infection could be a sequel of IFN induction and may have a role in antigen presentation by the infected cells.
TABLE 2.
Real-time RT-PCR analysis
| Gene namea | Real-time RT-PCR Ctb in:
|
Ct differencec | Fold change in expression by:
|
||
|---|---|---|---|---|---|
| APV-infected cells | Uninfected cells | PCR | Microarray analysis | ||
| Signal transducer and activator of transcription 1-α/β | 18.27 | 22.39 | 4.12 | 17.39 | 54.86 |
| IFN-induced 56-kDa protein | 15.24 | 23.1 | 7.86 | 232.32 | 20.90 |
| IFN-induced protein with TPR 4 | 16.14 | 24.76 | 8.62 | 393.44 | 6.63 |
| CXC chemokine 9E3 (human IL-8 homologue) | 24.82 | 26.38 | 1.56 | 2.95 | 53.84 |
| IFN-induced GTP-binding protein Mx | 17.55 | 26.36 | 8.81 | 448.82 | 61.38 |
| IFN-induced protein 6-16 precursor | 15.48 | 25.7 | 10.22 | 1192.69 | 30.64 |
| IFN-α-inducible protein P27-H | 16.01 | 26.2 | 10.19 | 1168.14 | 23.67 |
| Ubiquitin carboxyl-terminal hydrolase 18 (Ubp) | 16.9 | 22.89 | 5.99 | 63.56 | 11.83 |
| Complement C3F | 17.99 | 19.31 | 1.32 | 2.50 | 12.91 |
| CD80-like protein precursor | 20.68 | 22.4 | 1.72 | 3.29 | 4.67 |
| Ornithokinin receptor | 29.03 | 33.44 | 4.41 | 21.26 | 2.98 |
| Collagen alpha 2(I) chain precursor | 15.18 | 14.32 | −0.86 | −1.82 | −4.32 |
| Cullin homologue 1 (CUL-1) | 23.21 | 23.51 | 0.3 | 1.23 | 2.13 |
| Lumican precursor (keratan sulfate proteoglycan) | 15.81 | 14.35 | −1.46 | −2.75 | −4.93 |
| Thrombospondin 2 precursor | 16.91 | 14.45 | −2.46 | −5.50 | −8.24 |
| Similar to sequestosome 1 | 16.05 | 18.71 | 2.66 | 6.32 | 7.78 |
| TATA element modulatory factor (TMF) | 19.58 | 20.6 | 1.02 | 2.03 | 4.83 |
| Elongation factor 1-alpha 1 | 14.76 | 14.38 | −0.38 | −1.30 | −1.57 |
| Transcriptional repressor protein YY1 (Yin and Yang 1) | 21.84 | 22.59 | 0.75 | 1.68 | 1.89 |
| Ring finger protein 6 | 20.41 | 21.14 | 0.73 | 1.66 | 4.38 |
Genes validated by RT-PCR are also marked and listed in Table 1, except for the transcriptional repressor protein YY1 gene, which was found to be unaltered in expression by both microarray analysis and RT-PCR.
Threshold cycle (Ct) values are shown for the PCR amplification curves of a selected group of genes in APV-infected versus uninfected host cell mRNA at 96 h p.i. generated by a SYBR green-based RT-PCR detection system. Ct is defined as the fractional cycle number at which the fluorescence passes the fixed threshold, which is set at the start of exponential growth of the PCR product.
Cycle difference in the onset of exponential amplification of the target sequence.
Correlation between functional groups and their clustering patterns.
The entire microarray data set (>47,000 individual observations) for all SSH clones was clustered based on the expression profiles of all genes over the entire 96-h time course by using the hierarchical clustering algorithm (Spotfire Decision Site, version 6.5) (Fig. 1A) to help visualize the expression patterns that individual genes followed over time. For instance, the repression profiles of those genes that were downregulated by infection are shown in Fig. 1A, group I. Similarly, group III depicts the dynamics of the gene cluster that showed elevated expression, while those genes whose transcription levels did not change are shown as group II.
The differentially expressed genes were also analyzed by principal-component analysis to identify gene clusters based on the similarity of their expression patterns over time. Twelve distinct clusters of genes whose expression changed in a coordinated manner were obtained. Results for the cluster of IFN response genes and the cluster of chemokine and adhesion molecule genes are shown in Fig. 1B and C, respectively. There appears to be a strong correlation between functional groups and clusters, i.e., genes that perform similar functions share comparable expression profiles. For instance, the dynamics for all IFN response genes were very similar and clustered together as shown (Fig. 1B). This is not altogether surprising, since all IFN response genes are known to be coregulated by transcription factors activated by IFN-α/β or virus infection. Of great interest is the finding that several differentially regulated genes that have no sequence homology in the database and were thus designated “unknowns” followed expression profiles very similar to those of the IFN-responsive genes and clustered with them (Fig. 1B). Based on this observation, it is likely that these unknown transcripts could be (i) novel IFN response factors or (ii) novel chicken ISGs with only limited homology to their known orthologues in other species. Future detailed analysis of these genes in the avian system, particularly examination of their upstream promoter sequences for the presence of ISREs and the ability to be activated by IFN stimulation, would help determine if they belong to the IFN-activated group of genes. Similarly, the chemokine (9E3 and K60), complement C3, and adhesion molecule (CD80 and ornithokinin receptor) mRNAs also had very similar expression profiles and clustered together into a distinct group. Interestingly, 9E3, K60, and C3, all three of which are chemoattractants, have profiles similar to each other, whereas the surface adhesion molecules (CD80-like protein and ornithokinin receptor), though they have patterns of induction similar to those of 9E3, K60, and C3, show dynamics more closely clustered with each other than with 9E3, K60, and C3. Again, a common transcription coregulatory mechanism, possibly mediated by IFN and nuclear factor kappa B (NF-κB), may be responsible for a similar temporal pattern of these genes.
Real-time RT-PCR analysis.
RT-PCR analysis was performed on a set of 20 genes that were found to be either up-regulated, down-regulated, or unchanged in expression by microarray analysis of APV-infected cells. Results obtained by RT-PCR (Table 2) for the genes examined were in accordance with the microarray findings for 90% of the genes. The two genes showing discordance were ring finger protein 6 and CUL-1. They showed elevated expression by microarray analysis at 4.38- and 2.13-fold in APV-infected cells, respectively. Although both genes also showed up-regulation in expression at 1.66- and 1.23-fold, respectively, by RT-PCR, the change was not significant compared to that found by microarray analysis. The difference in the results of these two methods for 10% of the genes tested may possibly be due to differences in the kinetics and sensitivities of these two techniques for various target sequences.
A “synthesis” model of the host response to APV infection.
The availability of expression profiles of host cell genes during the course of infection with APV enables the development of a “synthesis” model depicting how the cell responds to viral infection (Fig. 3). What emerges is a picture of the changes unfolding at the gene expression level in cells infected with APV and a revelation of key genes and pathways with a role in the antiviral and pathophysiological response to APV infection. The model presents a powerful means of visualizing the global changes that occur in an APV-infected cell and provides a facile means of developing testable hypotheses and designing specific experiments to test the roles of individual genes and pathways in an infected cell. For instance, the model predicts that IFN and proinflammatory genes are a predominant immediate host response to infection and that proinflammatory molecules could be the basis of disease-related pathology and likely targets for developing disease intervention strategies. The model also predicts the involvement of the Vps pathway in pneumovirus maturation and budding and provides a mechanism for this process. Further investigation of this mechanism is expected to advance our knowledge of the biology of pneumovirus budding, which could also be extended to other paramyxoviruses.
In sum, the results of our investigations have shed considerable light on host cell molecular responses to APV infection and have provided insights into host defensive measures as well as putative virus-directed mechanisms at the cellular level with implications for disease pathogenesis. The data reported here provide a myriad of mRNAs whose levels change upon host-APV interaction and that suggest an array of testable hypotheses should serve as a foundation for future research to unravel the roles of these genes in the pathogenic mechanisms of APV. Our investigations also demonstrate that study of the molecular details of host-virus interaction is crucial to our understanding of the complexities of the host-virus relationship and is likely to facilitate the development of antiviral agents, immunological interventions, and strategies to alleviate respiratory diseases in avian species.
Acknowledgments
S.M. is supported by a postdoctoral fellowship from the Cargill, Inc., “Animal Genomics” research program. Research in the laboratory of V.K. is supported by competitive awards from the National Institutes of Health, the U.S. Department of Agriculture's NRI and IFAFS programs, the Minnesota Turkey Research and Promotion Council, and the Minnesota Agricultural Experiment Station.
REFERENCES
- 1.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook, J. K. 2000. Avian pneumovirus infections of turkeys and chickens. Vet. J. 160:118-125. [DOI] [PubMed] [Google Scholar]
- 3.Coronel, E. C., K. G. Murti, T. Takimoto, and A. Portner. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J. Virol. 73:7035-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowles, C. R., S. D. Emr, and B. F. Horazdovsky. 1994. Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J. Cell Sci. 107:3449-3459. [DOI] [PubMed] [Google Scholar]
- 5.Dar, A. M., S. Munir, S. M. Goyal, M. S. Abrahamsen, and V. Kapur. 2001. Sequence analysis of the nucleocapsid and phosphoprotein genes of avian pneumoviruses circulating in the U.S. Virus Res. 79:15-25. [DOI] [PubMed] [Google Scholar]
- 6.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 7.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 9.Diatchenko, L., Y. F. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, M. M., and L. F. Fries. 1991. The role of complement in inflammation and phagocytosis. Immunol. Today 12:322-326. [DOI] [PubMed] [Google Scholar]
- 12.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda, N., I. Toida, A. Iwasaki, K. Kawai, and H. Akaza. 2002. Surface antigen expression on bladder tumor cells induced by bacillus Calmette-Guerin (BCG): a role of BCG internalization into tumor cells. Int. J. Urol. 9:29-35. [DOI] [PubMed] [Google Scholar]
- 19.Kaiser, P., S. Hughes, and N. Bumstead. 1999. The chicken 9E3/CEF4 CXC chemokine is the avian orthologue of IL8 and maps to chicken chromosome 4 syntenic with genes flanking the mammalian chemokine cluster. Immunogenetics 49:673-684. [DOI] [PubMed] [Google Scholar]
- 20.Kaul, T. N., H. Faden, R. Baker, and P. L. Ogra. 1984. Virus-induced complement activation and neutrophil-mediated cytotoxicity against respiratory syncytial virus (RSV). Clin. Exp. Immunol. 56:501-508. [PMC free article] [PubMed] [Google Scholar]
- 21.Kochs, G., C. Janzen, H. Hohenberg, and O. Haller. 2002. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc. Natl. Acad. Sci. USA 99:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mastronarde, J. G., B. He, M. M. Monick, N. Mukaida, K. Matsushima, and G. W. Hunninghake. 1996. Induction of interleukin (IL)-8 gene expression by respiratory syncytial virus involves activation of nuclear factor (NF)-κB and NF-IL-6. J. Infect. Dis. 174:262-267. [DOI] [PubMed] [Google Scholar]
- 23.Mbow, M. L., G. K. DeKrey, and R. G. Titus. 2001. Leishmania major induces differential expression of costimulatory molecules on mouse epidermal cells. Eur. J. Immunol. 31:1400-1409. [DOI] [PubMed] [Google Scholar]
- 24.McBride, H. M., V. Rybin, C. Murphy, A. Giner, R. Teasdale, and M. Zerial. 1999. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell 98:377-386. [DOI] [PubMed] [Google Scholar]
- 25.Mochizuki, K., N. Hayashi, K. Katayama, N. Hiramatsu, T. Kanto, E. Mita, T. Tatsumi, N. Kuzushita, A. Kasahara, H. Fusamoto, T. Yokochi, and T. Kamada. 1997. B7/BB-1 expression and hepatitis activity in liver tissues of patients with chronic hepatitis C. Hepatology 25:713-718. [DOI] [PubMed] [Google Scholar]
- 26.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura, N., A. Yamamoto, Y. Wada, and M. Futai. 2000. Syntaxin 7 mediates endocytic trafficking to late endosomes. J. Biol. Chem. 275:6523-6529. [DOI] [PubMed] [Google Scholar]
- 28.O'Regan, M. N., K. R. Parsons, C. A. Tregaskes, and J. R. Young. 1999. A chicken homologue of the co-stimulating molecule CD80 which binds to mammalian CTLA-4. Immunogenetics 49:68-71. [DOI] [PubMed] [Google Scholar]
- 29.Page, R. K., O. J. Fletcher, P. D. Lukert, and R. Rimler. 1978. Rhinotracheitis in turkey poults. Avian Dis. 22:529-534. [PubMed] [Google Scholar]
- 30.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez, O. D., and G. P. Nolan. 2001. Resistance is futile: assimilation of cellular machinery by HIV-1. Immunity 15:687-690. [DOI] [PubMed] [Google Scholar]
- 32.Rossi, D., and A. Zlotnik. 2000. The biology of chemokines and their receptors. Annu. Rev. Immunol. 18:217-242. [DOI] [PubMed] [Google Scholar]
- 33.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 35.Schena, M., D. Shalon, R. Heller, A. Chai, P. O. Brown, and R. W. Davis. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1,000 genes. Proc. Natl. Acad. Sci. USA 93:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schimmoller, F., I. Simon, and S. R. Pfeffer. 1998. Rab GTPases, directors of vesicle docking. J. Biol. Chem. 273:22161-22164. [DOI] [PubMed] [Google Scholar]
- 37.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proeins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroeder, C., H. Beug, and W. Muller-Esterl. 1997. Cloning and functional characterization of the ornithokinin receptor. Recognition of the major kinin receptor antagonist, HOE140, as a full agonist. J. Biol. Chem. 272:12475-12481. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher, B., D. Bernasconi, U. Schultz, and P. Staeheli. 1994. The chicken Mx promoter contains an ISRE motif and confers interferon inducibility to a reporter gene in chick and monkey cells. Virology 203:144-148. [DOI] [PubMed] [Google Scholar]
- 40.Sheikh, H., H. Yarwood, A. Ashworth, and C. M. Isacke. 2000. Endo180, an endocytic recycling glycoprotein related to the macrophage mannose receptor is expressed on fibroblasts, endothelial cells and macrophages and functions as a lectin receptor. J. Cell Sci. 113:1021-1032. [DOI] [PubMed] [Google Scholar]
- 41.Shin, J. 1998. P62 and the sequestosome, a novel mechanism for protein metabolism. Arch. Pharm. Res. 21:629-633. [DOI] [PubMed] [Google Scholar]
- 42.Sick, C., K. Schneider, P. Staeheli, and K. C. Weining. 2000. Novel chicken CXC and CC chemokines. Cytokine 12:181-186. [DOI] [PubMed] [Google Scholar]
- 43.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takimoto, T., K. G. Murti, T. Bousse, R. A. Scroggs, and A. Portner. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J. Virol. 75:11384-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toth, T. E., and P. B. Siegel. 1986. Cellular defense of the avian respiratory tract: paucity of free-residing macrophages in the normal chicken. Avian Dis. 30:67-75. [PubMed] [Google Scholar]
- 46.Vadlamudi, R. K., I. Joung, J. L. Strominger, and J. Shin. 1996. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J. Biol. Chem. 271:20235-20237. [DOI] [PubMed] [Google Scholar]
- 47.Wang, S. Z., H. Xu, A. Wraith, J. J. Bowden, J. H. Alpers, and K. D. Forsyth. 1998. Neutrophils induce damage to respiratory epithelial cells infected with respiratory syncytial virus. Eur. Respir. J. 12:612-618. [DOI] [PubMed] [Google Scholar]
- 48.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.You, X., W. Teng, and Z. Shan. 1999. Expression of ICAM-1, B7.1 and TPO on human thyrocytes induced by IFN-α. Chin. Med. J. 112:61-66. [PubMed] [Google Scholar]
- 50.Yu, M., J. H. Tong, M. Mao, L. X. Kan, M. M. Liu, Y. W. Sun, G. Fu, Y. K. Jing, L. Yu, D. Lepaslier, M. Lanotte, Z. Y. Wang, Z. Chen, S. Waxman, Y. X. Wang, J. Z. Tan, and S. J. Chen. 1997. Cloning of a gene (RIG-G) associated with retinoic acid-induced differentiation of acute promyelocytic leukemia cells and representing a new member of a family of interferon-stimulated genes. Proc. Natl. Acad. Sci. USA 94:7406-7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, X., J. Shin, T. W. Molitor, L. B. Schook, and M. S. Rutherford. 1999. Molecular responses of macrophages to porcine reproductive and respiratory syndrome virus infection. Virology 262:152-162. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Y., B. A. Luxon, A. Casola, R. P. Garofalo, M. Jamaluddin, and A. R. Brasier. 2001. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J. Virol. 75:9044-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]