Abstract
An R638A mutation of the polymerase acidic protein (PA) subunit of the RNA polymerase of influenza A/WSN/33 virus results in severe attenuation of viral growth in cell culture by promoting the synthesis of defective interfering RNAs. We propose that R638A is an “elongation” mutant that destabilizes PA-RNA template interactions during elongation. A C453R mutation in PA can compensate for this defect, suggesting that amino acids C453 and R638 form part of the same domain.
The RNA-dependent RNA polymerase complex of influenza A virus consists of three subunits, polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), and polymerase acidic protein (PA). All three subunits are required for the transcription (viral RNA [vRNA] → mRNA) and replication (vRNA → cRNA → vRNA) of the eight segments of the negative single-stranded vRNA genome (3). The PB1 subunit forms the core of the complex and is responsible for polymerase activity. The PB2 subunit is involved in generation of capped RNA primers for the initiation of transcription by binding the cap structures of host pre-mRNAs prior to their endonucleolytic cleavage by PB1. The PA subunit is essential for both transcription and replication, but its exact role in the replication cycle of the virus is not clear. Recently, we reported that a histidine-to-alanine mutation at position 510 of PA (H510A) inhibited the endonucleolytic cleavage of capped RNAs, suggesting that PA might be directly involved in the generation of capped primers (4). PA can also induce a generalized proteolysis of both viral and host proteins, but the significance of this observation remains to be determined (9, 13, 17, 19).
In a recent report (4), a recombinant A/WSN/33 virus with an R638A mutation in the PA protein that produced pinhead-sized plaques on MDBK cells was described. The R638A PA protein was expressed at levels similar to those of the wild-type PA in transfected cells, suggesting that neither the stability nor the proteolytic activity, believed to be associated with PA, was significantly affected. In functional assays, a recombinant polymerase with the R638A mutation in its PA subunit was able to transcribe and replicate a vRNA-like chloramphenicol acetyltransferase (CAT) RNA as determined by measuring CAT activity and RNA levels in transfected cells. Intriguingly, in this transient assay system, the R638A mutant polymerase apparently synthesized increased amounts of CAT vRNA (about a fivefold increase compared to the wild type) but reduced amounts of CAT mRNA (about a fivefold decrease compared to the wild type). In the context of viral infection, however, we did not observe a significant difference in the ratios of vRNA, cRNA, and mRNA for PA, HA, or NA in a primer extension assay (results not shown), suggesting that this discrepancy might be due to the artificial nature of the transient assay used previously. Further studies, employing in vitro tests to assay the promoter binding, endonuclease, and transcription initiation (with ApG or capped RNA) activities of the R638A recombinant polymerase, revealed no significant differences in these activities between the recombinant and wild-type polymerases (reference 4 and results not shown).
The in vitro tests of the R638A mutant polymerase provided no clear indication of the reason for the attenuation, and therefore, we decided to characterize the R638A virus in more detail. While performing plaque assays of the R638A virus on MDBK cells, we observed a single plaque that was significantly larger than the remaining plaques, suggesting that it could be formed by a virus that either reverted to wild-type or acquired a compensatory mutation(s). The virus from this plaque (R638Arev) was isolated, purified by three cycles of plaquing on MDBK cells, and amplified on MDBK cells. As well as confirming the presence of the R638A mutation, we also found a single nucleotide change in the PA vRNA of R638Arev compared to the PA of the original R638A virus (from now on referred to as R638Aorig) that resulted in a cysteine-to-arginine mutation at position 453 of the PA protein. To test whether the C453R mutation was responsible for the improved growth properties of the R638Arev virus, we generated a recombinant virus containing both the R638A and C453R PA mutations by using the plasmid-based rescue method (5, 15).
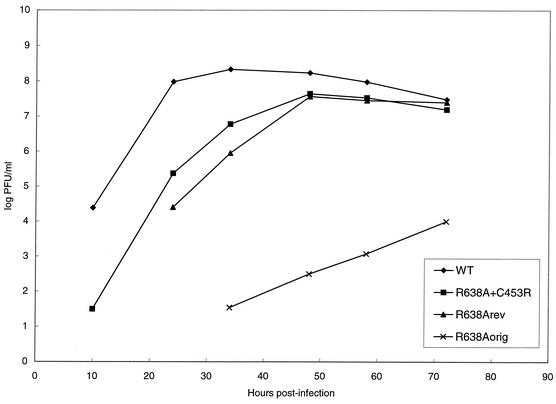
We compared the growth properties of the recombinant and wild-type viruses by infecting semiconfluent monolayers of MDBK cells and assaying the amount of infectious virus released into the medium at different time points postinfection by plaque assay on MDBK cells (Fig. 1). We confirmed that the R638Aorig virus was severely attenuated in MDBK cells. However, the R638Arev virus showed much improved growth properties. The rescued R638A+C453R double mutant exhibited growth properties similar to those of R638Arev, demonstrating that the C453R mutation is responsible for the improved growth of R638Arev. Although both R638Arev and R638A+C453R exhibited a decrease in growth kinetics, both reached maximum viral titers close to those of the wild-type virus. Thus, the C453R mutation resulted in nearly complete reversion of the phenotype of the R638A mutant. It is evident that the C453R mutation has a crucial effect on the growth properties of R638Arev. However, it should be noted that R638Arev might contain additional mutations in segments other than PA that might affect some of the properties of this virus.
FIG. 1.
Growth curves of recombinant influenza viruses on MDBK cells. Semiconfluent MDBK cells in 35-mm-diameter dishes were infected with wild-type (WT), R638Aorig, R638Arev, or R638A+C453R recombinant influenza A/WSN/33 viruses at a multiplicity of infection of 0.001. At the indicated time points, infectious particles present in the media were titrated by plaque assay in MDBK cells. The presented values are averages from duplicate experiments. The variation between the two values did not exceed 20%.
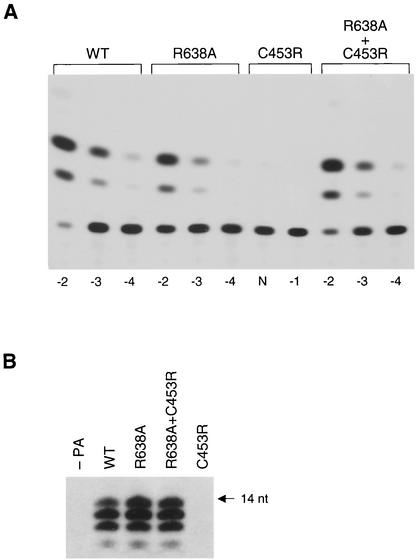
Interestingly, a recombinant virus with a C453R PA mutation (but without R638A) could not be generated. In agreement, the activity of a C453R polymerase in a CAT reporter assay was below detection levels (Fig. 2A). However, if both C453R and R638A mutations were present, we observed activities similar to those of the wild-type polymerase. In addition, a C453R recombinant polymerase produced only background levels of a 14-nucleotide (nt)-long RNA transcript in an ApG-primed in vitro transcription reaction (Fig. 2B). In contrast, both R638A and R638A+C453R polymerases were as active as the wild type.
FIG. 2.
Functionality of recombinant polymerases. (A) CAT expression in 293T cells transfected with pPOLI-CAT-RT and pcDNA expression vectors for PB1, PB2, PA (wild-type [WT] or mutants as indicated), and NP proteins. Transfections and CAT assays were performed as described earlier (4, 18) by using serially diluted cell extracts as indicated (N, undiluted; −1, 10-fold; −2, 100-fold; −3, 1,000-fold; and −4, 10,000-fold diluted). (B) Transcription initiation with ApG dinucleotide primer. Transcription reactions were carried out in vitro with nuclear extracts from cells expressing PB1, PB2, and PA (wild-type [WT] or mutants as indicated) by using a 14-nt 3′ end vRNA template in the presence of a 15-nt 5′ end vRNA as described earlier (4). The slowest-moving band represents the full-length (14-nt) transcription product. The faster-moving bands (12 and 13 nt) are most likely partial transcription products, as previously described (1). Longer exposure showed a low but detectable signal for the C453R mutant (not shown).
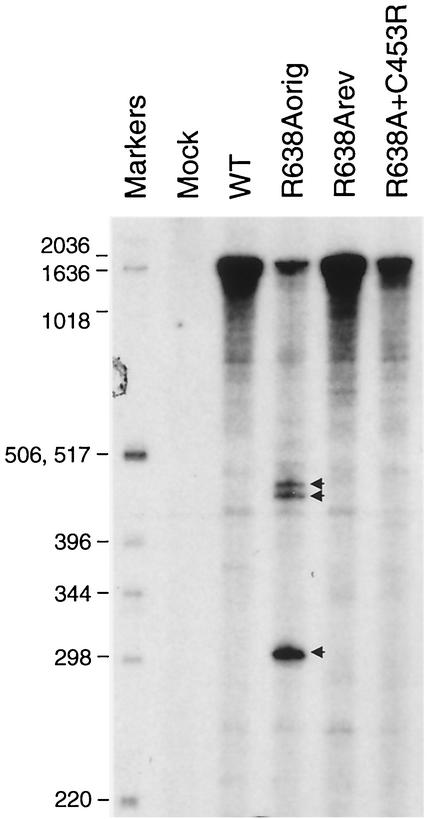
Having confirmed that the R638Aorig virus is severely attenuated compared to the wild type or, indeed, compared to R638Arev and R638A+C453R viruses, and that the initiation of transcription was not affected by the R638A mutation (see above), we analyzed vRNAs in infected cells qualitatively (Fig. 3) . Thus, MDBK cells were infected with wild-type, R638Aorig, R638Arev, or R638A+C453R recombinant viruses and the PA vRNA was analyzed by using a primer extension assay (4). We observed three major low-molecular-weight primer extension products for R638Aorig, which were absent for the other viruses. These results suggested that the R638Aorig virus synthesizes incomplete RNA molecules that might interfere with the replication of the virus.
FIG. 3.
Analysis of PA vRNA in MDBK cells infected with recombinant influenza viruses. MDBK cells in 35-mm-diameter dishes were infected with wild-type (WT) or mutant influenza A/WSN/33 viruses at a multiplicity of infection of approximately 0.01, and total RNA was isolated at 12 h postinfection. Primer extension assays were performed by using PA vRNA-specific primer 5′-CGCTCGAGCTTGCGGAAAAGGCA-3′ as described earlier (4, 6). The PA-specific sequence is underlined. The expected size of the primer extension products is 2,169 nt. Low-molecular-weight products are indicated by arrows. Primer extension products were analyzed on 4% polyacrylamide gels containing 7 M urea in Tris-borate-EDTA buffer. Markers, 32P-labeled 1-kb DNA ladder (Invitrogen); sizes are indicated.
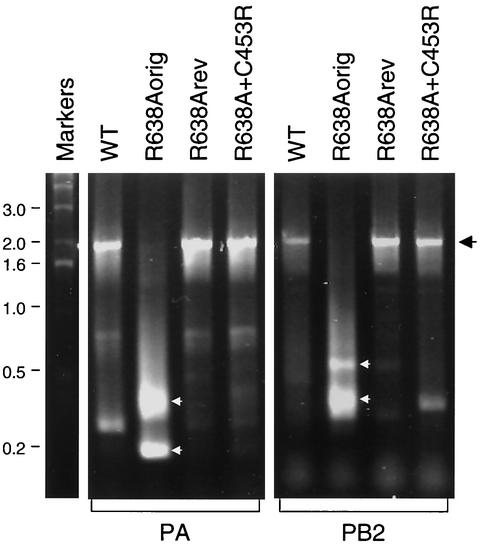
Incomplete RNAs, containing large, usually single internal deletions, have been described for influenza and are known as defective interfering (DI) RNAs (14). DI RNAs contain the 5′ and 3′ ends of the parent vRNA. They can be transcribed and replicated by the viral RNA polymerase and packaged into virions, often resulting in defective particles lacking the full-length parent segment. DI particles of influenza most often are produced by serial undiluted passages in cell culture that result in accumulating deletions in the polymerase genes. Therefore, we examined PA and PB2 vRNA, isolated from wild-type, R638Aorig, R638Arev, and R638A+C453R viruses, for the presence of DI RNAs by reverse transcriptase PCR (RT-PCR) (Fig. 4). We found very low levels of full-length products for the R638Aorig virus but found high levels of shorter products varying from about 0.2 up to about 1 kb long. In contrast, the wild-type, R638Arev, and R638A+C453R viruses produced mostly full-length products and only minor yields of shorter products. Direct sequencing of the PA-specific low-molecular-weight PCR products confirmed that these represented DI RNAs containing single internal deletions (Table 1). In addition, we cloned and sequenced DI RNAs derived from the PB1 segment of the R638Aorig virus. Five DI RNAs containing internal deletions were characterized (Table 1), providing further evidence that the R638Aorig virus contained polymerase segment-specific DI RNAs.
FIG. 4.
Analysis of vRNA in virions by RT-PCR. RNA was isolated from the viruses indicated by using the RNeasy mini Kit (Qiagen). RT-PCRs were performed with the following primers: 5′-GCGAATTCAATGCATGTGTGAGGA-3′ and 5′-CGCTCGAGCTTGCGGAAAAGGCA-3′ (for PA) or 5′-ATATGCGGCCGCATTGATGGCCATCCGAATTCT-3′ and 5′-ACGTGGATCCACCATGGAAAGAATAAAAGAACTA-3′ (for PB2). Underlining indicates the PA- and PB2-specific sequences. The expected size of the products is 2,125 bp for PA and 2,302 bp for PB2. Full-length PCR products are indicated by a black arrow on the right. The main PA- and PB2-specific DI RNAs in the R638Aorig sample are indicated by white arrows. The faint bands (not indicated) were not characterized in detail and most likely represent PCR artifacts or minor DI RNAs. PCR products were analyzed on 1% agarose gels. Markers, 1-kb DNA ladder (Invitrogen); sizes are indicated. WT, wild type.
TABLE 1.
Analysis of DI RNAs of the R638Aorig virus
Comparison of the hemagglutination titers of R638Aorig stock of 105 PFU/ml and the wild-type stock of 107 PFU/ml showed that both stocks contained similar numbers of physical particles as tested by a hemagglutination assay (results not shown). The observed low infectivity-to-particle ratio of the R638Aorig virus is consistent with the presence of DI RNAs.
Our results suggest that the R638A mutation in the PA subunit of the RNA polymerase leads to the generation of DI RNAs and, consequently, to the accumulation of defective viral particles. The wild-type, R638Aorig, and R638A+C453R viruses were generated by plasmid-based rescue on 293T cells, followed by a single amplification step on MDBK cells. As there were no apparent differences in growth conditions, the likeliest reason for the production of DI RNAs by R638Aorig is the presence of the mutation. How could a single amino acid mutation have such an effect? DI RNAs are believed to be generated by a “jumping” RNA polymerase (10). A plausible hypothesis is that the R638A mutation destabilizes polymerase-RNA template interactions during elongation that results in the release of the RNA template from the polymerase and, consequently, in incomplete RNA transcripts. Such incomplete transcripts, lacking the 3′ end, are not functional templates for replication or for packaging into RNPs and therefore are most likely quickly degraded by host nucleases. If, however, the polymerase reinitiates downstream on the template and completes elongation, the product would be stable due to the presence of both 5′ and 3′ terminal sequences that can bind to the viral RNA polymerase (1, 7, 20). These can be maintained by the virus as DI RNAs and, eventually, interfere with viral growth, if packaged instead of the full-length parent segment.
Odagiri and Tobita (16) reported that mutations in the NS2 gene could lead to the production of DI particles lacking the PA gene in a single cycle of virus replication. Since NS2 does not seem to be directly involved in the replication process of vRNA, in the light of our results with PA, it is likely that there are different mechanisms which can lead to the generation of incomplete vRNAs.
A previous study showed that PA can be cross-linked to promoter RNA (8). This interaction was sensitive to high salt, and therefore, it was suggested that the interaction was sequence independent. This previously identified RNA-binding activity of PA could be related to the role of PA in maintaining appropriate interactions between the RNA polymerase complex and the RNA template during elongation as suggested by this study. Thus, the PA subunit of the polymerase complex could act as an elongation factor. A recent finding that PA enhances the template binding activity of PB1 (11) is in agreement with this hypothesis. The fact that the loss of an arginine at position 638 could be compensated for by gaining another arginine at position 453 suggests that these two regions of PA are spatially close to one another, possibly forming part of the same RNA-binding domain. Basic amino acid side chains, in particular arginine residues, are known to play a significant role in RNA-protein interactions (2, 12).
Acknowledgments
We thank J. Robinson and A. Taylor for DNA sequencing. We also thank Andrew Catchpole and Pierre Fechter for helpful discussions.
This work was supported by the MRC (program grant G9523972 and cooperative component grant G9901312 to G.G.B. and senior nonclinical fellowship G117/457 to E.F.).
REFERENCES
- 1.Brownlee, G. G., and J. L. Sharps. 2002. The RNA polymerase of influenza A virus is stabilized by interaction with its viral RNA promoter. J. Virol. 76:7103-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615-621. [DOI] [PubMed] [Google Scholar]
- 3.Fodor, E., and G. G. Brownlee. 2002. Influenza virus replication, 1-29. In C. W. Potter (ed.), Influenza. Elsevier, Amsterdam, The Netherlands.
- 4.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fodor, E., L, Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fodor, E., P. Palese, G. G. Brownlee, and A. García-Sastre. 1998. Attenuation of influenza A virus mRNA levels by promoter mutations. J. Virol. 72:6283-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor, E., B. L. Seong, and G. G. Brownlee. 1993. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J. Gen. Virol. 74:1327-1333. [DOI] [PubMed] [Google Scholar]
- 9.Hara, K., M. Shiota, H. Kido, Y. Ohtsu, T. Kashiwagi, J. Iwahashi, N. Hamada, K. Mizoue, N. Tsumura, H. Kato, and T. Toyoda. 2001. Influenza virus RNA polymerase PA subunit is a novel serine protease with Ser624 at the active site. Genes Cells 6:87-97. [DOI] [PubMed] [Google Scholar]
- 10.Jennings, P. A., J. T. Finch, G. Winter, and J. S. Robertson. 1983. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza RNA? Cell 34:619-627. [DOI] [PubMed] [Google Scholar]
- 11.Lee, M. T., K. Bishop, L. Medcalf, D. Elton, P. Digard, and L. Tiley. 2002. Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lustig, B., S. Arora, and R. L. Jernigan. 1997. RNA base-amino acid interaction strengths derived from structures and sequences. Nucleic Acids Res. 25:2562-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naffakh, N., P. Massin, and S. van der Werf. 2001. The transcription/replication activity of the polymerase of influenza A viruses is not correlated with the level of proteolysis induced by the PA subunit. Virology 285:244-252. [DOI] [PubMed] [Google Scholar]
- 14.Nayak, D. P., T. M. Chambers, and R. K. Akkina. 1985. Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr. Top. Microbiol. Immunol. 114:103-151. [DOI] [PubMed] [Google Scholar]
- 15.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odagiri, T., and K. Tobita. 1990. Mutation in NS2, a non-structural protein of influenza A virus, extragenically causes aberrant replication and expression of the PA gene and leads to generation of defective interfering particles. Proc. Natl. Acad. Sci. USA 87:5988-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perales, B., J. J. Sanz-Ezquerro, P. Gastaminza, J. Ortega, J. F. Santaren, J. Ortín, and A. Nieto. 2000. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol. 74:1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pleschka, S., S. R. Jaskunas, O. G. Engelhardt, T. Zürcher, P. Palese, and A. García-Sastre. 1996. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 70:4188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanz-Ezquerro, J. J., S. de la Luna, J. Ortín, and A. Nieto. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J. Virol. 69:2420-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiley, L. S., M. Hagen, J. T. Matthews, and M. Krystal. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68:5108-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]