Abstract
Lentiviral vectors derived from human immunodeficiency virus type 1 (HIV-1) show great promise as gene carriers for future gene therapy. Insertion of a fragment containing the central polypurine tract (cPPT) in HIV-1 vector constructs is known to enhance transduction efficiency drastically, reportedly by facilitating the nuclear import of HIV-1 cDNA through a central DNA flap. We have studied the impact of the cPPT on the kinetics of HIV-1 vector transduction by real-time PCR. The kinetics of total HIV-1 DNA, two-long-terminal-repeat (2-LTR) circles, and, by an Alu-PCR, integrated proviral DNA were monitored. About 6 to 12 h after transduction, the total HIV-1 DNA reached a maximum level, followed by a steep decrease. The 2-LTR circles peaked after 24 to 48 h and were diluted upon cell division. Integration of HIV-1 DNA was first detected at 12 h postinfection. When HIV-1 vectors that contained the cPPT were used, DNA synthesis was similar but a threefold higher amount of 2-LTR circles was detected, confirming the impact on nuclear import. Moreover, a 10-fold increase in the amount of integrated DNA was observed in the presence of the cPPT. Only in the absence of the cPPT was a saturation in 2-LTR circle formation seen at a high multiplicity of infection, suggesting a role for the cPPT in overcoming a barrier to the nuclear import of HIV-1 DNA. A major effect of the central DNA flap on the juxtaposition of both LTRs is unlikely, since transduction with HIV-1 vectors containing ectopic cPPT fragments resulted in increased amounts of 2-LTR circles as well as integrated DNA. Inhibitors of transduction by cPPT-containing HIV vectors were also studied by real-time PCR. The reverse transcriptase inhibitor azidothymidine (AZT) and the nonnucleoside reverse transcriptase inhibitor α-APA clearly inhibited viral DNA synthesis, whereas integrase inhibitors such as the diketo acid L-708,906 and the pyranodipyrimidine V-165 specifically inhibited integration.
In contrast to oncoretroviruses, human immunodeficiency virus type 1 (HIV-1) and other lentiviruses have the capacity to replicate in nondividing cells such as macrophages. Lentiviral integration requires transport of the preintegration complex (PIC) through the nuclear pores by an active, energy-dependent process (6). At least three components of the HIV-1 PIC seem to contribute to nuclear transport: Vpr (23, 27, 28, 42, 45), matrix protein (7, 24, 48), and integrase (3, 25, 38). These different proteins interact with each other and with the cellular import machinery (14). However, the respective contribution of each protein to the karyophilic properties of HIV-1 PICs has not been completely elucidated (17, 18, 22, 31). Nuclear import is not determined by viral proteins only. Recently, it was shown that the central DNA flap of HIV-1 acts as a cis determinant of HIV-1 DNA nuclear import (21, 51). During the second-strand DNA synthesis of reverse transcription, initiation of DNA synthesis at the central polypurine tract (cPPT) results in a strand displacement event. The cPPT, an exact copy of the PPT at the 3′ end, serves as the second initiation site of second-strand DNA synthesis, while the central termination sequence (CTS) marks the termination of strand displacement synthesis. The final product of HIV-1 reverse transcription is a linear DNA molecule with a central 99-nucleotide-long plus-strand overlap, the central DNA flap (10, 11). The central DNA flap is believed to act as a cis determinant for HIV-1 DNA nuclear import: in the presence of the DNA flap, the HIV linear DNA is efficiently imported into the nucleus; in its absence, HIV cDNA accumulates in infected cells as unintegrated linear DNA in the vicinity of the nuclear membrane (51).
HIV-1-derived lentiviral vectors have become efficient tools to transfer genes into nondividing cells and are therefore promising candidate vectors for future gene therapy (32, 47). In the original lentiviral vector constructs, the cPPT-CTS fragment was removed (32). Subsequently, it became clear that HIV-1 vectors devoid of the DNA flap exhibit a strong defect in nuclear import (21, 51). It was demonstrated by Southern blotting that facilitated nuclear import results in an increased amount of integrated vector DNA and thus in a higher transduction efficiency of the lentiviral vector containing the cPPT. The concomitant increase in the mean expression level of the reporter gene EGFP, as measured by flow cytometry, was attributed to the increased amount of integrated DNA, reflecting the integration of multiple vector copies per cell. The extent of the increase in transduction efficiency by including the cPPT-CTS was cell type specific but, on average, 5- to 10-fold.
The current treatment for patients infected with HIV-1 is based on combination therapy including potent reverse transcriptase and protease inhibitors. These drug regimens are effective in reducing viral load and decreasing morbidity and mortality, but the emergence of multiple-drug-resistant virus strains in treated patients necessitates the development of new antiviral agents, preferentially targeting other replication steps (16). A crucial step in the HIV-1 replication cycle is the integration of viral DNA in the chromosome of the target cell after the reverse transcriptase (RT) of viral RNA into a double-stranded DNA copy in the cytoplasm of the infected cell. Integration is a multistep process catalyzed by integrase, a 32-kDa protein encoded by the pol gene. In the first step of the integration reaction, called 3′ processing, the dinucleotide pGT is removed from each 3′ end adjacent to a conserved CA dinucleotide (for a review on integrase, see reference 5). Next, the viral DNA is transported as a nucleoprotein particle or PIC into the nucleus, where the strand transfer reaction takes place. This is a concerted DNA cleavage-ligation reaction whereby the host DNA is cleaved and both 3′ ends of the viral DNA are covalently linked to the 5′ ends of the host DNA. Repair of the 5-base single-stranded DNA gaps between the 5′ ends of the viral DNA and the 3′ ends of the host DNA is likely carried out by host cell enzymes (4, 49). A possible role of integrase in DNA splicing (13) has also been proposed. In a nonproductive pathway, a minor fraction of the linear HIV DNA is circularized to give rise to one-long-terminal-repeat (1-LTR) or 2-LTR circles that are lost for integration (20, 44, 50).
Integration is required for productive HIV infection (19, 30, 43). Therefore, integration defines a point of no return in the life cycle of HIV and is an attractive target for the development of new antiviral drugs (for reviews, see references 39 and 41). The search for HIV integrase inhibitors has been facilitated by the availability of recombinant integrase, which can be assayed in vitro by using DNA oligonucleotides that correspond to the ends of the LTRs (8). By using these oligonucleotide-based assays, a vast arsenal of integrase inhibitors has been discovered, but unfortunately, most of these inhibitors seem to be toxic or inactive in cell culture or do not selectively target integrase (12, 15, 40). Before an integrase inhibitor can be included in a potent combination regimen of antiviral drugs, it is essential to corroborate its selectivity towards the integration step of the viral replication cycle. Recently, a direct approach to demonstrate the authenticity of an integrase inhibitor in cell culture, based on a quantitative real-time Alu-PCR, has been proposed (9). By amplifying HIV-1 DNA that is colinear with an Alu repeat, the absolute amount of integrated proviral DNA after HIV-1 infection was determined. To date, two classes of compounds have been reported as authentic HIV integrase inhibitors, namely, the diketo acids (26) and the pyranodipyrimidines (PDPs) (34). Diketo acids can be considered reference compounds for the evaluation of integrase inhibitors in cell culture (37). Whereas diketo acids specifically inhibit the DNA strand transfer reaction, PDPs apparently interfere with the binding of HIV integrase to the DNA substrate (26, 34).
In the present study, we used HIV-1-based lentiviral vectors to study the kinetics of the early steps of the lentiviral replication cycle by quantitative real-time PCR. We analyzed the kinetics of the different viral DNA forms during HIV-1-based vector transduction. The impact of the cPPT on the amount of each DNA species formed was investigated, and the effect of the known inhibitors of HIV-1 replication on these kinetics was determined as well.
MATERIALS AND METHODS
Compounds.
The PDP derivative V-165 was obtained from Ampharm, Inc., Ramsey, N.J. (34). Azidothymidine (AZT) was synthesized in our laboratory at the Rega Institute for Medical Research. Loviride (α-APA R89439) was obtained from the Janssen Research Foundation (Beerse, Belgium). The diketo acid L-708,906 (26) was kindly provided by T. R. Burke (National Institutes of Health, Bethesda, Md.).
Cells.
293T cells, E1A-transformed human embryonic kidney cells expressing SV40 large T antigen, were obtained from O. Danos (Génethon, Evry, France). 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco BRL) supplemented with 10% fetal calf serum (FCS; Harlan Sera-Lab Ltd.), 2 mM glutamine (Gibco BRL), and 20 μg of gentamicin (Gibco BRL)/ml at 37°C in a 5% CO2 humidified atmosphere.
Lentiviral vector production.
The HIV-1-based lentiviral vector particles were produced by using the three-plasmid transient-transfection system as originally described by Baekelandt et al. (1) and Naldini et al. (32). The various pHR′-derived transfer plasmids used all encode for EGFP as the reporter gene, allowing the measurement of the transduction efficiency by fluorescence-activated cell sorter (FACS) analysis. The second-generation packaging plasmid used, pCMVΔR8.91, lacks the vif, vpr, vpu, and nef genes. Vectors were pseudotyped with vesicular stomatitis virus G glycoprotein expressed from pMDG. For the transfection of a 10-cm-diameter dish of 293T cells, a 700-μl mixture of three plasmids was made in 150 mM NaCl: 20 μg of transfer plasmid, 10 μg of packaging construct, and 5 μg of the envelope plasmid pMDG. To this DNA solution, 700 μl of a polyethyleneimine solution (110 μl of a 10 mM stock solution in 150 mM NaCl) was added. After 15-min incubation at room temperature, the DNA-polyethyleneimine complex was added dropwise to the 293T cells in DMEM-1% FCS. After 5 h of incubation, medium was replaced with medium containing 10% FCS and 25 mM HEPES. Supernatants were collected on days 2, 3, and 6 posttransfection after low-speed centrifugation and filtered through a 0.45-μm-pore-size filter. Next, the vector particles were sedimented by ultracentrifugation at 48,000 × g for 5 h at 4°C in a fixed-angle rotor (Biofuge Stratos; Heraeus Instruments GmbH&Co, Hanau, Germany). Pellets were redissolved in phosphate-buffered saline, resulting in an 800-fold concentration. Different viral stocks were normalized based on p24 antigen content (HIV-1 p24 core profile enzyme-linked immunosorbent assay; DuPont, Dreieich, Germany). Transduction titers were determined by limiting dilution in CHO cells and FACS analysis.
Description of the transfer plasmids used.
The plasmid pHGFPWS (1) was derived from pHR′ but contains the SIN-18 deletion (53) and the woodchuck hepatitis posttranscriptional regulatory element (54). The plasmid pCHGFPWS contains, in addition, a fragment encompassing the HIV-1 cPPT and CTS fragments, located in front of the hCMV promoter (1). The plasmid pCHGFPW is similar to pCHGFPWS but contains an intact LTR.
Construction of novel transfer plasmids with central and ectopic cPPT-CTS.
The constructs were derived from pHRGFPW, originally provided by D. Trono (University of Geneva, Geneva, Switzerland) (54). The cPPT was amplified by PCR from the packaging plasmid pCMVΔR8.91 (32) with the forward primer 5′-GGGATCGATAGACAGCAGTACAAATGG-3′ and the reverse primer 5′-GGGATCGATTTCCAAACTGGATCTCTG-3′. The resulting 207-bp fragment was restricted with ClaI and cloned in sense orientation into the ClaI site of pHRGFPW, in front of the internal hCMV promoter, resulting in pCHGFPW. Before the cloning of cPPT-CTS into the KpnI or NotI site, located respectively at the 3′ and 5′ ends of the HIV-1 sequence in pHRGFPW, the restricted PCR fragment was blunted by T4 DNA polymerase. Cloning of this fragment in sense orientation at the respective sites resulted in p3CHGFPW and p5CHGFPW.
Transduction of 293T cells with HIV-1-based lentiviral vectors.
The day prior to transduction, 293T cells were seeded in 24-well plates at ∼105 cells per well. Transductions with the lentiviral vectors were carried out at a multiplicity of infection (MOI) of 3 or 10. Vector was added to the cells in the presence of 4 μg of polybrene/ml in DMEM-1% FCS. After 4 h of incubation, medium was replaced by DMEM containing 10% FCS. In each 24-well plate, 293T cells that were not transduced were incubated in parallel. Each time a sample was prepared for PCR analysis, an aliquot of untransduced cells was prepared as well. The latter samples served as no-amplification controls (NACs). Inhibitors of lentiviral transduction were added to the cells 1 h prior to transduction. When transduction medium was replaced by DMEM-10% FCS, fresh inhibitors were added.
DNA preparation.
At different time points after transduction, cells were lysed by the addition of a 10 mM Tris-Cl (pH 8.0) buffer containing 0.5% sodium dodecyl sulfate, 100 mM NaCl, 25 mM EDTA (pH 8.0), and 0.2 mg of proteinase K/ml and incubated for at least 6 h at 56°C. After lysis, DNA was extracted by phenol-chloroform and DNA concentration was measured by absorption spectroscopy.
Quantification of different HIV-1 DNA species in the cell by real-time PCR.
A specifically designed set of TaqMan probe and primers was used to quantify the amount of each specific HIV-1 DNA form in the cell lysate. For the quantification of late reverse transcripts (total HIV-1 DNA), the forward primer 5′-TGTGTGCCCGTCTGTTGTGT-3′, the reverse primer 5′-GAGTCCTGCGTCGAGAGAGC-3′, and the probe 5′-(FAM)-CAGTGGCGCCCGAACAGGGA-(TAMRA)-3′ were used (9). For the quantification of 2-LTR circles, the forward primer 5′-GTGCCCGTCTGTTGTGTGACT-3′, the reverse primer 5′-CTTGTCTTCTTTGGGAGAGAATTAGC-3′, and the probe 5′-(FAM)-TCCACACTGACTAAAAGGGTCTGAGGGATCTCT-(TAMRA)-3′ were employed. Quantification of integrated proviruses (Alu-PCR) was done with the 2-LTR forward primer and probe; as the reverse primer, 5′-TGCTGGGATTACAGGCGTGAG-3′ was used. For each real-time PCR analysis, a standard curve was generated by using dilutions of a representative standard of the amplicon being measured. We used the pHR′ transfer plasmid as standard for the total HIV DNA PCR and a plasmid containing the joined U3-U5 region as standard for the 2-LTR PCR. These standards were diluted in an amount of uninfected cellular DNA equivalent to that present in the unknown samples (200 ng of DNA for late reverse transcripts; 400 ng of DNA for 2-LTR) to create a similar PCR mixture for both the unknown samples and the standards. The standard curve ranged from 102 to 107 copies of DNA and was run in duplicate. DNA extracts from 293T cells that were transduced with HIV-1-derived vector and passaged six times were used as standard for the Alu-PCR. For the Alu-PCR standard curve, the copy number of integrated proviruses in the standard sample was first determined by performing a late RT-PCR by using the standard of the late RT-PCR. After determination of the absolute amount of the integrated proviruses, the Alu-PCR DNA standard was diluted and used to generate the Alu-PCR standard curve.
No-template controls (no DNA added to the PCR mixture) and NACs (DNA extracted from cells that were not transduced) were run with each experiment. Reactions contained 1× TaqMan universal master mix (Applied Biosystems, Lennik, Belgium), 300 nM forward primer (for Alu-PCR, 150 nM forward primer), 300 nM reverse primer (for Alu-PCR, 600 nM reverse primer), 100 nM probe, and 200 ng (late reverse transcripts), 400 ng (2-LTR circles), or 1 μg (integrated proviral DNAs) of template DNA in a 50-μl volume. After initial incubations at 50°C for 2 min and at 95°C for 10 min, 40 cycles of amplification were carried out at 95°C for 15 s (for Alu-PCR, 30 s), followed by 1 min at 60°C (for Alu-PCR, 1 min 30 s). Reactions were analyzed by using the ABI Prism model 7700 sequence detection system (Applied Biosystems). In duplicate transduction experiments, standard deviations on PCR data were below 10%. However, the multistep procedure and the variability between vector preparations confounded statistical analysis on the absolute values of independent transductions. Therefore, copy numbers are shown relative to those obtained with a control vector transduction run in parallel.
RESULTS
Kinetics of HIV DNA forms during HIV-1-based vector transduction.
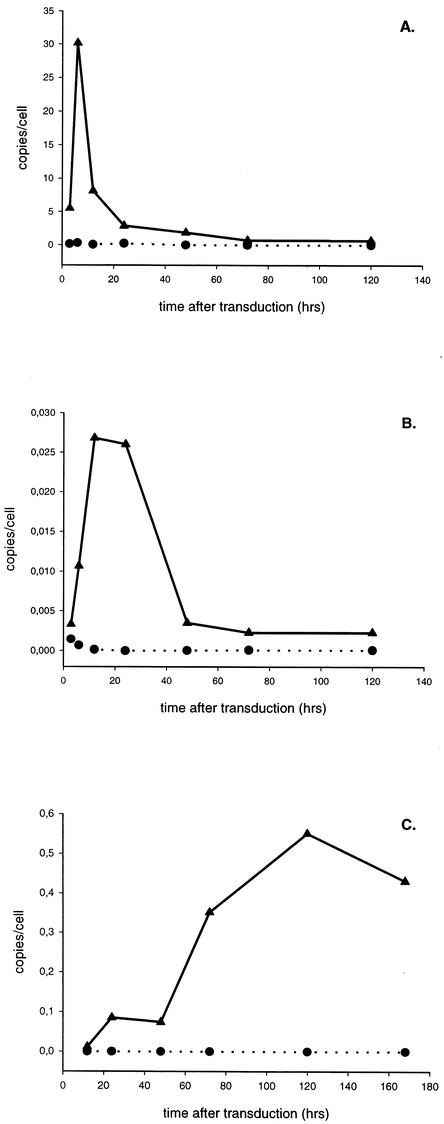
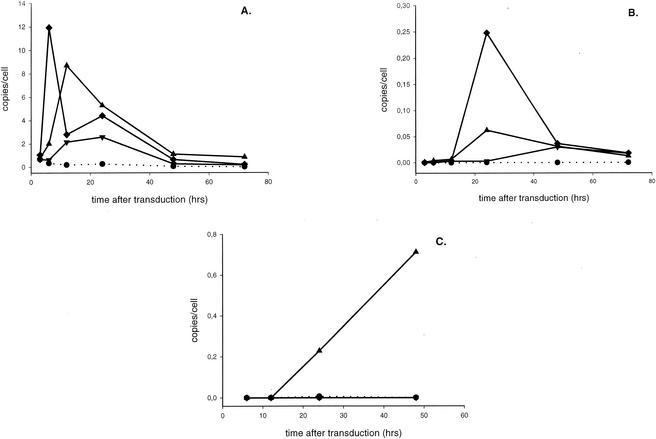
After reverse transcription of the viral RNA genome into double-stranded DNA, the DNA is imported into the nucleus to be inserted into the chromosome by a concerted strand transfer reaction mediated by integrase. Through the unproductive pathways of DNA circularization, 1- and 2-LTR circles are made in the nucleus. Little is known about the kinetics of formation and degradation of the different viral DNA species during HIV infection. By using three real-time PCRs, we quantified the amount of total HIV DNA (also referred to as late-reverse-transcript products), 2-LTR circles, and integrated proviral DNA during vector transduction as a function of time. To confine HIV replication to a single round, we used HIV-1-based vectors instead of wild-type HIV. We transduced 293T cells with HIV-1-based vectors at an MOI of 3 and took aliquots at different time points after infection for DNA extraction. DNA samples were also obtained after passaging of the cells. The vectors contained the HIV-1 cPPT that is known to enhance transduction efficiency by increasing nuclear import. Real-time quantitative PCR analysis of total HIV DNA revealed peak DNA synthesis 6 to 12 h after transduction, followed by a marked decrease over the next 20 to 30 h (Fig. 1A). This decline points to the degradation of an important fraction of the viral DNA, probably by cellular enzymes. The 2-LTR circles (Fig. 1B) reached a maximum level at 24 to 48 h and were then diluted upon cell division, as recently reported (36). This maximal amount of 2-LTR circles represented only 0.7% ± 0.5% of the amount of total HIV-1 DNA at the same time point. By quantitative Alu-PCR, the first integrated proviral DNA was detected at 12 h postinfection (p.i.), whereupon the amount increased slowly (Fig. 1C). From the integration rate, it was calculated that it took, on average, 15.4 h to integrate one HIV vector DNA copy. The integrated proviral DNAs, measured by quantitative Alu-PCR, accounted for about 5.34% ± 2.52% of the initial maximal amount of viral DNA made.
FIG. 1.
Kinetics of formation of the different HIV-1 DNA species during lentiviral vector transduction. 293T cells were transduced with HIV-1-based vector CHGFPWS, containing the cPPT (MOI = 3). Cells were lysed at different time points p.i., and total DNA was extracted. Each DNA sample was analyzed by three quantitative PCRs. The kinetics of the formation of total HIV-1 DNA (A), 2-LTR circles (B), and integrated proviral DNA (C) are shown for cells transduced with the cPPT vector (▴). For each time point, an aliquot of nontransduced cells (•) (NAC) was analyzed as well.
Impact of cell division on the kinetics of HIV-1 vector DNA production.
Next, we investigated the effect of cell division on the kinetics of the viral DNA. If cells are allowed to divide before linear viral DNA is integrated, cDNA may be diluted and lost for integration in one of the daughter cells. Therefore, transduction of 293T cells at confluency was compared with that of actively dividing cells. All cells were transduced with HIV-1 vectors at an MOI of 10. The rate and the overall amount of DNA synthesis were indistinguishable between both cell populations. Although the overall integration efficiency, as measured by Alu-PCR, was comparable, the initial integration rate tended to be two- to fourfold higher in nondividing cells (data not shown). The amount of 2-LTR circles was at least twofold lower in the dividing cells 48 h after transduction (data not shown), likely due to the dilution of those circular DNA species upon cell division, as reported recently (36). Our standard transduction experiments were carried out with actively dividing 293T cells.
Impact of the cPPT on the kinetics of HIV-1 vector transduction.
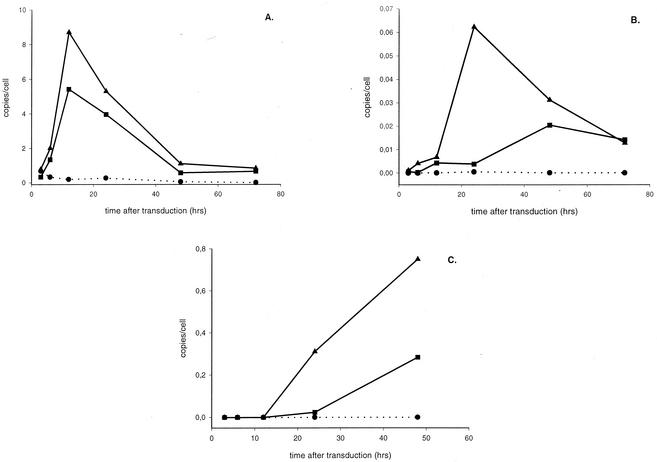
The kinetics of transduction were originally studied with lentiviral vectors containing the HIV-1 cPPT (Fig. 1). Since it is generally believed that the cPPT enhances nuclear import and thereby stimulates lentiviral vector transduction efficiency (51), we studied the influence of the cPPT on the kinetics of the formation of three distinct HIV DNA species. Therefore, we transduced 293T cells in parallel by using vectors with or without cPPT at an MOI of 10. Real-time PCR analysis revealed clear differences between both vectors (Fig. 2 and Table 1). Late reverse transcripts of vectors either with or without cPPT reached a maximum level at 12 h that was followed by a steep decrease (Fig. 2A). No significant difference in the rate and extent of viral DNA synthesis could be observed (Table 1). The 2-LTR circles were detected earlier and were present in a significantly larger amount when the cPPT was present (Fig. 2B). The 2-LTR circles peaked already after 24 h in the presence of the cPPT, whereas in the absence of the cPPT, a steady increase until 48 h after transduction was observed. As summarized in Table 1, a threefold increase in the maximal amount of circular DNA was detected in the presence of the cPPT. Since 2-LTR circles are formed in the nucleus, these quantifications confirm the stimulatory effect of the cPPT on the nuclear import of PICs in the infected cell.
FIG. 2.
Impact of the cPPT on the amounts of different viral DNA species in transduced cells. Kinetics of the formation of total HIV-1 DNA (A), 2-LTR circles (B), and integrated proviral DNA (C) are shown. Transductions of 293T cells were carried out in parallel with HIV-1 vectors CHGFPWS and HGFPWS, respectively, with or without cPPT (MOI = 10). Quantitative real-time PCR analysis was performed on the DNA extracts of 293T cells transduced with vector without cPPT (▪) or with cPPT (▴). For each time point, an aliquot of nontransduced cells (•) (NAC) was analyzed as well.
TABLE 1.
Impact of the cPPT on the kinetics of HIV-1 vector transduction
| DNA form | Relative amount of HIV-1 DNAa
|
||
|---|---|---|---|
| 3′ cPPT vector | cPPT vector | 5′ cPPT vector | |
| Total HIV-1 DNA | 0.62 ± 0.4 | 1.14 ± 0.53 | 0.85 ± 0.34 |
| 2-LTR circles | 1.67 ± 0.49 | 2.84 ± 0.62 | 3.28 ± 0.44 |
| Integrated proviral DNA | 10.4 ± 5.0 | 10.6 ± 0.7 | 9.65 ± 5.2 |
Maximal amounts of each DNA form relative to the maximal amounts obtained with an HIV-1 vector lacking cPPT. Data are average values ± standard deviations from three to five separate experiments.
The differences in integration efficiency between the cPPT and non-cPPT vectors were evidenced by Alu-PCR (Fig. 2C). As expected, more integrated proviral DNA was detected in the presence of the cPPT. For each integrated DNA copy per cell with a non-cPPT vector, an equivalent of 10.6 ± 0.7 copies/cell was integrated when the cPPT was present in the lentiviral vector. Moreover, distinct kinetics of integration were observed. Whereas the first integration events occurred after 24 h in the absence of the cPPT, we could already detect proviral DNA at 12 h p.i. when vectors with cPPT were used. These results are consistent with a more efficient nuclear import by the presence of the cPPT, resulting in earlier and greater integration of DNA. The impact on integration was thereby more pronounced than the impact on 2-LTR formation.
Impact of MOI on HIV-1 DNA formation.
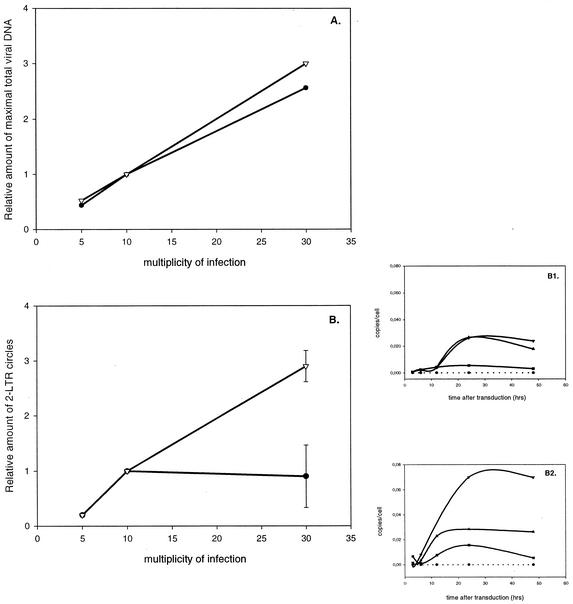
Next, we analyzed the effect of the MOI on transductions with HIV-1 vectors with or without cPPT. Therefore, 293T cells were transduced at a different MOI and the DNA species were again quantified over time (Fig. 3). The amounts of late reverse transcripts increased proportionally with the MOI, both in the presence and in the absence of the cPPT (Fig. 3A). However, when quantifying 2-LTR circles, an important difference between the cPPT and the non-cPPT vectors was apparent (Fig. 3B). In the absence of the cPPT, a saturation of circle formation around an MOI of 10 was evidenced in contrast to vectors with cPPT. Since 2-LTR circle formation is a measure of nuclear import, our data suggest that the cPPT can overcome a barrier for DNA import. As a result of the facilitated nuclear import by the cPPT at a high MOI, there was less saturation of DNA integration at a high MOI, as measured by Alu-PCR (data not shown).
FIG. 3.
Impact of MOI on the amounts of 2-LTR circles during HIV-1-based vector transduction in the absence or presence of the cPPT. 293T cells were transduced in parallel by using vectors CHGFPWS and HGFPWS, respectively, with (▿) or without (•) cPPT at different MOIs. The maximal amount of total HIV DNA produced was calculated for the different MOIs by quantitative PCR. The 2-LTR circles were quantified as an estimate of nuclear import. (A) The maximal amount of total HIV DNA relative to the amount at an MOI of 10 is shown at three MOIs (5, 10, and 30). (B) The number of 2-LTR circles measured at different MOIs is shown relative to the amount at an MOI of 10. (B1) Time course of the formation of 2-LTR circles during transduction in the absence of the cPPT at MOIs of 5 (▪), 10 (▴), and 30 (▾). (B2) Time course of the formation of 2-LTR circles during transduction with cPPT-containing vectors at MOIs of 5 (▪), 10 (▴), and 30 (▾). For each time point, a NAC (•) was analyzed as well.
Impact of the position of the cPPT in the viral genome on the formation of 2-LTR circles.
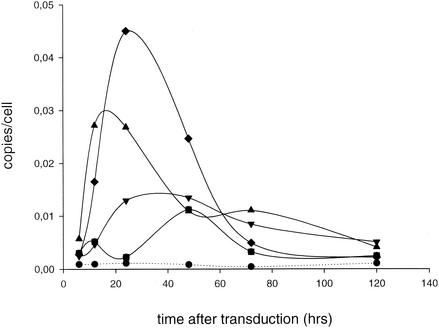
In theory, the circularization of the linear viral DNA in the nucleus could be favored by a central DNA flap. By relieving topological constraints, a centrally located DNA flap may facilitate the juxtaposition of both LTRs and the ligation of the two ends in the nucleus (51). In that perspective, the increase in 2-LTR circles would not reflect improved nuclear import. To rule out this possibility, we cloned the cPPT-CTS fragment at the 5′ and 3′ ends of the lentiviral vector construct pHGFPW. We reasoned that an ectopic location of the DNA flap would not favor DNA ligation. Transductions of 293T cells were carried out with HIV-based vectors lacking the cPPT (using pHGFPW) or containing the cPPT, either in the center (pCHGFPW) or at the 5′ or 3′ end (p5CHGFPW or p3CHGFPW, respectively). The quantitative analysis revealed no clear difference in the maximal amount of 2-LTR circles obtained with 5CHGFPW, as is shown in Fig. 4 and Table 1. RT of the vector carrying cPPT-CTS at the 3′ end was less efficient; still, the number of 2-LTR circles was 1.7-fold higher than that in the absence of the cPPT. All cPPT-containing vectors resulted in a dramatic increase in the integration efficiency (Table 1).
FIG. 4.
Impact of an ectopic PPT on the amounts of 2-LTR circles formed. Kinetics of the formation of 2-LTR circles is shown. Transductions of 293T cells were carried out in parallel with the HIV-1 vectors derived from pCHGFPW (▴), p3CHGFPW (▾), or p5CHGFPW (⧫) (MOI = 10). These vectors carry the cPPT-CTS fragment in the center, at the 3′ end, or at the 5′ end, respectively. A vector without cPPT (derived from pHGFPW) was used in parallel (▪). Quantitative real-time PCR analysis was performed on the DNA extracts of transduced 293T cells. For each time point, an aliquot of nontransduced cells (•) (NAC) was analyzed as well.
Impact of HIV inhibitors on HIV DNA kinetics.
Prior to the development of any new authentic inhibitor of HIV-1 integration, experimental evidence for inhibition of the integration process in cell culture has to be obtained (15). The HIV DNA quantification method described in combination with HIV vector-based single-round infection should facilitate the identification of new antiviral targets for HIV-1 inhibitors (9). This method particularly allows the discrimination between inhibitors of HIV-1 RT and integration. Since the lentiviral vectors are pseudotyped with the vesicular stomatitis virus G glycoprotein, inhibition of HIV entry is not assessed for. To confirm the usefulness of this assay to demonstrate that a candidate integrase inhibitor targets the integration step in cell culture, we tested the diketo acid L-708,906 (26) and the PDP V-165 (34) as representatives of both classes of the authentic integrase inhibitors described so far. In parallel, we tested AZT as a prototype of the nucleoside reverse transcriptase inhibitors and α-APA as a nonnucleoside reverse transcriptase inhibitor (35). Therefore, 293T cells were transduced with lentiviral vectors encoding EGFP (MOI = 10) in the absence or presence of 187 nM AZT, 570 nM α-APA, 25 μM L-708,906, or 25 μM V-165. FACS analysis was performed on transduced cells before and after passaging to quantify the extent of the inhibition of transduction (Table 2). In the presence of AZT or α-APA, the transduction efficiency decreased by 60 to 70%. The inhibition of transduction efficiency by the diketo acid and by the PDP was more pronounced after passaging of the cells (70 to 80% versus 35 to 52%). The results of DNA quantification are presented in Fig. 5 and Table 2. The nucleoside reverse transcriptase inhibitor AZT inhibited HIV DNA synthesis (Fig. 5A) and 2-LTR circle formation (Fig. 5B). The number of integrants was about 10% of that of the uninhibited control (Fig. 5C). A similar inhibition profile was observed for α-APA (Table 2). When the diketo acid L-708,906 was added during lentiviral transduction, no significant inhibition of the late reverse transcripts was observed (Fig. 5A) whereas the amount of 2-LTR circles clearly increased (Fig. 5B) in accordance with the inhibition of DNA strand transfer subsequent to nuclear import. As expected, almost no integrated proviral DNA could be detected by Alu-PCR (Fig. 5C). In the presence of V-165, no significant inhibition of DNA synthesis was detected; there was no clear effect on 2-LTR circle formation but a pronounced inhibition of proviral DNA (Table 2).
TABLE 2.
Inhibition of HIV-1 vector transduction
| 293T cells with: | Relative transduction efficiencya
|
Relative amount of DNAb
|
|||
|---|---|---|---|---|---|
| Before passaging | After passaging | Total HIV-1 DNA | 2-LTR circles | Integrated proviral DNAs | |
| AZT (187 nM) | 0.32 ± 0.20 | 0.32 ± 0.37 | 0.64 ± 0.25 | 0.35 ± 0.27 | 0.13 ± 0.10 |
| α-APA (570 nM) | 0.365 ± 0.090 | 0.38 ± 0.11 | 0.39 ± 0.19 | 0.22 ± 0.15 | 0.12 ± 0.10 |
| L-708,906 (25 μM) | 0.475 ± 0.035 | 0.22 ± 0.13 | 1.0 ± 0.4 | 3.1 ± 0.6 | 0.003 ± 0.002 |
| V-165 (25 μM) | 0.65 ± 0.40 | 0.31 ± 0.04 | 0.92 ± 0.49 | 0.88 ± 0.55 | 0.20 ± 0.13 |
Transduction efficiency of 293T cells by HIV-1 vectors before (72 h after transduction) and after (120 h after transduction) passaging of transduced cells in the presence of inhibitors was measured by FACS analysis and is shown relative to the transduction efficiency obtained in the absence of inhibitors. Average values ± standard deviations from two separate experiments are shown.
Total DNA was extracted from transduced cells at different intervals after transduction. HIV DNA was quantified with quantitative PCR using specific primers and probe for total HIV-1 DNA, 2-LTR circles, or integrated proviral DNA. For each DNA species, the maximal amount relative to the maximal amount obtained in the absence of inhibitor is represented. Data are average values ± standard deviations from two to five separate experiments.
FIG. 5.
Kinetics of HIV-1 vector transduction in the presence of HIV-1 inhibitors. 293T cells were transduced with the cPPT vectors CHGFPWS at an MOI of 10 in the absence (▴) or presence (▾) of 187 nM AZT or 25 μM l-708,906 (⧫). For each time point, the formation of total HIV-1 DNA (A), 2-LTR circles (B), and integrated proviral DNA (C) was quantified. For each time point, a NAC (•) was run in parallel.
DISCUSSION
Real-time PCR methodology to quantify HIV-1 integration.
The HIV-1 integration process is an attractive target for the development of new antiviral therapy. Moreover, efficient integration is crucial for the future success of lentiviral vectors for gene therapy. There is an active ongoing search to identify lead compounds that inhibit integrase. Most efforts have been focused on the discovery of integrase inhibitors by using oligonucleotide-based enzymatic assays. Although these assays are widely used, authentic integrase inhibition has to be corroborated in HIV-infected cells. Although the cPPT is now inserted in most lentiviral vector constructs, the mechanism whereby it increases transduction efficiency is not well understood. Recently, Butler et al. described an elegant method to quantify HIV integration (9). Using real-time Alu-PCR, HIV-1 integration in cell culture was quantified relative to a total HIV DNA standard. In our experiments, we used a similar real-time quantitative PCR analysis to determine the kinetics of the formation and degradation of different viral DNA species during HIV-1 vector transductions. Using this technology, we studied the effect of the cPPT and HIV inhibitors on the transduction process.
Real-time PCR using TaqMan probes offers the advantage of an absolute quantification of input DNA in contrast to semiquantitative end point PCR or Southern blotting. A representative standard for each amplicon to be quantified and a specific probe annealing to the correct strand are required. In our hands, the quantitative total HIV DNA PCR (detection limit, 0.01 copy/cell) and the 2-LTR quantitative PCR (detection limit, 0.001 copy/cell) proved highly sensitive and yielded perfect standard curves (slope = −3.2), pointing to optimal PCR efficiency. Amplification of HIV-1 integrants by Alu-PCR is hampered by the frequency of the Alu repeats in the human genome (one Alu repeat every 5,000 bp). Likely, not all integrants will be located near such an Alu repeat. Although this quantitative Alu-PCR is highly specific for detecting integration events, it is not as sensitive (0.1 copies/cell) and efficient as the other quantitative PCRs (slopes varying from −2.3 to −3.4). From a direct comparison between linker-primer PCR and Alu-PCR for the detection of HIV-1 integrants, it was estimated that the latter only amplifies half of all integrated proviral DNAs (46). We estimated the efficiency of our quantitative Alu-PCR at 47% by direct comparison with the DNA copy numbers obtained with total HIV-1 DNA quantitative PCR. The number of circles present at that time was calculated to be less than 0.1% of the total viral DNA. The efficiency of the quantitative Alu-PCR may also be confounded by interfering Alu-Alu amplifications. Theoretically, performing a total HIV-1 DNA PCR after integration has been completed should result in a copy number similar to that obtained with Alu-PCR. Although after one passage, circular and linear nonintegrated viral DNAs are detected as well, they account for only about 0.015 ± 0.005 DNA copies per cell, based on the assumption that the ratio of 1-LTR to 2-LTR circles is about 9 to 1 (9). Still, for kinetic studies of single-round infection, Alu-PCR was preferred. While this work was under revision, a nested quantitative Alu-PCR was described with a 10- to 100-fold increased sensitivity for quantifying HIV integration (33). We plan to verify this reported sensitivity in the near future.
Kinetics of HIV-1 vector integration.
The kinetics of HIV-1 DNA during HIV-based vector transduction are characterized by an initial steep increase in the linear HIV-1 DNA synthesized during the RT process, with a maximum at 12 h, but followed by a significant degradation of the HIV-1 DNA. This phenomenon has been described by Butler et al. and Vandegraaff et al. (9, 46). According to Vandegraaff et al., 10% of the cDNA made is integrated; based on the data obtained by Butler et al., this efficiency is about 5% for transductions in 293T cells. We calculated a DNA integration efficiency in the presence of the cPPT to be 5.34% ± 2.52%. In our hands, the rate and extent of the degradation of the DNA was not dependent on either the cell division or MOI of the vector. It has been speculated that HIV DNA degradation results from a cellular defense mechanism in the cytoplasm, although the underlying mechanism remains to be identified (2, 9, 46). Exploiting the cellular defense mechanism may yield gene therapeutic approaches against HIV replication, whereas circumventing DNA degradation may increase lentiviral vector performance.
It is known that 2-LTR circles represent only a minor fraction of the circular DNA made during HIV infection (20, 44, 50). At 24 to 48 h after transduction, the 2-LTR circles represented 0.7% of the total viral DNA present. The subsequent decrease in their number is likely related to dilution upon cell division, as recently reported (36). Our numbers are consistent with those in recent reports where 2-LTR circles accounted for 1.0 to 1.5% of the total viral DNA synthesized (9, 46). 2-LTR circles were first detected between 3 and 6 h after transduction, but integrated proviral DNAs were detected only by quantitative Alu-PCR from 12 h after transduction. The apparent time difference can probably be attributed to the difference in the sensitivities of the PCRs used. From our quantitative Alu-PCR data, a vector integration rate was calculated. Based on this rate and on the transduction efficiency, as measured by FACS analysis, an average provirus would take about 15 h to integrate after DNA synthesis is complete (at 12 h p.i.). Based on the data of Butler et al. (9), we calculated integration to take 16 h, in accordance with the results of our study.
We have determined the kinetics of the HIV-1-derived lentiviral vector transduction of 293T cells. How does this relate to the established kinetics of HIV-1 infection in lymphocytes? When analyzing the kinetics of cell-free HIV infection with a linker-primer PCR assay, it was concluded that total HIV DNA peaked at 14 h p.i. (46), which is consistent with our results. The first integrated proviral DNAs were detected at 4 h p.i., and the 2-LTR DNA circles started to accumulate from 7 h p.i. In a synchronous cell-to-cell transmission model of HIV, the first integrated HIV DNA was detected by Southern blotting at 8 h p.i. (29). The integration process was completed at 72 h p.i. In time-of-addition experiments with HIV-1, the timing of the consecutive replication steps can be pinpointed by the loss of the inhibitory activity of known inhibitors (16). Reverse transcriptase inhibitors lose activity if addition is postponed for more than 4 h p.i.; the diketo acid L-708,906 loses activity if it is added later than 8 h p.i. It follows that in these single-round infections with free virus at a high MOI, the RT step occurs at 4 h p.i. and DNA strand transfer occurs at 8 h. Even if our quantitative Alu-PCR detects only half of the integrated proviral DNAs, this analysis suggests that HIV-1 vector transduction proceeds at a lower rate than the HIV-1 infection of T-cell lines. It remains to be investigated whether transduction rates are cell dependent and whether viral proteins (e.g., accessory proteins) may increase transduction rates. Identification of the parameters responsible for the lower integration rate of vectors with respect to virus may lead to improved lentiviral vectors.
Impact of HIV-1 cPPT on the kinetics of vector transduction.
It is generally accepted that insertion of the HIV-1 cPPT in HIV-1 vectors considerably improves the transduction efficiency (21, 51, 52). Vector improvement has been attributed to facilitated nuclear import in the presence of the cPPT (51). We analyzed the effect of the cPPT on vector kinetics by quantitative PCR. In keeping with what has been described previously, the cPPT apparently had no significant effect on the rate of initial DNA synthesis nor on the amount of the reverse transcripts synthesized. The impact on nuclear import was evidenced by the threefold increase in the amount of 2-LTR circles after transduction with cPPT vectors (Table 1). The rate of DNA circularization was also much higher in the presence of the cPPT (Fig. 2B). Transduction with cPPT vectors resulted in a 10-fold increase in integrated vector DNA, and the integration rate was clearly stimulated. The increased integration efficiency of cPPT vectors can at least partially be attributed to facilitated nuclear import. As an indirect consequence, the HIV-1 DNA may also be protected to some extent from degradation in the cytoplasm and from dilution upon cell division. If there is a greater and faster nuclear import of HIV cDNA in the presence of the cPPT, this may serve as protection against the degradation and dilution of nonintegrated DNA.
A new aspect of the role of the cPPT during nuclear import was evidenced in our experiments using high multiplicities of vector. Whereas the nuclear import, estimated by the amount of 2-LTR circles, was saturated at an MOI of 10 in the absence of the cPPT, no saturation was observed with cPPT vectors. Apparently, the threshold for nuclear import can be overcome by the cPPT. Next, we constructed lentiviral vectors carrying the cPPT-CTS fragment on ectopic locations (3′ or 5′ end). Remarkably, there was only a limited effect on DNA synthesis. Ectopic cPPT-CTS stimulated 2-LTR circle formation and integration as well. These results are at odds with the hypothesis that the central DNA flap relieves topological constraints for efficient translocation through the nucleopore (51). More likely, the central DNA flap serves as a signal facilitating nuclear import with respect to both rate and maximal capacity. By itself, this would already lead to increased integration. Still, it is possible that the DNA flap has a direct stimulatory effect on integration that needs further investigation.
Quantitative PCR methodology can be used to corroborate the inhibition of DNA integration in cell culture.
In analogy with Butler et al. (9), we performed transductions with cPPT vectors in the presence of various HIV inhibitors. The diketo acid L-708,906 was shown to inhibit integration without an effect on viral DNA synthesis (Fig. 5). Interestingly, inhibition of strand transfer was accompanied by an increase in the number of 2-LTR circles. The methodology can thus be used to corroborate the authenticity of integrase inhibition in cell culture. L-708,906 is a specific inhibitor of DNA strand transfer. The recently described PDP V-165 inhibits integrase-DNA complex formation rather than DNA strand transfer. This was evidenced by an inhibition of integration without an increase in 2-LTR circle formation. Our assay format includes data on transduction efficiency as measured by FACS analysis to document the extent of inhibition at the concentrations used (Table 2). Interestingly, the inhibitory effect of both integrase inhibitors was more pronounced after passaging of the cells. EGFP may be expressed from the nonintegrated DNA forms (1-LTR and 2-LTR circles, half-site integrants, or linear DNA) present before passaging of the cells and detected by FACS analysis. These nonintegrated DNA forms are diluted upon cell division. The decreased EGFP expression after passaging is thus consistent with an inhibition of the integration process. In conclusion, our kinetic analysis of the impact of the cPPT on HIV-1 vector transduction will be helpful for the detailed study of the HIV-1 integration process, the optimization of HIV-1 vectors, and the development of authentic integrase inhibitors.
Acknowledgments
The HIV-1-derived lentivirus vector constructs were kind gifts from O. Danos (Généthon), D. Trono (University of Geneva), and P. Charneau (Institut Pasteur, Paris, France). We thank Martine Michiels and Sofie Willems for the production of lentiviral vectors. Anje Claeys is acknowledged for critical reading of the manuscript.
B.V.M. and J.D.R. are both funded by a grant from the Flemish Institute supporting Scientific-Technological Research in Industry (IWT). Z.D. is a postdoctoral fellow of the Flemish Fund for Scientific Research (FWO).
REFERENCES
- 1.Baekelandt, V., A. Claeys, P. Cherepanov, E. De Clercq, B. De Strooper, B. Nuttin, and Z. Debyser. 2000. DNA-dependent protein kinase is not required for efficient lentivirus integration. J. Virol. 74:11278-11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa, P., P. Charneau, N. Dumey, and F. Clavel. 1994. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res. Hum. Retrovir. 10:53-59. [DOI] [PubMed] [Google Scholar]
- 3.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 4.Brin, E., J. Yi, A. M. Skalka, and J. Leis. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275:39287-39295. [DOI] [PubMed] [Google Scholar]
- 5.Brown, P. O. 1997. Integration of retroviral DNA. Curr. Top. Microbiol. Immunol. 157:19-47. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman, F. D., and R. Craigie. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. USA 88:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 10.Charneau, P., and F. A. Clavel. 1991. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J. Virol. 65:2415-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charneau, P., M. Alizon, and F. Clavel. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherepanov, P., J. A. Este, R. F. Rando, J. O. Ojwang, G. Reekmans, R. Steinfeld, G. David, E. De Clercq, and Z. Debyser. 1997. Mode of interaction of G-quartets with the integrase of human immunodeficiency virus type 1. Mol. Pharmacol. 52:771-780. [DOI] [PubMed] [Google Scholar]
- 13.Chow, S. A., K. A. Vincent, V. Ellison, and P. O. Brown. 1992. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 255:723-726. [DOI] [PubMed] [Google Scholar]
- 14.Cullen, B. R. 2001. Journey to the center of the cell. Cell 105:697-700. [DOI] [PubMed] [Google Scholar]
- 15.Debyser, Z., P. Cherepanov, B. Van Maele, E. De Clercq, and M. Witvrouw. 2002. In search of authentic inhibitors of HIV-1 integration. Antivir. Chem. Chemother. 13:1-15. [DOI] [PubMed] [Google Scholar]
- 16.De Clercq, E. 2001. New developments in anti-HIV chemotherapy. Curr. Med. Chem. 8:1529-1558. [DOI] [PubMed] [Google Scholar]
- 17.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 18.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 19.Englund, G., T. S. Theodore, E. O. Freed, A. Engleman, and M. A. Martin. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnet, C. M., and W. A. Haseltine. 1991. Circularization of human immunodeficiency virus type 1 DNA in vitro. J. Virol. 65:6942-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Follenzi, A., L. E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217-222. [DOI] [PubMed] [Google Scholar]
- 22.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, A. V. Albright, F. Gonzalez-Scarano, and M. H. Malim. 1998. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J. Virol. 72:6004-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallay, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 25.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV- 1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 27.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins, Y., M. McEntee, K. Weis, and W. C. Greene. 1998. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J. Cell Biol. 143:875-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kok, T., P. Li, and C. J. Burrel. 2001. HIV DNA integration during cell-to-cell transmission of infection: evidence for partially integrated DNA structures in acutely infected cells. Arch. Virol. 146:1963-1978. [DOI] [PubMed] [Google Scholar]
- 30.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limon, A., E. Devroe, R. Lu, H. Z. Ghory, P. A. Silver, and A. Engelman. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 33.O'Doherty, U., W. J. Swigaard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76:10942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pannecouque, C., W. Pluymers, B. Van Maele, V. Tetz, P. Cherepanov, E. De Clercq, M. Witvrouw, and Z. Debyser. 2002. New class of HIV integrase inhibitors that block viral replication in cell culture. Curr. Biol. 12:1169-1177. [DOI] [PubMed] [Google Scholar]
- 35.Pauwels, R., K. Andries, Z. Debyser, P. Van Daele, D. Schols, P. Stoffels, K. De Vreese, R. Woestenborghs, A.-M. Vandamme, C. G. M. Janssen, J. Anné, G. Cauwenbergh, J. Desmyter, J. Heykants, M. A. C. Janssen, E. De Clercq, and P. A. J. Janssen. 1993. Potent and highly selective human immunodeficiency virus type 1 (HIV-1) inhibition by a series of alfa-anilinophenylacetamide derivatives targeted at HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierson, T. C., T. L. Kieffer, C. T. Ruff, C. Buck, S. J. Gange, and R. F. Siliciano. 2002. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pluymers, W., G. Pais, B. Van Maele, C. Pannecouque, V. Fikkert, T. R. Burke, E. De Clercq, M. Witvrouw, N. Neamati, and Z. Debyser. 2002. Inhibition of human immunodeficiency virus type 1 integration by diketo derivatives. Antimicrob. Agents Chemother. 46:3292-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 39.Pluymers, W., E. DeClercq, and Z. Debyser. 2001. HIV-1 integration as a target for antiretroviral therapy: a review. Curr. Drug Targets 1:133-149. [DOI] [PubMed] [Google Scholar]
- 40.Pluymers, W., N. Neamati, C. Pannecouque, V. Fikkert, C. Marchand, T. R. Burke, Jr., Y. Pommier, D. Schols, E. De Clercq, Z. Debyser, and M. Witvrouw. 2000. Viral entry as the primary target for the anti-HIV activity of chicoric acid and its tetra-acetyl esters. Mol. Pharmacol. 58:641-648. [DOI] [PubMed] [Google Scholar]
- 41.Pommier, Y., C. Marchand, and N. Neamati. 2000. Retroviral integrase inhibitors year 2000: update and perspectives. Antivir. Res. 47:139-148. [DOI] [PubMed] [Google Scholar]
- 42.Popov, S., M. Rexach, L. Ratner, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J. Biol. Chem. 273:13347-13352. [DOI] [PubMed] [Google Scholar]
- 43.Sakai, H., M. Kawamura, J. Sakuragi, S. Sakuragi, R. Shibata, A. Ishimoto, N. Ono, S. Ueda, and A. Adachi. 1993. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 67:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shank, P. R., S. H. Hughes, H. J. Kung, J. E. Majors, N. Quintrell, R. V. Guntaka, J. M. Bishop, and H. E. Varmus. 1978. Mapping unintegrated avian sarcoma virus DNA: termini of linear DNA bear 300 nucleotides present once or twice in two species of circular DNA. Cell 15:1383-1395. [DOI] [PubMed] [Google Scholar]
- 45.Sherman, M. P., C. M. de Noronha, M. I. Heusch, S. Greene, and W. C. Greene. 2001. Nucleocytoplasmic shuttling by human immunodeficiency virus type 1 Vpr. J. Virol. 75:1522-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandegraaff, N., R. Kumar, C. J. Burrell, and P. Li. 2001. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 75:11253-11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vigna, E., and L. Naldini. 2000. Lentiviral vectors: excellent tools for experimental gene transfer and promising candidates for gene therapy. J. Gene Med. 2:308-316. [DOI] [PubMed] [Google Scholar]
- 48.von Schwedler, U., R. S. Kornbluth, and D. Trono. 1994. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T-lymphocytes. Proc. Natl. Acad. Sci. USA 91:6992-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimura, F. K., and R. A. Weinberg. 1979. Restriction endonuclease cleavage of linear and closed circular murine leukemia viral DNAs: discovery of a smaller circular form. Cell 16:323-332. [DOI] [PubMed] [Google Scholar]
- 51.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 52.Zennou, V., C. Serguera, C. Sarkis, P. Colin, E. Perret, J. Mallet, and P. Charneau. 2001. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat. Biotechnol. 19:446-450. [DOI] [PubMed] [Google Scholar]
- 53.Zufferey, R., T. Dull, R. J. Mandel, A. Bukovsky, D. Quiroz, L. Naldini, and D. Trono. 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72:9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zufferey, R., J. E. Donello, D. Trono, and T. J. Hope. 1999. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]