Abstract
As a step toward understanding the assembly pathway of the porcine reproductive and respiratory syndrome virus (PRRSV), the oligomeric properties of the nucleocapsid (N) protein were investigated. In this study, we have demonstrated that under nonreducing conditions the N protein forms disulfide-linked homodimers. However, inclusion of an alkylating agent (N-ethylmaleimide [NEM]) prevented disulfide bond formation, suggesting that these intermolecular disulfide linkages were formed as a result of spurious oxidation during cell lysis. In contrast, N protein homodimers isolated from extracellular virions were shown to have formed NEM-resistant intermolecular disulfide linkages, the function of which is probably to impart stability to the virion. Pulse-chase analysis revealed that N protein homodimers become specifically disulfide linked within the virus-infected cell, albeit at the later stages of infection, conceivably when the virus particle buds into the oxidizing environment of the endoplasmic reticulum. Moreover, NEM-resistant disulfide linkages were shown to occur only during productive PRRSV infection, since expression of recombinant N protein did not result in the formation of NEM-resistant disulfide-linked homodimers. Mutational analysis indicated that of the three conserved cysteine residues in the N protein, only the cysteine at position 23 was involved in the formation of disulfide linkages. The N protein dimer was shown to be stable both in the presence and absence of intermolecular disulfide linkages, indicating that noncovalent interactions also play a role in dimerization. Non-disulfide-mediated N protein interactions were subsequently demonstrated both in vitro by the glutathione S-transferase (GST) pull-down assay and in vivo by the mammalian two-hybrid assay. Using a series of N protein deletion mutants fused to GST, amino acids 30 to 37 were shown to be essential for N-N interactions. Furthermore, since RNase A treatment markedly decreased N protein-binding affinity, it appears that at least in vitro, RNA may be involved in bridging N-N interactions. In cross-linking experiments, the N protein was shown to assemble into higher-order structures, including dimers, trimers, tetramers, and pentamers. Together, these findings demonstrate that the N protein possesses self-associative properties, and these likely provide the basis for PRRSV nucleocapsid assembly.
Porcine reproductive and respiratory syndrome is a widespread disease causing respiratory disorders in young pigs and reproductive failure in sows and gilts. The disease was first recognized in North America in 1987 (reviewed in reference 15), and shortly thereafter, the etiological agent was isolated from pigs in Europe (45) and in North America (2, 7). North American and European isolates of porcine reproductive and respiratory syndrome virus (PRRSV) correspond to two distinct genotypes (25, 27, 30), which exhibit significant antigenic differences (31, 46). Consequently, PRRSV has been divided into two subspecies, European (type I) and North American (type II), which show ∼60.4% nucleotide similarity.
PRRSV is an enveloped, positive-sense RNA virus belonging to the family Arteriviridae in the order Nidovirales (5). The PRRSV genome is ∼15 kb long (28) with a 3′ polyadenylated tail (36, 50) and, by extrapolation from what is known of the simian hemorrhagic fever virus—another member of the family Arteriviridae—possesses a 5′ cap (37). The replicase proteins, which are expressed from the genomic RNA, are encoded by two large open reading frames (ORFs) (1a and 1b) located at the 5′ end of the genome. The structural proteins are encoded by seven downstream ORFs and are translated from a nested set of 3′-coterminal subgenomic mRNAs (28, 51). The coding region of the genome is flanked by 5′ and 3′ nontranslated regions of 189 and 151 nucleotides, respectively.
ORF7, located at the 3′ terminus of the genome, encodes the nucleocapsid (N) protein, the sole protein believed to form the viral capsid. The nucleotide and predicted amino acid sequences of the N protein are well conserved among PRRSV isolates within a given genotype, whereas N proteins from the North American and European genotypes show only 56% identity. The N protein is a small, basic, 123-amino-acid protein that is highly immunogenic. Accordingly, numerous panels of monoclonal antibodies (MAbs) have been generated, the major epitopes for which lie predominantly in the central region of the protein (34, 49) in an area of high surface probability (16). The carboxy terminus is important for maintaining the antigenic structure of the N protein, as deletion or mutation in this region destroys most conformational epitopes (29, 47, 49). The amino-terminal half of the protein contains two stretches of basic amino acids that are presumed to be involved in binding to genomic RNA (8). In addition, these basic amino acids were identified as putative nuclear localization signals (35). Recent studies have demonstrated that the PRRSV N protein is phosphorylated on serine residues; however the function of this modification has yet to be determined (48).
PRRSV is assembled by the budding of preformed nucleocapsids into the lumen of the endoplasmic reticulum (ER) and/or Golgi region (for a review, see reference 41). Vesicles containing enveloped nucleocapsids are then released by means of exocytosis or cell lysis (9). The oligomeric process of PRRSV core particle assembly has not yet been elucidated; however, the prominence of a dimeric form of the N protein has been noted (10, 24). As the viral coat protein, the N protein likely possesses the ability to interact with itself as well as to multimerize. Furthermore, since the maturation process following icosahedral-virus capsid assembly often involves the formation of disulfide bonds, or other covalent linkages (reference 23 and references therein), it is possible that disulfide linkages may be involved in N protein interactions. The purpose of this study, therefore, was to elucidate the mechanism by which N proteins interact and to evaluate the role of disulfide bonds in this interaction. Our results suggest that the N protein dimerizes shortly after synthesis via noncovalent interactions and that this association occurs independently of other PRRSV proteins. Following exposure to an oxidizing environment, such as that encountered in the ER during egress from the cell, the N protein dimer becomes disulfide linked, the function of which is probably to impart stability to virions.
MATERIALS AND METHODS
Cells and viruses.
HeLa, BSC, TK-143, and Marc-145 (a subclone of MA104 cells [18]) cells were grown in Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum (CanSera). The cells were maintained at 37°C with 5% CO2. Stocks of PRRSV (strain PA8), wild-type vaccinia virus, recombinant vaccinia viruses (vTF7-3 [14] and VV-N expressing the PRRSV N protein) were prepared in Marc-145 or BSC cells as described previously (49, 50).
Antibodies.
Antibodies utilized in this study included a mixture of N-specific MAbs described previously (31, 47, 49). The monospecific polyclonal rabbit antiserum raised to the N protein was a generous gift from Serge Dea, Institute Armand-Frappier, Laval, Quebec, Canada.
Plasmid construction.
cDNA cloning of the PRRSV N gene to produce pGEM3zf-ORF7; pCITE-N; pCITE-GST-N; and the carboxy-terminal deletion mutants pGEX-C-11, pGEX-C-50, pGEX-C-66, pGEX-C-86, and pGEX-C-98 was described previously (47, 49). To construct the amino-terminal deletion mutants pGEX-N-18, pGEX-N-30, pGEX-N-52, and pGEX-N-69, N gene fragments from the respective deletion mutants in pCITE-2c (described in reference 49) were first digested with PstI, blunted with T4 DNA polymerase, and then digested with BglII. The resultant fragments were subcloned into the BamHI-SmaI site of pGEX-2T (Pharmacia). For the construction of recombinant vaccinia virus expressing the PRRSV N protein (VV-N), the N gene excised from pGEM3zf-ORF7 using BamHI was inserted into the BglII site of the vaccinia virus transfer vector pVVSL1, producing pVVSL1-N. Linearized pVVSL1-N was electroporated into BSC cells previously infected with wild-type vaccinia virus (Western Reserve) to allow for homologous recombination in vivo. Recombinant vaccinia virus was plaque purified by two rounds of plaque assays on TK−143 cells in the presence of 25 μg of 5-bromodeoxyuridine (Sigma)/ml. The recombinant vaccinia virus expressing the PRRSV N protein was confirmed by PCR and radioimmunoprecipitation. General procedures for DNA manipulation, cloning, and PCR are described elsewhere (38). The Escherichiacoli strains XL1-Blue (Stratagene) and DH5α were used as hosts for generating mutant genes and for general-purpose cloning, respectively.
Mutagenesis.
The desired cysteine mutations were incorporated into the N gene using the following primer pairs: C23S-Fwd, 5′-GTCAATCAGCTGaGCCAGATGCTG-3′; C23S-Rv, 5′-CAGCATCTGGCtCAGCTGATTGAC-3′; C75S-Fwd, 5′-GCGGCAATTGaGTCTGTCGTCAATC-3′; C75S-Rv, 5′-GATTGACGACAGACtCAATTGCCGC-3′; C90S-Fwd, 5′-CGCTGGGACTaGCACCCTGTCAG-3′; and C90S-Rv, 5′-CTGACAGGGTGCtAGTCCCAGCG-3′ (lowercase letters represent mutated nucleotides). PCR mutagenesis and screening were carried out as described previously (47).
Protein expression, radiolabeling, immunoprecipitation, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
For proteins expressed using the T7 vaccinia system, HeLa cells grown to 90% confluence were infected for 1 h at 37°C with vTF7-3 at a multiplicity of infection of 10. Following infection, fresh medium containing 10% fetal calf serum was added, and incubation continued for an additional 1 h. The cells were washed twice in OPTI-MEM and then transfected for 12 h using Lipofectin (Invitrogen) according to the manufacturer's instructions. The transfection solution was removed, and the cells were starved for 1 h in methionine-deficient medium. The cells were then labeled for 5 h in the presence of 50 μCi of EasyTag EXPRESS protein labeling mix ([35S]methionine and [35S]cysteine; specific activity, 407 MBq/ml) (Perkin-Elmer)/ml. For coimmunoprecipitation experiments, plasmids pCITE-N and pCITE-GST-N were cotransfected into HeLa cells and labeled as described above. For pulse-chase experiments, PRRSV-infected cells seeded on 35-mm-diameter dishes were starved for 1 h in methionine-deficient medium 36 h postinfection and pulse-labeled for 30 min with 150 μCi of [35S]methionine/ml. The cells were washed and replenished with normal Dulbecco's modified Eagle medium for the duration of the chase period.
At the end of the labeling period, the cells were harvested, washed twice with cold phosphate-buffered saline (PBS), and lysed with RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 150 mM NaCl, 50 mM Tris-HCl [pH 7.4], 10 mM EDTA, 0.1% SDS) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) with or without 20 mM N-ethylmaleimide (NEM) (Fisher). After incubation on ice for 20 min, the cell lysates were centrifuged at 14,000 rpm for 30 min in a microcentrifuge (model 5415; Eppendorf), and the supernatants were recovered. For immunoprecipitation, cell lysates equivalent to 1/15 of a 100-mm-diameter dish were adjusted with RIPA buffer to a final volume of 100 μl and incubated for 2 h at room temperature with 1 μl of a mixture of N-specific MAbs. The immune complexes were adsorbed to 7 mg of protein A-Sepharose CL-4B beads (Amersham Biosciences) for 16 h at 4°C. The beads were collected by centrifugation at 6,000 rpm (model 5415; Eppendorf) for 2 min and washed twice with RIPA buffer and once with wash buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). The beads were resuspended in 20 μl of SDS-PAGE sample buffer (10 mM Tris-HCl [pH 6.8], 25% glycerol, 10% SDS, 0.12% [wt/vol] bromophenol blue) with or without 10% β-mercaptoethanol (βME), boiled for 5 min, and analyzed by SDS-12% PAGE. The gels were dried on filter paper, and radiographic images were obtained using a PhosphorImager (Molecular Dynamics PhosphorImager SI). In the case of the coimmunoprecipitation experiment, coimmunoprecipitation lysis buffer (50 mM Tris HCl [pH 8.0], 120 mM NaCl, 0.5% NP-40, 1 mM PMSF) was used in place of RIPA buffer throughout the procedure.
Virus purification.
Culture supernatant from PRRSV-infected cells labeled with [35S]methionine was centrifuged at 2,500 rpm (model TJ-6; Beckman) for 15 min to remove cellular debris. The virus particles were purified through a 20% (wt/vol) sucrose cushion prepared in TE buffer (10 mM Tris HCl [pH 8], 1 mM EDTA) at 35,000 rpm for 2 h at 4°C in an SW41 rotor (model XL-90; Beckman). The pellets were resuspended in 80 μl of RIPA buffer containing 1 mM PMSF with or without 20 mM NEM and used in immunoprecipitation. For sucrose gradient fractionation of the N protein, cell lysates prepared in the presence or absence of 20 mM NEM were layered on top of a 10 to 40% sucrose gradient prepared in STE buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 2 mM EDTA, 0.1% NP-40). Centrifugation was performed at 45,000 rpm for 20 h in an SW55 rotor (Beckman), and a total of 20 fractions were collected from the top to the bottom of the gradient.
Protein expression in E. coli.
Glutathione S-transfererase (GST) fusion proteins were expressed in E. coli strain DH5α. One hundred milliliters of Luria-Bertani medium containing 100 μg of ampicillin/ml was inoculated with 1/100 of an overnight culture and grown to an optical density at 600 nm of 0.6. The cultures were induced with 1 mM IPTG (isopropylthio-β-d-galactoside), and after 3 h, the bacteria were pelleted for 10 min at 6,000 × g. The pellet was resuspended in 5 ml of PBS and sonicated on ice three times for 30 s each time with 2-s intervals (model W-385; Heat Systems-Ultrasonics, Inc.). Triton X-100 was added to a final concentration of 1%, and the proteins were solubilized for 30 min at 4°C with constant agitation. The insoluble fraction and cell debris were removed by centrifugation at 10,000 × g for 10 min at 4°C. The supernatants that were recovered were incubated with 100 μl of a 50% slurry of glutathione Sepharose 4B beads (Amersham Pharmacia) for 30 min at 4°C with constant agitation. The beads complexed to the GST fusion proteins were collected at 1,000 × g, washed five times in PBS containing 1% Triton X-100, and resuspended in a final volume of 250 μl of binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 5 mM dithiothreitol, 0.5% NP-40, 1 mM PMSF, 5% glycerol), resulting in a 20% slurry for use in subsequent GST pull-down assays.
GST pull-down assay.
To produce radiolabeled recombinant N protein, plasmid pCITE-N was transcribed in vitro using T7 RNA polymerase (T7 mMESSAGE mMACHINE; Ambion), and the resulting RNA was translated in rabbit reticulocyte lysate (Promega) in the presence of 50 μCi of [35S]methionine/ml according to the manufacturer's instructions. To produce radiolabeled authentic N protein, PRRSV-infected Marc-145 cells were labeled 36 h postinfection with 50 μCi of [35S]methionine/ml for 8 h, after which cell lysates were prepared using RIPA buffer as described above. Approximately equal amounts, as judged by Coomassie blue staining, of the various GST fusion proteins complexed to glutathione-Sepharose beads in a 20% slurry were incubated with [35S]methionine-labeled N proteins in binding buffer in a final volume of 400 μl overnight at 4°C with constant agitation. The beads were rinsed four times in binding buffer and boiled for 5 min in SDS-PAGE sample buffer containing 10% βME, and the proteins were analyzed by electrophoresis in 12% polyacrylamide gels. The gels were dried and exposed to a PhosphorImager to obtain radiographic images.
RNase A treatment.
The effect of RNase A treatment on protein binding in vitro was examined according to the protocol of Burniston et al. (3). Briefly, GST fusion proteins were bound to glutathione-Sepharose beads as described above and washed once with binding buffer containing 5 mM EDTA. The beads were resuspended in the same buffer containing 50 or 100 μg of RNase A/ml and incubated for 1 h at 37°C. Simultaneously, cell lysate containing radiolabeled N protein was diluted at a ratio of 1:100 in binding buffer supplemented with 5 mM EDTA containing 50 or 100 μg of RNase A/ml, followed by incubation for 1 h at 37°C. The glutathione-Sepharose beads coupled to the GST fusion protein were pelleted and resuspended in the RNase-treated cell lysate and incubated overnight at 4°C. The glutathione-Sepharose beads were washed four times with binding buffer, resuspended in SDS-PAGE sample buffer containing βME, and boiled for 5 min. The bound proteins were resolved by SDS-PAGE and visualized using a PhosphorImager.
Immunoblot analysis.
Cell lysates were separated on an SDS- 12% polyacrylamide gel under reducing conditions, followed by transfer to a nitrocellulose membrane in 10 mM 3-[cyclohexamino]-1-propanesulfonic acid buffer (pH 10.4; Sigma) prepared in 20% methanol. The membrane was blocked with 5% (wt/vol) skim milk in TBS (10 mM Tris HCl [pH 8], 150 mM NaCl) overnight and washed three times for 5 min each time in TBS containing 0.05% Tween 20. The membrane was then incubated with N-specific rabbit antiserum at a dilution of 1:5,000 in 1% bovine serum albumin-TBS for 2 h. The membrane was washed again and incubated with the alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (H+L) (Bio-Rad) at a dilution of 1:5,000 in 1% bovine serum albumin-TBS for 1 h. Color was developed with BCIP (5-bromo-6-chloro-3-indolyl phosphate; p-toluidine salt) and nitro blue tetrazolium chloride (Bio-Rad) in 100 mM Tris-HCl- 100 mM NaCl- 5 mM MgCl2 (pH 9.5) according to the manufacturer's instructions.
Mammalian two-hybrid assay.
The N gene was subcloned into the BamHI site of pM and pVP16 (Clontech) in frame with the Gal4 DNA-binding domain or the herpes simplex virus VP16 activation domain, respectively. The reporter construct p5xGal4SV40-luc, which contains the luciferase reporter gene and five copies of the Gal4 DNA-binding site upstream of the simian virus 40 (SV40) promoter, was generously provided by Milo Vassallo and Naoko Tanese (New York University School of Medicine, New York, N.Y.). The pM and pVP16 N gene constructs, along with either p5xGal4SV40-luc or pG5CAT, were cotransfected into HeLa cells at 50% confluence in a ratio of 5:5:1 and assayed for reporter gene activity 48 h posttransfection. Luciferase (Dual-Luciferase Reporter Assay; Promega) and chloramphenicol acetyltransferase (CAT) (Flash CAT Nonradioactive CAT assay kit; Strategene) activities were determined according to the instructions of the manufacturer.
N protein cross-linking.
The membrane-permeable and thiol-cleavable cross-linker 3,3′-dithiobis(succinimidylpropionate) (DSP) (Pierce) was prepared at 2 mM in 10% (vol/vol in PBS) dimethyl sulfoxide according to the manufacturer's instructions. PRRSV- or VV-N-infected Marc-145 cells (100-mm-diameter dish) were harvested 36 or 18 h postinfection, respectively. Virus particles were purified as described above. Samples were resuspended in 0.8 ml of PBS containing 2 mM DSP and incubated at room temperature for various periods up to 1 h. The reaction was quenched with 50 mM Tris-HCl [pH 7.5] and incubated for an additional 15 min. The cells were lysed in RIPA buffer containing 1 mM PMSF, denatured under nonreducing conditions, and separated by SDS-PAGE followed by transfer to nitrocellulose for immunoblot analysis.
RESULTS
Dimerization of the PRRSV N protein.
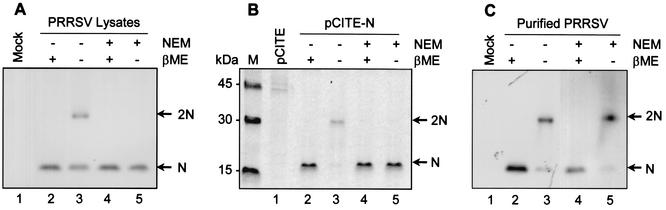
Fundamental to the process of virus particle assembly is the ability of the nucleocapsid protein to interact with itself. The purpose of this study, therefore, was to elucidate the mechanism by which N proteins interact. The N proteins of all North American-type PRRSV sequenced to date contain three conserved cysteine residues at positions 23, 75, and 90, whereas N proteins from the European-type PRRSV contain only two conserved cysteine residues at positions 27 and 76. Given the conserved nature and locations of these cysteine residues, at least in the PRRSV N protein, and given that disulfide bonds often form between capsid proteins during the maturation process (23), it was of interest to investigate whether disulfide bonds might participate in N-N interactions. To explore this possibility, N protein expressed from PRRSV-infected cells was immunoprecipitated and resolved on an SDS-PAGE gel under reducing and nonreducing conditions. A protein with an approximate molecular mass of 30 kDa was identified under nonreducing conditions (Fig. 1A, lane 3), and this was thought to be the N protein dimer. To address the possibility that the disulfide linkage had formed as a result of oxidation during sample preparation, the alkylating agent NEM was included in both the wash and lysis buffers to prevent aberrant disulfide bond formation. The results showed that under nonreducing conditions, disulfide bonds ceased to form between cellular N proteins in the presence of NEM (Fig. 1A, lane 5). These data indicated that the disulfide bond formation observed between N proteins in Fig. 1A, lane 3, likely occurred as a result of the oxidizing conditions encountered during cell lysis. Germane to this observation, however, was the efficiency with which N proteins became disulfide linked, raising the possibility that cytoplasmic N protein may already be present as a noncovalent dimer or higher-order oligomer.
FIG. 1.
Dimerization of the PRRSV N protein. Virus-infected or N gene-transfected cells were radiolabeled with [35S]methionine and solubilized in the absence (−) (lanes 2 and 3) or presence (+) (lanes 4 and 5) of 20 mM NEM. The lysates were immunoprecipitated with a mixture of N-specific MAbs and resolved by SDS-PAGE under reducing (+βME) (lanes 2 and 4) or nonreducing (−βME) (lanes 3 and 5) conditions. (A) PRRSV-infected cells. Lane 1, mock infected; lanes 2 to 5, virus infected. (B) vTF7-3-infected and N gene-transfected cells. Lane 1, pCITE transfected; lanes 2 to 5, pCITE-N transfected. (C) PRRSV particles purified from the culture supernatant. Lane 1, mock infected; lanes 2 to 5, PRRSV infected. The radiographic images were obtained using a PhosphorImager. The arrows indicate monomers (N) and dimers (2N) of the N protein.
To investigate whether the same phenomenon occurred in an environment devoid of other PRRSV proteins, disulfide bond formation between recombinant N proteins was examined. Under nonreducing conditions in the absence of NEM, recombinant N protein shifted from the monomeric form (Fig. 1B, lane 2) to the dimeric form (Fig. 1B, lane 3). However, in the presence of NEM, disulfide-linked recombinant N protein dimers ceased to form (Fig. 1B, lane 5), an observation similar to that made with the N protein from virus-infected cells (Fig. 1A, lane 5).
To examine the status of disulfide-linked dimer formation in the virion, virus particles were purified and lysed in either the presence or absence of NEM and examined by immunoprecipitation and SDS-PAGE under reducing and nonreducing conditions. Contrary to what was observed for cytoplasmic N protein, in the virus particle, the dimeric form of N was present under nonreducing conditions in both the absence (Fig. 1C, lane 3) and the presence (Fig. 1C, lane 5) of NEM. These results suggest that disulfide bond formation between virion N proteins is specific. The presence of both the monomeric and the dimeric form of the N protein under nonreducing conditions suggests that not all of the N protein is dimerized within the virion. The existence of three cysteine residues within the N protein could theoretically allow both intra- and intermolecular disulfide bond formation, and therefore the possibility remains that some of the monomeric N protein observed under nonreducing conditions may have undergone intramolecular disulfide bonding.
NEM-resistant disulfide bond formation.
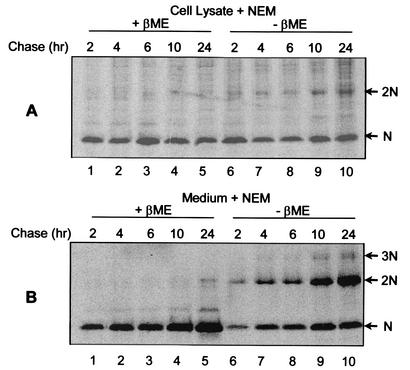
It was evident from the previous experiments that the majority of N protein in the virion formed disulfide-linked homodimers despite the presence of a high concentration of NEM during lysis (Fig. 1C, lane 5). Therefore, at some stage during virus assembly, N proteins become covalently disulfide linked. Since N is a cytoplasmic protein, disulfide bonds are not likely to form until the protein migrates out of the reducing environment of the cytoplasm (13). Therefore, pulse-chase studies were performed on PRRSV-infected cells to determine the length of time before disulfide-linked N proteins were formed. PRRSV-infected cells were pulse-labeled with [35S]methionine and chased for up to 24 h. At specified times, cells and virus particles (purified from the supernatant) were lysed in the presence of NEM and analyzed under reducing and nonreducing conditions. It was possible to observe the formation of NEM-resistant covalent dimers at the earliest time point of 2 h (Fig. 2A, lane 6). Although NEM-resistant dimers accumulated over time, the majority of N protein in the cell remained predominantly in the noncovalently linked form (lanes 6 to 10). The inefficient accumulation of NEM-resistant dimers in the cell may have been due to the rapid removal of specifically disulfide-linked homodimers that would accompany the release of mature virions, since in the extracellular virus a significant amount of the N protein population was in the form of covalently linked dimers (Fig. 2B, lanes 6 to 10). Moreover, a small fraction of the virion N protein was found to form higher-order NEM-resistant structures (lanes 7 to 10).
FIG. 2.
NEM-resistant disulfide bond formation between PRRSV N proteins. PRRSV-infected cells were pulse-labeled for 30 min with 150 μCi of [35S]methionine/ml and chased for the indicated times, after which the cells were solubilized in the presence of 20 mM NEM. After 2, 4, 6, 10, and 24 h of chase, the medium was collected and the virus was pelleted by centrifugation at 35,000 rpm (model XL-90; Beckman) for 2 h through a 20% sucrose cushion. The virus pellet was solubilized in the presence of 20 mM NEM. The N protein was immunoprecipitated from cell lysates (A) and purified virus particles (B), separated by SDS-PAGE under reducing (+βME) (lanes 1 to 5) or nonreducing (−βME) (lanes 6 to 10) conditions, and visualized using a PhosphorImager. The arrows indicate monomers (N), dimers (2N), and trimers (3N) of the N protein.
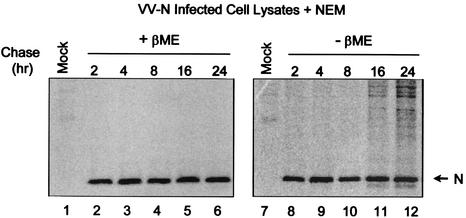
To determine whether the appearance of disulfide-linked N proteins required the presence of other viral components, cells infected with recombinant vaccinia virus expressing the PRRSV N protein were pulse-labeled with [35S]methionine and chased for up to 24 h. The cells were lysed at the indicated times in the presence of NEM, and NEM-resistant disulfide linkage formation was examined (Fig. 3). Unlike the situation in PRRSV-infected cells, at no point during the time course were NEM-resistant disulfide-linked recombinant N protein homodimers observed (lanes 8 to 12). Since it is likely that the process of preformed capsids budding into the ER is what allows NEM-resistant disulfide linkages to form, our observations suggest that the N protein alone may not be sufficient to promote capsid budding into the ER and that viral RNA or other viral proteins are required for this process. Therefore, although disulfide bonds form spontaneously between N proteins with or without virion formation and in the presence or absence of other viral proteins, it is likely that the formation of NEM-resistant disulfide linkages occurs only during productive PRRSV infection.
FIG. 3.
NEM-resistant disulfide-linked homodimer formation between recombinant N proteins. Marc-145 cells infected with the vaccinia virus recombinant VV-N were pulse-labeled with [35S]methionine and chased for the indicated times. The cells were solubilized in the presence of 20 mM NEM, and the N protein was immunoprecipitated using N-specific MAbs. The precipitated proteins were resolved by SDS-PAGE under reducing (+βME) (lanes 1 to 6) and nonreducing (−βME) (lanes 7 to 12) conditions and visualized using a PhosphorImager.
Sucrose gradient analysis of N protein dimers.
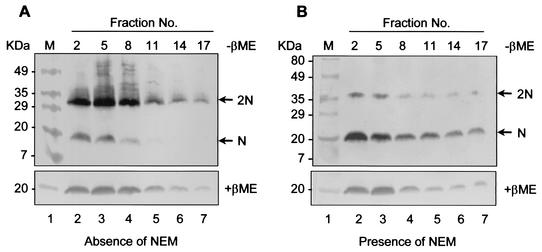
The results from the above-mentioned experiments (Fig. 1) suggest that the N protein in the cytoplasm may exist as a noncovalently linked homodimer regardless of whether disulfide bridges mediate this initial interaction. Therefore, to determine the size of the native oligomer, sucrose velocity gradient analysis was performed. PRRSV-infected cells were lysed in either the presence or absence of NEM and fractionated through a 10 to 40% sucrose gradient. The fractionated samples were separated under reducing or nonreducing conditions and subjected to immunoblot analysis (Fig. 4). In the absence of NEM, the majority of N protein formed disulfide-linked homodimers which sedimented predominantly in fractions 1 to 8 (Fig. 4A, lanes 2 to 4). In NEM-treated cell lysates, where the majority of the N protein would be expected to be in a non-disulfide-linked form, the N protein sedimented primarily in the same fractions as the disulfide-linked N protein dimer (Fig. 4B, lanes 2 to 4). Since the N protein migrated in the same relative position in the sucrose gradient regardless of whether disulfide bonds were allowed to form, this suggests that the dimer is stable. Therefore, it is possible that N proteins dimerize, at least initially, through noncovalent interactions. The strong tendency for dimeric interactions between N protein monomers also suggests that the noncovalent dimers could be the major intermediates in PRRSV capsid assembly.
FIG. 4.
Sucrose gradient analysis of N protein dimers in the presence or absence of NEM. PRRSV-infected cells were solubilized either in the absence (A) or presence (B) of 20 mM NEM, centrifuged at 14,000 rpm to remove insoluble materials, and then separated on a 10 to 40% linear sucrose gradient at 45,000 (model XL-90; Beckman) rpm for 20 h. Samples from designated fractions were run under nonreducing (−βME) (top) or reducing (+βME) (bottom) conditions and then transferred to nitrocellulose membranes. The membranes were probed with N-specific rabbit antibody. Sedimentation is from left to right. The arrows indicate monomers (N) and dimers (2N) of the N protein.
Analysis of intermolecular disulfide linkages in cysteine mutants.
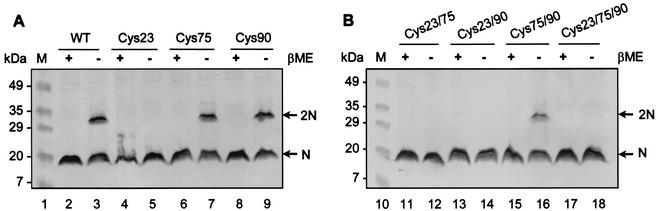
Having determined that intermolecular disulfide bond formation occurs between N proteins, we wanted to identify the cysteine residue(s) involved in this interaction. Individual cysteines at positions 23, 75, and 90 were mutated to serine to produce all seven possible combinations of single, double, and triple cysteine mutants. The cysteine mutants were expressed in HeLa cells using the T7 vaccinia virus system, and the cell lysates were resolved by SDS-PAGE under reducing and nonreducing conditions, followed by immunoblot analysis. Under nonreducing conditions, the wild-type N protein formed dimers as well as monomers (Fig. 5, lane 3). The single mutants Cys75 and Cys90 and the double mutant Cys75/90 exhibited the same pattern as the wild-type N protein (Fig. 5, lanes 7, 9, 16), indicating that Cys75 and Cys90 do not participate in intermolecular disulfide linkages. In contrast, Cys23, Cys23/75, Cys23/90, and Cys23/70/90 did not form dimers but rather remained as monomers under nonreducing conditions (Fig. 5, lanes 5, 12, 14, 18). These studies indicate that cysteine at position 23 is responsible for disulfide-linked dimerization of the N protein.
FIG. 5.
Analysis of intermolecular disulfide linkages in N protein cysteine mutants. HeLa cells infected with vTF7-3 were transfected with plasmids expressing N protein single cysteine mutants (A) and double and triple cysteine mutants (B). The cells were radiolabeled with [35S]methionine, lysed with RIPA buffer, and separated under reducing (+βME) or nonreducing (−βME) conditions. The proteins were transferred to a nitrocellulose membrane and subjected to immunoblot analysis using an N-specific rabbit antibody. The arrows indicate monomers (N) and dimers (2N) of the N protein. Molecular mass markers are shown on the left.
Homotypic interaction of the N protein in vivo.
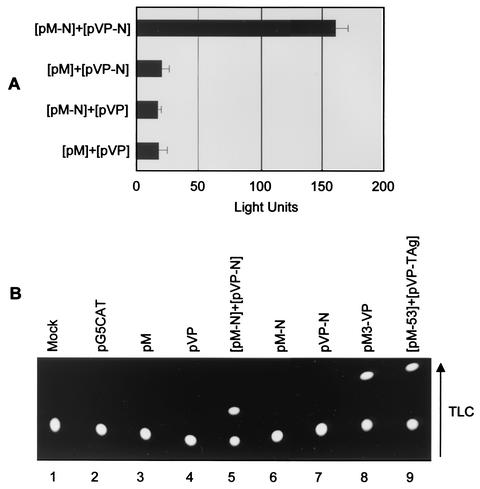
To investigate N-N interactions in vivo, the mammalian two-hybrid assay was employed. The N gene was cloned downstream of either the herpes simplex virus VP16 activation domain to generate pVP-N or the Gal4 DNA-binding domain to generate pM-N. The premise of the assay is that interaction between the two fusion proteins will tether the transcription activation and DNA-binding domains together upstream of either the SV40 (luciferase) or the adenovirus E1b (CAT) promoter, thereby enabling transcription of the downstream reporter gene. Expression of the N fusion proteins was confirmed by immunoblot analysis (data not shown). The pM and pVP constructs, along with the reporter construct p5xGal4SV40-luc or pG5CAT, were transfected into HeLa cells. A Renilla reporter construct (pRL-TK; Promega) was included in the transfection mixture to allow the normalization of transfection efficiencies. When the N protein was transfected along with the opposing empty vector, very little or no activation was observed (Fig. 6A and B, lanes 6 and 7), whereas when the interaction between the two N fusion proteins was examined, a significant increase in luciferase activation (Fig. 6A) or CAT expression (Fig. 6B, lane 5) ensued. These experiments confirmed that N proteins interact in vivo and further support the notion that the N protein displays a natural ability to homodimerize under otherwise reducing conditions.
FIG. 6.
Analysis of N protein interactions in vivo by the mammalian two-hybrid assay. HeLa cells seeded on 35-mm-diameter dishes were cotransfected with 100 ng of the reporter plasmid p5xGal4SV40-luc (A) or pG5CAT (B), and 500 ng of pM or pM-N together with 500 ng of pVP16 or pVP16-N as indicated. Renilla or β-galactosidase reporter constructs were included in the transfection mixture to allow normalization of transfection efficiencies in the luciferase (A) and CAT (B) assays, respectively. The luciferase and CAT assays were carried out 48 h posttransfection. For the luciferase assays, data from three independent experiments are shown; the error bars indicate standard deviations. For the CAT assay, the arrow indicates the direction of migration by thin-layer chromatography (TLC).
Homotypic interaction of the N protein in vitro.
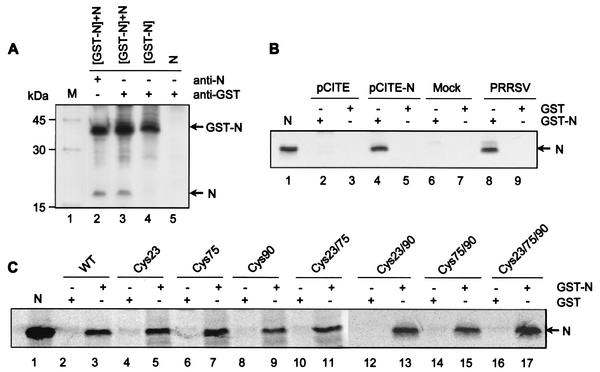
To study the ability of the N protein to interact with itself in vitro, a coimmunoprecipitation assay was performed (Fig. 7A). HeLa cells were cotransfected with plasmids expressing the N gene alone and the N gene as a GST fusion (GST-N), both of which were expressed using the T7 vaccinia virus system. The cotransfected cells were radiolabeled with [35S]methionine, and the cell lysates were immunoprecipitated with either an N-specific MAb or an anti-GST antibody in coimmunoprecipitation buffer. The N-specific MAbs precipitated both the N protein and the GST-N fusion protein (lane 2), indicating that each protein was efficiently expressed. In the coimmunoprecipitation experiment, both the N and GST-N proteins were immunoprecipitated with the GST antibody (lane 3), indicating that N and GST-N interact in this assay. The GST antibody did not immunoprecipitate the N protein alone (lane 5), which confirmed that N was precipitated due to a specific interaction with the N protein portion of the GST fusion.
FIG. 7.
Homotypic interactions between N proteins in vitro. (A) Coimmunoprecipitation of the N protein. HeLa cells infected with vTF7-3 were cotransfected with plasmids expressing the recombinant N protein alone (N) or as a GST fusion protein (GST-N). The cells were radiolabeled with [35S]methionine and lysed using a nonionic detergent (see Materials and Methods). The cell lysates were immunoprecipitated under reducing conditions in the presence (+) of nonionic detergents with either a mixture of N-specific MAbs (lane 2) or an anti-GST polyclonal antibody (lanes 3, 4, and 5) and analyzed by SDS-PAGE. Lanes 2 and 3, cotransfection of both GST-N and N gene constructs; lane 4, transfection of GST-N gene construct; lane 5, transfection of N gene construct. (B) GST pull-down assay. Bacterially expressed GST-N (lanes 2, 4, 6, and 8) or GST (lanes 3, 5, 7, and 9) were bound to glutathione-Sepharose beads and incubated with either in vitro-translated N protein labeled with [35S]methionine (lanes 4 and 5) or virus-infected cell lysates metabolically labeled with [35S]methionine (lanes 8 and 9). The beads were washed four times, and the bound proteins were eluted by boiling them in reducing sample buffer, followed by SDS-PAGE and autoradiography. The N protein immunoprecipitated from virus-infected cells was run in lane 1 as a marker. In vitro translation products from empty plasmid (lanes 2 and 3) and from mock-infected cell lysates (lanes 6 and 7) were incubated with the bead-bound proteins as a control. (C) GST pull-down assay examining the interaction between N protein cysteine mutants and the wild-type (WT) N protein in vitro. Bacterially expressed GST and GST-N proteins coupled to glutathione-Sepharose beads and incubated with individual cysteine mutants of the N protein synthesized by in vitro translation are shown. N protein immunoprecipitated from PRRSV-infected cells was run as a marker (lane 1).
The GST pull-down assay was conducted to further confirm the homodimeric property of the N protein in vitro. The N protein expressed as a GST fusion protein in E. coli was coupled to glutathione-Sepharose beads and incubated with either in vitro-translated N protein or N protein from radiolabeled PRRSV-infected cell lysates (Fig. 7B). After extensive washing of the bead-bound complex, the bound proteins were denatured under reducing conditions and analyzed by SDS-PAGE and autoradiography. In all binding experiments, approximately equal amounts of protein were used, as verified by Coomassie blue staining (data not shown). As shown in Fig. 7B, the N and GST-N proteins interacted efficiently in this assay (lanes 4, 8) whereas no N protein was precipitated in association with GST alone (lanes 5, 9), suggesting that the interaction between N and GST-N was specific. These results indicate clearly that the N protein interacts with itself in vitro.
To determine if mutation of cysteine residues would compromise homotypic N protein interactions in vitro, each of the single, double, and triple cysteine mutants was translated in vitro and used as prey in the GST pull-down assay. All of the mutants bound to GST-N with approximately equal affinity (Fig. 7C), thereby supporting the hypothesis that disulfide bonds, while important for stabilization of N protein dimers, are not the driving force behind this homotypic interaction. Therefore, cysteine residues are not required for N-N interactions in vitro.
Identification of the N protein homodimerization domain.
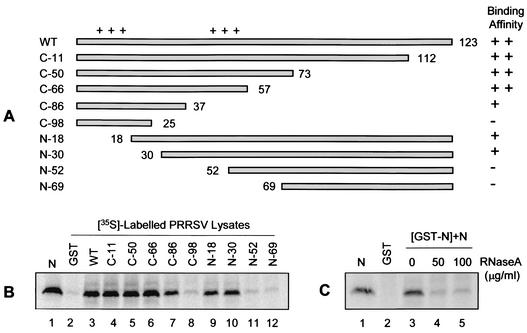
Given that the N protein interacts with itself even in the absence of disulfide bond formation (Fig. 7B, lane 17, shows a triple cysteine mutant), it was of interest to identify the region of the N protein mediating noncovalent and/or ionic interactions. To define the primary sequence required for N-N interactions, a series of GST-N fusion constructs were generated by progressively deleting amino acids from either the amino or carboxy terminus of the N protein (Fig. 8A). Each deletion mutant was expressed as a GST fusion protein and, following coupling to glutathione-Sepharose beads, was tested for its ability to interact with N protein from PRRSV-infected cell lysates (Fig. 8B). Expression levels were comparable for all of the GST-N deletion mutants examined (data not shown). The relative binding intensity was determined by densitometry analysis and normalized with respect to the wild-type N protein. As shown in Fig. 8B, deletion of up to 66 amino acids from the carboxy terminus (C-66 [lane 6]) did not compromise the strength of N-N interaction relative to that of the wild-type N (lane 3). Deletion of up to 86 carboxy-terminal amino acids (C-86 [lane 7]) or up to 30 amino-terminal amino acids (N-30 [lane 10]) resulted in a slight but similar reduction in the strength of N-N interactions (∼75 to 80% of wild-type activity). Further deletion of 98 amino acids from the carboxy terminus (C-98 [lane 8]) and 52 to 69 amino acids from the amino terminus (N-52 and N69 [lanes 11 and 12]) severely reduced N-N interactions (<15% of wild-type activity) to background levels (lane 2). Therefore, it appears that amino acids 30 to 37 are important for mediating N protein homotypic interactions in vitro.
FIG. 8.
Identification of the N protein homodimerization domain. (A) Schematic presentation of various deletion mutants of the N protein. The horizontal bars represent the residues expressed by the mutants, and the numbers indicate the residues where deletions begin (in the case of amino-terminal deletions) or end (in the case of carboxy-terminal deletions). The plus signs above the bars represent the two stretches of basic amino acids. N binding activity was quantitated densitometrically and is presented at the right of the figure: ++, activity >90% of wild type; +, activity up to 80% of wild type; −, activity <15% of wild type. (B) Autoradiographs illustrating the N protein-binding activity of amino- and carboxy-terminal deletion mutants. Immunoprecipitated N protein was included as a marker (lane 1). In all cases, equal amounts of protein bound to beads were used, as estimated by Coomassie blue staining. (C) Effect of RNase A treatment on N protein self-association in vitro. GST and GST-N bound to glutathione-Sepharose beads were incubated for 1 h at 37°C with the indicated concentrations of RNase A. The beads were washed three times and used in binding experiments with [35S]methionine-labeled PRRSV-infected cell lysates that had been treated with various concentrations of RNase A for 1 h at 37°C. Following overnight incubation at 4°C, the beads were washed three times and the bound proteins were visualized by SDS-PAGE and PhosphorImager analysis. N protein immunoprecipitated from PRRSV-infected cells was run as a marker (lanes 1).
The deletion mutants C-86, N-18, and N-30 all exhibited slightly reduced binding affinities for the N protein. While these deletion mutants possess the homodimerization domain, they each encode only one of the two clusters of positively charged residues, whether amino acids 10 to 13 or 43 to 48. Given the close proximity of the clusters of positively charged amino acids to the putative homodimerization domain, we wanted to investigate whether these highly basic residues might contribute to N-N interactions in vitro through RNA bridging. Accordingly, the effect of RNase treatment on N-N interactions was examined. In this modified binding assay (3), both bead-bound proteins and cell lysates were treated with RNase A prior to coincubation. Upon examination of the bound proteins, it was evident that RNase A treatment markedly decreased N-N interactions (Fig. 8C, lanes 4 and 5) relative to the control (lane 3). These results suggest that, at least in vitro, RNA may participate in bridging N-N interactions.
Higher-order oligomerization of the N protein.
The oligomeric properties of the PRRSV N protein, in both the presence and absence of other viral constituents, were further examined by cross-linking studies. The membrane-permeable cross-linking reagent DSP was applied to PRRSV- or VV-N-infected cells, and the resultant oligomeric forms of the N protein were examined by immunoblot analysis. When the N protein from purified virions was subjected to DSP cross-linking in a time course experiment, a number of higher-order oligomers of N were observed (Fig. 9A). It appeared as though each successive oligomer resulted from the addition of a single monomer of N, suggesting that the building blocks for nucleocapsid assembly may in fact be a combination of monomers and dimers of N, which then associate into higher-order structures. Similarly, numerous oligomeric forms of the N protein were identified in virus-infected cells (Fig. 9B, lane 2). Interestingly, these oligomeric forms of the N protein were also readily observed in cells expressing recombinant N protein (Fig. 9B, lane 3), thereby indicating that the oligomeric properties of the N protein are inherent and independent of other viral proteins.
FIG. 9.
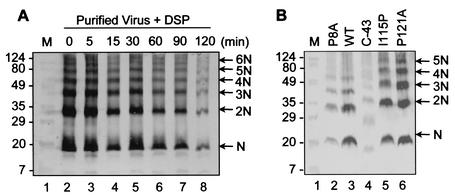
Analysis of N protein multimerization by DSP cross-linking. Virus particles or cells expressing either viral or recombinant N proteins were cross-linked with 2 mM DSP for up to 1 h and solubilized in RIPA buffer. The lysates were boiled in standard SDS sample buffer under nonreducing conditions and were separated by SDS-PAGE followed by immunoblot analysis using N-specific rabbit antibody and the alkaline phosphatase-conjugated secondary antibody against rabbit immunoglobulin G. (A) Kinetic studies of purified virus samples treated with DSP and cross-linked for increasing periods of time. (B) Cross-linked N protein point and deletion mutants probed with N-specific rabbit antibody. The arrows indicate oligomeric forms of the N protein. WT, wild type. Molecular mass markers (M) in kilodaltons are shown on the left.
Given that the recombinant N protein (Fig. 9B, lane 3) produced the same cross-linked products as the authentic N protein from PRRSV-infected cells (lane 2), DSP cross-linking could serve as a useful assay for investigating N protein oligomerization. Using this assay, we examined the oligomerization profile of the deletion mutant C-43, which encodes amino acids 1 to 80 and encompasses both the putative RNA-binding and N-N interaction domains (Fig. 9B, lane 4). This deletion mutant was able to oligomerize and form all of the higher-order structures that were observed for the wild-type N protein, suggesting that amino acids 81 to 123 are not essential for N protein oligomerization. Additionally, two carboxy-terminal point mutants, I115P (mutation of isoleucine at position 115 to proline) and P121A (mutation of proline at position 121 to alanine), which were previously shown to compromise N protein conformational antigenic epitopes (47), were assessed for the ability to multimerize. In both cases, the mutants formed the same higher-order oligomers (Fig. 9B, lanes 5 and 6) as the wild-type N protein (lane 3). These observations suggest that although the C terminus is important for maintaining the antigenic structure of the N protein, it may not be necessary for mediating N protein homotypic interactions. We conclude from these multimerization experiments that (i) both cytoplasmic and virion N proteins form at least monomers, dimers, trimers, tetramers, and pentamers and (ii) the carboxy-terminal amino acids, although critical for the antigenic structure of N, are not required for oligomerization.
DISCUSSION
One of the fundamental roles of the viral capsid protein is to provide a protective enclosure for the viral genome during the extracellular phase of the virus life cycle. It would follow, then, that capsid proteins would exhibit some potential for multimerization. At present, very little is known about the structure of the PRRSV virion, except for its overall size and shape. Extracellular virions appear as pleomorphic or spherical enveloped particles, 50 to 72 nm in diameter, with an isometric core (41) ∼25 to 30 nm in diameter (9). Since the formation or release of virus-like particles and the assembly of detectable structures intracellularly or in vitro has not been reported upon expression of recombinant N protein, we set out to analyze whether N proteins, or segments thereof, interact with themselves and, if so, to map the putative interaction domains. The findings in this report indicate that the N protein is capable of forming dimers and higher-order multimers, providing the beginnings of a structural basis for particle formation in PRRSV and for arteriviruses in general. Here, we have demonstrated that the N polypeptide exhibits specific protein-protein interactions with itself both in vivo and in vitro and that the protein-binding capacity resides in the ability of the N protein to form both covalent (in the form of disulfide linkages) and noncovalent interactions.
Role of disulfide bonds.
We have demonstrated that N proteins interact to form dimers and that these appear to be the major oxidative products. It was found that NEM-resistant dimers, and therefore specifically disulfide-linked dimers, are formed within the virus particle. Additionally, pulse-chase experiments indicated that in the later stages of infection, NEM-resistant disulfide linkages form between N proteins within the virus-infected cell. It appears, therefore, that naturally occurring disulfide bonds form between N proteins when the virus particle egresses from the cell, suggesting that exposure to oxidizing conditions is what enables these dimers to be stabilized by disulfide linkages, a phenomenon common to many icosahedral viruses (23). Recombinant N protein, while able to form dimers, was unable to form NEM-resistant disulfide linkages, thereby supporting the notion that virus assembly is a necessary prerequisite for specific disulfide bond formation. However, the formation of homodimers means that in mechanistic terms N proteins exhibit a strong tendency to dimerize and that dimerization occurs faster than diffusion. It appears, therefore, that the N protein has an intrinsic ability to dimerize.
Since virions exist as metastable structures, they must be efficiently assembled, resistant to environmental degradation, and yet capable of disassembly upon infection. Disulfide bonds offer one mechanism for viruses to switch between various degrees of stability by taking advantage of the difference in oxidizing conditions between the intra- and extracellular environments (13, 23). Disulfide bonds are reported to have been involved in the assembly and stabilization of the viral capsid structure of a number of viruses with icosahedral nucleocapsids (1, 17, 21, 22, 53), and it is frequently the case that these disulfide bonds form as the virus exits the cell.
Since the N protein lacks a signal sequence, it is presumed to be a soluble protein and therefore should exist in a reduced state while in the cytoplasm. During the viral life cycle, however, the PRRSV nucleocapsid enters the oxidizing environment of the ER as it acquires its outer envelope. As the virus particle enters the secretory pathway, it will be exposed to oxidizing conditions and will continue to be exposed when released from the host cell. Under these conditions, disulfide bonds would serve to strengthen the structure of the PRRSV nucleocapsid and hence protect the genome against the harsh conditions of the extracellular environment. Conversely, upon infection, exposure of the capsid to the reducing environment of the cytoplasm could destabilize disulfide bonds and lead to capsid disassembly. Therefore, it appears that, as with many viruses possessing icosahedral nucleocapsids, PRRSV takes advantage of the difference in redox potential that exists between the intra- and extracellular compartments of the cell in an effort to secure safe passage and ease of disassembly of the virus in the presence and absence of disulfide bonds, respectively.
While the principles of virus assembly would be expected to be similar for all arteriviruses, it is necessary to point out that the N proteins from equine arteritis virus and simian hemorrhagic fever virus, both members of the family Arteriviridae, completely lack cysteine residues. Our experimental data, however, do not suggest that disulfide bonds are the only, or even the primary, force mediating the initial N-N interactions. In fact, disulfide bonds do not appear to be relevant until the virus enters the secretory pathway (ER and Golgi) and/or egresses from the cell, both of which occur following core particle assembly. Furthermore, for many viruses with disulfide-linked capsid proteins, including hepatitis B virus (17), SV40 (22), and human papillomavirus (21), it appears that disulfide bonds function primarily to impart stability to the virion and/or to finalize the maturation process, as is the case for many icosahedral viruses (23). As such, the principles of arterivirus assembly may very well be the same for all family members, the only difference being that PRRSV has adapted an additional mechanism for stabilizing N-N interactions and, in turn, core particles.
Systematic mutation of the three cysteine residues at positions 23, 75, and 90, either alone or in combination, revealed that the cysteine at position 23 is involved in the formation of intermolecular disulfide bonds between N proteins. Therefore, cysteine 23 would be expected to be oriented within the N protein dimer so that, in the presence of an oxidizing environment, disulfide bond formation would be possible. Interestingly, cysteine 23 is located within the basic amino-terminal half of the protein, close to the region that is presumed to be involved in RNA binding (8). Topographical arrangement of cysteine 23 within the virus interior is consistent with this putative function and therefore suggests that intermolecular disulfide bonds may serve to stabilize RNA-protein interactions. Moreover, in GST pull-down assays, clusters of positively charged amino acids were shown to contribute to N-N interactions, thereby placing cysteine 23 in the most advantageous position of the three cysteine residues to participate in disulfide bridging. In contrast, cysteines at positions 75 and 90 do not appear to be involved in intermolecular disulfide bond formation, at least not within N protein homodimers. This, however, does not exclude the possibility that these residues participate in stabilization of higher-order oligomers or multimers of the N protein.
Primary sequence requirement for N-N interactions.
The highly reducing environment of the cytoplasm precludes the formation of disulfide bonds; therefore, it is unlikely that PRRSV nucleocapsid assembly depends solely upon disulfide bond formation. We demonstrated interaction between wild-type N proteins using a variety of different assays, including the GST pull-down assay and the mammalian two-hybrid assay, both of which essentially examine N-N interactions independent of disulfide bond formation. Using a series of deletion mutants fused to GST, amino acids 30 to 37 (IAQQNQSR) were shown to contribute most strongly to N-N interactions. Due to the high concentration of amide containing the residues asparagine (N) and glutamine (Q) in this stretch of amino acids, it is possible that N-N interactions occur partly through hydrogen bonding (40, 52). This domain corresponds to a relatively hydrophilic region of the N protein and therefore is likely to be exposed to the aqueous environment rather than sequestered internally. It is important to note, however, that while a specific binding domain may have been delineated, the PRRSV N protein is highly conformational, and without the crystallographic structure to refer to, conclusions about whether a mutation has a legitimate effect on N-N interactions or simply disrupts N protein structure await confirmation.
Additionally, N protein-binding affinity varied with respect to the presence or absence of two stretches of basic amino acids situated in the amino terminus. Removal of either one of these basic hydrophilic regions led to similarly reduced interaction capability. Presumably, N protein-RNA interactions are required for encapsidation of viral genomic RNA, and therefore it is possible that RNA promotes N-N interactions by neutralizing charge repulsions between the two stretches of basic amino acids (3). Examination of N-N interactions under conditions expected to eliminate RNA indicated that N binding activity is, to some extent, RNA dependent. This, however, does not rule out the possibility that RNA is required for the formation of higher-order multimers formed by the aggregation of N protein dimers. After all, each monomer must have at least two interfaces, one mediating dimerization of monomers and the other mediating multimerization of dimers (19).
Whether close proximity of the protein interaction and putative RNA-binding domains has any functional significance is not known. In this regard, however, it has been shown that the protein-binding domains in the influenza NS1 (32) and the human immunodeficiency virus Rev (33) proteins overlap with their RNA-binding domains. Similarly, the coronavirus nucleocapsid protein interacts with itself, and both the protein-protein- and protein-RNA-binding domains reside in similar locations between amino acids 163 and 229 (44). One potential outcome of this overlap of functional domains is that multimerization of the capsid protein may alter conformation of the RNA-binding domain, or vice versa, to facilitate RNA-protein interactions in virus assembly (4, 12, 20).
Multimerization of the PRRSV N protein.
Biological processes often require that a single gene product participate in multiple types of molecular interactions. Accordingly, viral capsids of multiple copies of identical gene products are formed so as to minimize the genetic information required for virus assembly. In many cases, capsid protein dimers serve as building blocks for the assembly of plant and animal viruses (1, 6, 11, 26, 39, 42, 43). Under our experimental conditions, it was possible to resolve at least four discrete N protein oligomers, including dimers, trimers, tetramers, and pentamers. These same oligomers were detected in virus-infected cells and N gene-transfected cells, as well as in the purified virion, suggesting that this assay reflects functional oligomeric contacts. These results suggest that nucleocapsid assembly occurs via direct polymerization of monomeric and dimeric forms of N or that these oligomers are reflective of the N protein configurations with the strongest contacts. It is also possible that both the protein-protein- and protein-RNA-binding domains mediate contacts between different N protein multimers during the various stages of oligomerization, just as cysteine at position 23 is relevant after certain contacts have been made that allow for virus egress from the cell.
In summary, we have demonstrated that the N protein possesses self-associative properties which likely provide the basis for PRRSV nucleocapsid assembly. We have shown that N proteins form dimers that progress from nondisulfide linked to disulfide linked via cysteine at position 23 as the virus buds from the cell. Finally, our experimental data suggest that amino acids 30 to 37 and RNA bridging may be involved in the stabilization of N-N interactions.
Acknowledgments
This work was supported by grants from the Ontario Ministry of Agriculture Food and Rural Affairs (OMAFRA) and Ontario Pork. S.K.W. is a recipient of the Ontario Graduate Scholarship.
REFERENCES
- 1.Baron, M. D., and K. Forsell. 1991. Oligomerization of the structural proteins of rubella virus. Virology 185:811-819. [DOI] [PubMed] [Google Scholar]
- 2.Benfield, D. A., E. Nelson, J. E. Collins, L. Harris, S. M. Goyal, D. Robinson, W. T. Christianson, R. B. Morrison, D. E. Gorcyca, and D. W. Chladek. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Investig. 4:127-133. [DOI] [PubMed] [Google Scholar]
- 3.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens, S. 1997. Principles of virion structure, function, and assembly, p. 3-37. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 5.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 6.Choi, J., and L. S. Loesch-Fries. 1999. Effect of C-terminal mutations of alfalfa mosaic virus coat protein on dimer formation and assembly in vitro. Virology 260:182-189. [DOI] [PubMed] [Google Scholar]
- 7.Collins, J. E., D. A. Benfield, W. T. Christianson, J. E. Harris, L. Hennings, J. C. Shaw, D. P. Goyal, S. M. McCullough, S. Morrison, R. B. Joo, H. S. Gorcyca, and D. W. Chladek. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Investig. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 8.Daginakatte, G. C., and S. Kapil. 2001. Mapping of the RNA-binding domain of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. Adv. Exp. Med. Biol. 494:547-552. [DOI] [PubMed] [Google Scholar]
- 9.Dea, S., N. Sawyer, R. Alain, and R. Athanassious. 1995. Ultrastructural characteristics and morphogenesis of porcine reproductive and respiratory syndrome virus propagated in the highly permissive MARC-145 cell clone. Adv. Exp. Med. Biol. 380:95-98. [DOI] [PubMed] [Google Scholar]
- 10.Delputte, P. L., N. Vanderheijden, H. J. Nauwynck, and M. B. Pensaert. 2002. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 76:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, Z., Q. R. Yang, J.-S. Twu, and A. H. Sherker. 1999. Specific in vitro association between the hepatitis C viral genome and core protein. J. Med. Virol. 59:131-134. [PubMed] [Google Scholar]
- 13.Freedman, R. B., B. E. Brockway, and N. Lambert. 1984. Protein disulphide-isomerase and the formation of native disulphide bonds. Biochem. Soc. Trans. 12:929-932. [DOI] [PubMed] [Google Scholar]
- 14.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill, H. 1990. Overview of the history of mystery swine disease (swine infertility respiratory syndrome), p. 29-31. In Proceedings of the Mystery Swine Disease Committee Meeting. Livestock Conservation Institute, Madison, Wis.
- 16.Janin, J., and S. Wodak. 1978. Conformation of amino acid side-chains in proteins. J. Mol. Biol. 125:357-386. [DOI] [PubMed] [Google Scholar]
- 17.Jeng, K. S., C. P. Hu, and C. M. Chang. 1991. Differential formation of disulfide linkages in the core antigen of extracellular and intracellular hepatitis B virus core particles. J. Virol. 65:3924-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 19.Konig, S., G. Beterams, and M. Nassal. 1998. Mapping of homologous interaction sites in the hepatitis B virus core protein. J. Virol. 72:4997-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel, M., M. Lorinczi, R. Rijnbrand, S. M. Lemon, and S. J. Watowich. 2001. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J. Virol. 75:2119-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, M., P. Beard, P. A. Estes, M. K. Lyon, and R. L. Garcea. 1998. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J. Virol. 72:2160-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, P. P., A. Nakanishi, S. W. Clark, and H. Kasamatsu. 2002. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc. Natl. Acad. Sci. USA 99:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liljas, L. 1999. Virus assembly. Curr. Opin. Struct. Biol. 9:129-134. [DOI] [PubMed] [Google Scholar]
- 24.Mardassi, H., B. Massie, and S. Dea. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221:98-112. [DOI] [PubMed] [Google Scholar]
- 25.Mardassi, H., S. Mounir, and S. Dea. 1994. Identification of major differences in the nucleocapsid protein genes of a Quebec strain and European strains of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 75:681-685. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto, M., S. B. Hwang, K.-S. Jeng, N. Zhu, and M. M. C. Lai. 1996. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology 218:43-51. [DOI] [PubMed] [Google Scholar]
- 27.Meng, X.-J., P. S. Paul, P. G. Halbur, and M. A. Lum. 1995. Phylogenetic analysis of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch. Virol. 140:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meulenberg, J. J. M., M. M. Hulst, E. J. de Meijer, P. J. M. Moonen, A. den Besten, E. P. De Kluyver, G. Wensvoort, and R. J. M. Moormann. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meulenberg, J. J. M., A. P. van Nieuwstadt, A. van Essen-Zandbergen, J. N. A. Bos-de-Ruijter, J. P. M. Langeveld, and R. H. Meloen. 1998. Localization and fine mapping of antigenic sites on the nucleocapsid protein of porcine reproductive and respiratory syndrome virus with monoclonal antibodies. Virology 252:106-114. [DOI] [PubMed] [Google Scholar]
- 30.Murtaugh, M. P., M. R. Elam, and L. T. Kakach. 1995. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch. Virol. 140:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson, E. A., J. Christopher-Hennings, T. Drew, G. Wensvoort, J. E. Collins, and D. A. Benfield. 1993. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 31:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemeroff, M. E., X. Y. Qian, and R. M. Krug. 1995. The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 212:422-428. [DOI] [PubMed] [Google Scholar]
- 33.Olsen, H. S., A. W. Cochrane, P. J. Dillon, C. M. Nalin, andC. A. Rosen. 1990. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 4:1357-1364. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, M. J., J. Sarraseca, J. Garcia, A. Sanz, J. Plana-Duran, and J. I. Casal. 1997. Epitope mapping of the nucleocapsid protein of European and North American isolates of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 78:2269-2278. [DOI] [PubMed] [Google Scholar]
- 35.Rowland, R. R., R. Kervin, C. Kuckleburg, A. Sperlich, and D. A. Benfield. 1999. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 64:1-12. [DOI] [PubMed] [Google Scholar]
- 36.Sagripanti, J. L. 1985. Polyadenylic acid sequences in the genomic RNA of the togavirus of simian hemorrhagic fever. Virology 145:350-355. [DOI] [PubMed] [Google Scholar]
- 37.Sagripanti, J. L., R. O. Zandomeni, and R. Weinmann. 1986. The cap structure of simian hemorrhagic fever virion RNA. Virology 151:146-150. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Seifer, M., and D. N. Standring. 1995. Ribonucleoprotein complex formation by the human hepatitis B virus polymerase. Intervirology 38:295-303. [DOI] [PubMed] [Google Scholar]
- 40.Shimoni, L., and J. P. Glusker. 1995. Hydrogen bonding motifs of protein side chains: descriptions of binding of arginine and amide. Protein Sci. 4:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 42.Tellinghuisen, T., and R. J. Kuhn. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, J. G., and S. M. Lemon. 1993. Hepatitis delta virus antigen forms dimers and multimeric complexes in vivo. J. Virol. 67:446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Y., and X. Zhang. 1999. The nucleocapsid protein of coronavirus mouse hepatitis virus interacts with the cellular heterogeneous nuclear ribonucleoprotein A1 in vitro and in vivo. Virology 265:96-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wensvoort, G., C. Tepstra, J. M. A. Pol, E. A. ter Laak, M. Bloemraad, E. P. de Kluyver, C. Kragten, L. van Buiten, A. den Besten, F. Wagenaar, J. M. Broekhuijsen, P. L. J. M. Moonen, T. Zestra, E. A. de Boer, H. J. Tibead Bindingen, M. F. de Jong, P. van Veld, G. J. R. Groenland, J. A. van Gennep, M. T. Voets, J. H. M. Verheijden, and J. Braamskamp. 1991. Mystery swine disease in the Netherlands: the isolation of Lelystad virus. Vet. Q. 13:121-130. [DOI] [PubMed] [Google Scholar]
- 46.Wensvoort, G., E. P. de Kluyver, E. A. Luijtze, A. den Besten, L. Harris, J. E. Collins, W. T. Christianson, and D. Chladek. 1992. Antigenic comparison of Lelystad virus and swine infertility and respiratory syndrome (SIRS) virus. J. Vet. Diagn. Investig. 4:134-138. [DOI] [PubMed] [Google Scholar]
- 47.Wootton, S., G. Koljesar, L. Yang, K. J. Yoon, and D. Yoo. 2001. Antigenic importance of the carboxy-terminal beta-strand of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. Clin. Diagn. Lab. Immunol. 8:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wootton, S., R. R. Rowland, and D. Yoo. 2002. Phosphorylation of the porcine reproductive and respiratory syndrome virus (PRRSV) nucleocapsid protein. J. Virol. 76:10569-10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wootton, S. K., E. A. Nelson, and D. Yoo. 1998. Antigenic structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 5:773-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wootton, S. K., D. Rogan, and D. Yoo. 2000. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch. Virol. 145:2297-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, W. H., Y. Fang, R. Farwell, M. Steffen-Bien, R. R. Rowland, J. Christopher-Hennings, and E. A. Nelson. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology 287:183-191. [DOI] [PubMed] [Google Scholar]
- 52.Xu, D., C. J. Tsai, and R. Nussinov. 1997. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng. 10:999-1012. [DOI] [PubMed] [Google Scholar]
- 53.Zweig, M., C. J. Heilman, and B. Hampar. 1979. Identification of disulfide-linked protein complexes in the nucleocapsids of herpes simplex virus type 2. Virology 94:442-450. [DOI] [PubMed] [Google Scholar]