Abstract
The biochemical mechanism of template switching by human immunodeficiency virus type 1 (HIV-1) reverse transcriptase and the role of template dimerization were examined. Homologous donor-acceptor template pairs derived from the HIV-1 untranslated leader region and containing the wild-type and mutant dimerization initiation sequences (DIS) were used to examine the efficiency and distribution of transfers. Inhibiting donor-acceptor interaction was sufficient to reduce transfers in DIS-containing template pairs, indicating that template dimerization, and not the mere presence of the DIS, promotes efficient transfers. Additionally, we show evidence that the overall transfer process spans an extended region of the template and proceeds through a two-step mechanism. Transfer is initiated through an RNase H-facilitated acceptor invasion step, while synthesis continues on the donor template. The invasion then propagates towards the primer terminus by branch migration. Transfer is completed with the translocation of the primer terminus at a site distant from the invasion point. In our system, most invasions initiated before synthesis reached the DIS. However, transfer of the primer terminus predominantly occurred after synthesis through the DIS. The two steps were separated by 60 to 80 nucleotides. Sequence markers revealed the position of primer terminus switch, whereas DNA oligomers designed to block acceptor-cDNA interactions defined sites of invasion. Within the region of homology, certain positions on the template were inherently more favorable for invasion than others. In templates with DIS, the proximity of the acceptor facilitates invasion, thereby enhancing transfer efficiency. Nucleocapsid protein enhanced the overall efficiency of transfers but did not alter the mechanism.
Recombination is an important process that occurs during retroviral reverse transcription (25). It serves to disperse mutations introduced by the highly error-prone reverse transcriptase (RT) (3, 28, 53, 56, 69) and to correct for damage and defects in the viral genome (34, 38, 67). More importantly, recombination leads to the reassortment of genetic traits in the progeny, thereby contributing to genomic heterogeneity and evolution of the virus (5, 15, 21, 24, 54, 57, 65, 68). At least 10% of the infectious human immunodeficiency virus type 1 (HIV-1) strains identified to date are the result of recombination between different subtypes (36, 48, 60). Additionally, recombination has been shown to contribute to the development of progeny with enhanced or dual drug resistance (18, 30, 39, 76). The ability to maintain such remarkable genetic diversity enables viruses such as HIV to survive in a hostile environment where it is constantly challenged by the host immune system and antiretrovirals. Although selection pressures ultimately influence the genetic heterogeneity of the surviving population (54), the recombination event provides the raw material for selection. Evaluating recombination rates and break points within the genome should generate valuable insights into the mechanisms and factors that promote the process. Systems in vitro enable high-resolution analysis of the mechanism, helping to explain the basis of recombination products in cell culture systems and in vivo.
Template switching occurs when the nascent primer, after being displaced from the original template, reanneals and initiates synthesis on the copackaged RNA. In retroviruses, recombination has been shown to occur predominantly during RNA-templated or minus-strand synthesis (23, 62, 75, 77) and is promoted in general through a copy choice mechanism (73). An average of three crossovers per genome per replication cycle has been measured in HIV-1 (27, 75), demonstrating the high frequency of template switching. As with the obligatory minus strong-stop transfer, RT-associated RNase H activity and template homology are also essential to strand transfer events that result in recombination (4, 8, 12, 22, 35, 50, 51, 63, 64). Reduced levels of RT-RNase H activity in the virus have been shown to lower the frequency of template switching, as determined by direct repeat deletion assays in Moloney murine leukemia virus (MLV)-based cell culture systems (4, 26, 51, 63). Mechanistic studies in vitro show that RNase H cleavage of the initial template (donor) exposes single-stranded regions on the nascent DNA, where a homologous template (acceptor) can then base pair and interact to facilitate the transfer (11-13, 58). This constitutes a primary mechanistic step for the copy choice (73), forced copy choice (6), and dynamic copy choice (8, 63) models of template switching, all of which address template switching during RNA-templated synthesis.
A strong correlation has also been detected between the frequency of template switching and conditions that promote RT pausing, such as sequence and secondary structures within the template, misincorporation, and reduced deoxynucleoside triphosphate (dNTP) pools (11, 12, 14, 20, 31, 45, 47, 52, 58, 63, 71, 72). Stalling of synthesis concentrates RT-RNase H cleavages on the donor template, facilitating cDNA-acceptor interaction. Additionally, the viral chaperone protein nucleocapsid (NC), which coats the viral RNA, promotes template switching during both minus-strand strong-stop transfer and internal transfer events (1, 7, 10, 19, 31, 43, 49, 58, 59). The NC is thought to affect transfers through one or more mechanisms. On the one hand, the NC appears to stimulate RT-RNase H activity, thereby enhancing donor template degradation (49, 70). On the other hand, the nucleic acid chaperone activities of the NC promote strand exchange, allowing the acceptor to displace the cleaved donor and capture the nascent primer (32, 66, 74).
The retroviral genome consists of two homologous copies of positive-sense RNA strands that are noncovalently linked near their 5′ ends and exist within the mature virion as a stable dimer (16, 17, 55). While the diploid nature of the genome makes recombination possible (25, 46, 62), template dimerization and the dimerization site have been shown to facilitate the mechanism (2, 34, 37, 38). In HIV-1, the dimerization initiation sequence (DIS) within the 5′ untranslated leader region (UTR) has been identified as the primary linkage site and is presented as a hairpin motif that exposes an auto-complementary loop sequence (33, 41, 44, 61). Dimerization is initiated through loop-loop kissing interactions between the DISs of the two genomes. The DIS-associated recombination events observed in MLV in cell culture prompted us to examine the HIV-1 DIS and its effect on recombination. Recently, Balakrishnan et al. observed a high efficiency of template switching associated with templates containing the DIS (2). Transfer efficiencies in the systems comprising subgenomic RNA fragments in vitro showed a strong correlation between the ability of the RNAs to dimerize and the efficiency of template switching.
The present study examined the biochemical mechanism of template switching by HIV-1 RT and the role of template dimerization promoted by the DIS in stimulating the process. Our analysis shows that while the change in genetic markers revealed the site of primer terminus transfer, the initial events in the transfer process occurred well before the primer terminus transfer. We also observe that certain regions of the template were more favorable for acceptor invasion than others. The combined evidence suggests that the transfer process spans an extended region of the template and proceeds through a two-step mechanism. The detailed evidence for the transfer mechanism is presented.
MATERIALS AND METHODS
Reagents.
Recombinant HIV-1 RT (specific activity, 20,000 U/mg) was supplied by the Genetics Institute (Cambridge, Mass.). HIV-1 NCp7 (72 amino acids [aa]) was prepared by solid-phase chemical synthesis, as described by de Rocquigny et al. (9). HIV-1 NCp7 (55 aa) was generously provided by Robert J. Gorelick. NC stocks were diluted in buffer containing 50 mM Tris-HCl (pH 7.5) and 5 mM dithiothreitol and stored at −80°C in 5- to 10-μl aliquots. T7 RNA polymerase, T4 polynucleotide kinase, DNase I, restriction endonucleases, dNTPs, ribonucleoside triphosphates, and RNase inhibitor were purchased from Roche (Indianapolis, Ind.), and Escherichia coli RNase H was purchased from Invitrogen (Carlsbad, Calif.).
Plasmid constructs.
The HIV-1 5′ UTR constructs pNL187-384, pΔDIS-NL43, and pJRC150-363 and the pol constructs pNL-RT3612-3773, pJRC-RT3541-3753, pNL-DIS-2-pol, and pJRC-DIS-2-pol have been described previously (2). The SNP-2 and SNP-3 templates designed to inhibit DIS-induced dimerization contain nonpalindromic sequences in the DIS loop. The SNP-2 donor and acceptor contain 5′ AAGACUGCA DIS loop sequences, while the SNP-3 donor and acceptor contain 5′ AACUAGCAA DIS loop sequences. The SNP constructs were generated using an overlap PCR approach as previously described (2). The internal overlap primers carrying the mutations were as follows: for pNL-NP2, NL-NP2(+) 5′ GCTTGCTGAAGACTGCACGGCAAGAGGC and NL-NP2(−) 5′ CCTCTTGCCGTGCAGTCTTCAGCAAGCC; for pJRC-NP2, JRC-NP2(+) 5′ GGCTTGCTGAAGACTGCACAGCAAGAGGC and JRC-NP2(−) 5′ GCCTCTTGCTGTGCAGTCTTCAGCAAGCC; for pNL-NP3, NL-NP3(+) 5′ GCTTGCTGAACTAGCAACGGCAAGAGGC and NL-NP3(−) 5′ CCTCTTGCCGTTGCTAGTTCAGCAAGCC; and for pJRC-NP2, JRC-NP2(+) 5′ GGCTTGCTGAACTAGCAACAGCAAGAGGC and JRC-NP2(−) 5′ GCCTCTTGCTGTTGCTAGTTCAGCAAGCC. Mutations are indicated in bold. The overlap primers were used in combination with the end primers to generate full-length PCR fragments which were then cloned into pBluescript II KS(+) (2). Constructs were transformed into E. coli DH5α cells and sequenced.
Substrate preparation.
Using T7 RNA polymerase as previously described (2), RNA templates were generated by runoff transcription in vitro from BamHI-linearized plasmids. Templates were purified on 6% denaturing polyacrylamide gel. DNA primers, complementary to the 3′ terminus of the donor RNA templates and 5′ end labeled, were used to initiate synthesis. Primer MB20 (5′ CCCATTTATCTAATTCTCCC) was used for all of the UTR-based donors, and primer MB22 (5′ GCTTGCCAATACTCTGTCC) was used for all of the pol-based donor templates. Unless described otherwise, labeled primers and donor and acceptor RNAs were incubated at a 1.5:1:2 ratio for 1 min at 80°C and slowly cooled to room temperature to facilitate primer annealing and formation of donor-acceptor heterodimers (2). To inhibit the formation of donor-acceptor heterodimers before initiation of the reaction, the acceptor template was excluded from the annealing reaction and added just prior to initiation of the transfer reaction.
Dimerization assays.
Dimerization assays were performed as previously described (2). Briefly, 15 ng of internally labeled RNA in RT reaction buffer at a final volume of 10 μl was incubated at 80°C for 1 min and slowly cooled to room temperature. Samples were mixed with 2 μl of 50% glycerol, resolved on 4% nondenaturing polyacrylamide gels, and visualized and analyzed using a Storm PhosphorImager (Molecular Dynamics) and ImageQuant software (version 1.2). To examine heterodimer formation under transfer assay conditions, 50 fmols of internally labeled donor RNA was mixed with specified amounts of unlabeled acceptor RNA under buffer and heat-annealing conditions identical to those used for the transfer assays.
RT assays.
Standard transfer assays were performed using a 12-μl final reaction volume containing 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 1 mM dithiothreitol, 6 mM MgCl2, 50 μM dNTPs, 2 U of HIV-1 RT (50 ng), 4 nM primer-donor template, and 8 nM acceptor. Substrates were preincubated with RT for 2 min at room temperature, and reactions were initiated by the addition of MgCl2 and dNTPs. Reactions were terminated at the indicated times by addition of 12 μl of 2× termination buffer (90% formamide, 10 mM EDTA [pH 8.0], 0.1% each xylene cyanole and bromophenol blue). Reaction products were resolved on 6% polyacrylamide-urea gels, visualized using a PhosphorImager, and quantitated using ImageQuant software. For reactions with NC, sufficient NC was added to achieve 200% coating (1 NC per 3.5 nucleotides [nt] of template) of donor and acceptor RNAs. The mixture was incubated for 15 min at room temperature before the addition of RT. In experiments with blocking DNA oligomers and E. coli RNase H, 0.5 pmol of the oligomer or the appropriate amount of RNase H was added 1 min after initiation of the reaction.
Analysis of transfer products.
Transfer products were polyacrylamide gel electrophoresis (PAGE) purified, amplified, and cloned into pBluescript II KS(+) as previously described (2). Individual clones were sequenced to identify crossover sites within the recombinants.
RESULTS
Based on the initial analysis by Balakrishnan et al. of the dimerizing HIV-1 5′ UTR sequence and its ability to promote efficient template switching (2), we hypothesized that proximity of the donor and acceptor templates creates a favorable environment for transfers. Transfer efficiency was severely compromised upon deletion of the DIS hairpin from the UTR sequence and was enhanced when this element was inserted within a non-UTR sequence. Most strikingly, the ability of the templates to dimerize correlated with high transfer efficiency. The following report is focused on the determination of the biochemical mechanisms that cause the dimerization-enhanced transfers.
DIS-induced dimerization enhances transfer efficiency.
We speculated that the enhanced transfers observed with the DIS-containing templates were related to the ability of the sequence to promote dimerization rather than to the mere presence of the sequence itself. Using the various UTR and pol template sets from the previous study by Balakrishnan et al. (2), we therefore examined transfer efficiencies under controlled dimerization conditions as defined in Materials and Methods. The wild-type (WT) UTR donor and acceptor templates share a 177-nt region of homology, from the primer binding site to position +363, which includes the DIS hairpin (Fig. 1A). The nondimerizing ΔDIS donor contains a 23-nt deletion which removes the upper stem-loop of the DIS hairpin. The pol donor and acceptor templates span a region of the RT coding sequence and share a 140-nt region of homology. The 35-nt DIS hairpin sequence was inserted within the templates to generate the DIS-2-pol donor and acceptor RNAs. The DIS-containing UTR and DIS-2-pol templates dimerized efficiently (20 to 30%), while the dimerization efficiency of the ΔDIS and pol donors ranged from 1 to 3% (Fig. 1B) (2).
FIG. 1.
RNA templates used in the study and their dimerization efficiencies. (A) Schematic of the UTR, ΔDIS, pol, and DIS-2-pol templates used in this study. A schematic representation of the HIV-1 genome highlighting some of the prominent regions is included at the top of the panel. Donor (black horizontal lines) and acceptor (grey horizontal lines) templates were generated from NL4-3 and JRCSF strains of HIV-1, respectively (2). Nucleotide positions in the genomic RNA that correspond to the donor and acceptor RNA templates are indicated, along with the positions of the primer binding site (PBS) and DIS. The ΔDIS donor contains a 23-nt deletion which removes the upper stem-loop of the DIS hairpin, while the DIS-2-pol donor and acceptor RNAs contain the 35-nt DIS hairpin sequence inserted within the pol sequence (2). Synthesis was primed from 20-nt DNA primers (black arrows) annealed to the 3′ end of the donor template. (B) Dimerization of the various UTR- and pol-based donor and acceptor templates. Internally labeled RNAs were individually folded under standard reaction conditions and resolved by PAGE under native conditions (see Materials and Methods for details). Dimerization efficiency was calculated as the percentage of dimer from the total of dimer and monomer. Values were averaged from a minimum of three independent experiments. The inset depicts a representative gel showing dimerization of the SNP-2 donor (DSNP-2; lane 1) and acceptor (ASNP-2; lane 2) and SNP-3 donor (DSNP-3; lane 3) and acceptor (ASNP-3; lane 4) templates. The slower-migrating bands correspond to RNA homodimers (D), while the faster-migrating bands correspond to the monomer form (M) of the template.
If donor-acceptor dimerization were responsible for the high efficiency of transfers, then inhibiting this interaction in the DIS-containing templates should reduce the transfer efficiency. The acceptor RNA was therefore excluded from the initial primer-annealing step and was instead added just prior to initiation of the reaction. This limited the percentage of preformed donor-acceptor heterodimers at the start of the reaction. In parallel reactions, the donor and acceptor templates were allowed to predimerize during the primer-annealing step. Reactions were performed using the UTR and pol templates having or lacking the DIS, and the accumulation of transfer products over time was monitored (Fig. 2). Under dimerizing conditions with the WT UTR template set, transfers were efficient and transfer products accumulated at a rapid rate (Fig. 2A). A transfer efficiency of 4% was detectable as early as 2.5 min, and transfers steadily increased with time. However, in reactions in which the acceptor was inhibited from predimerizing with the donor, the transfers were less efficient and accumulated at a slower rate. In contrast, conditions that favored donor-acceptor dimerization had minimal effect on transfer efficiency when the ΔDIS UTR donor was used. Under either set of reaction conditions with DIS-deleted donor, transfer efficiency remained low and was similar to that supported by the WT donor under the nondimerizing conditions. In all of the cases, transfer products were not detected until 2 to 5 min of reaction time. This delay is consistent with the results of earlier work (50) and suggests that the transfer process involves slow steps that do not occur during simple primer extension.
FIG. 2.
Time course of transfers promoted by the UTR- and pol-based templates. Transfer assays were performed at a 1:2 donor/acceptor ratio under conditions in which donor and acceptor were predimerized (D) or acceptor was added at the start of the reaction (ND). Reactions were terminated at 2.5, 5, 10, 15, 30, 45, and 60 min. Samples were resolved by denaturing PAGE and quantitated using a PhosphorImager. Transfer efficiency was calculated as the percentage of transfer product over the combined donor extension and transfer products. (A) Plots of transfer efficiency versus time for the WT UTR donor (triangles) and ΔDIS donor (circles). (B) Transfer efficiency was plotted against time for the pol (circles) and DIS-2-pol (triangles) templates. Values were averaged from a minimum of three independent experiments.
A comparable trend was observed with the pol templates (Fig. 2B). Transfers promoted by the native pol templates showed characteristics similar to those promoted by the ΔDIS donor and were unaffected by the two different reaction conditions. Transfers promoted by the DIS-2-pol templates, in contrast, were greatly enhanced when the donor and acceptor templates were predimerized. Overall, the results demonstrate clearly that it is not the mere presence of the DIS but the actual donor-acceptor dimerization that promotes efficient transfer.
Mutations in the DIS loop sequence inhibit dimerization and lower transfer efficiency.
Since the ΔDIS UTR donor contained a 23-nt deletion, it had the potential to fold in conformations different from those assumed by the original donor RNA. Additionally, the deletion also restricted donor-acceptor homology in the region prior to the transfer peak. It was therefore possible that these factors contributed to the reduced transfers observed with the ΔDIS donor. To obviate such problems, we generated nondimerizing UTR templates by introducing point mutations in the DIS loop sequence in both donor and acceptor templates. The SNP-2 and SNP-3 donor-acceptor template pairs contain modifications within the DIS loop sequence such that the palindromic nature of the loop sequence is disrupted (Fig. 3A). The overall folding of the templates, as determined by mfold (78), remained unaltered. As expected, dimerization assays showed that the SNP templates dimerized poorly (Fig. 1A). The SNP-2 and SNP-3 donor templates exhibited 3 to 4% dimerization efficiency, while the corresponding acceptor templates dimerized with 8 to 10% efficiency. The SNP templates therefore served as effective control templates with minimum sequence and homology length alterations.
FIG. 3.
The SNP templates and their transfer efficiencies. (A) DIS loop sequences of the WT UTR, SNP-2, and SNP-3 templates. Loop sequences are indicated in uppercase, and adjoining stem sequences are indicated in lower case. The palindromic sequence within the loop is underlined in the WT, while mutations within the loop in the SNP templates are highlighted in bold. (B) Time course of transfers promoted by the SNP templates. Reaction details are same as described for Fig. 2. Transfer efficiency was plotted against time for the SNP-2 (triangles) and SNP-3 (circles) template pairs. Reactions were performed under conditions in which donor and acceptor were predimerized (D) or acceptor was added at the start of the reaction (ND). Values were averaged from a minimum of three independent experiments.
To address the role of template dimerization in template switching, transfer efficiencies promoted by the SNP-2 and SNP-3 donor-acceptor pairs were compared under the dimerizing and nondimerizing conditions described above (Fig. 3B). As expected, the characteristics of the transfer reaction reflected those exhibited by the ΔDIS UTR and the pol templates (Fig. 2). For both SNP template pairs, transfer products accumulated at a slow rate (Fig. 3B). Transfer efficiency ranged from 9 to 11% at 60 min under dimerizing conditions, while transfer efficiency was 6 to 8% under nondimerizing conditions. In contrast to those of the WT UTR templates, the transfer efficiencies of the SNP templates remained low and were not significantly affected by the dimerizing conditions. The overall results strongly suggest that the ability of the template to dimerize enhances the efficiency of transfer.
Acceptor titration highlights local concentration effects.
We hypothesized that in templates that dimerize, the close proximity between the donor and acceptor RNAs stimulates a high local acceptor concentration. This improves the opportunity for productive cDNA-acceptor interactions, thereby facilitating template switching. Using the various template sets to test this argument, we examined the effect of acceptor concentration on transfer efficiency (Fig. 4). The anticipation was that increasing acceptor concentration would have a stimulatory effect on transfers with DIS-lacking templates but only a minimal effect on the DIS-containing templates.
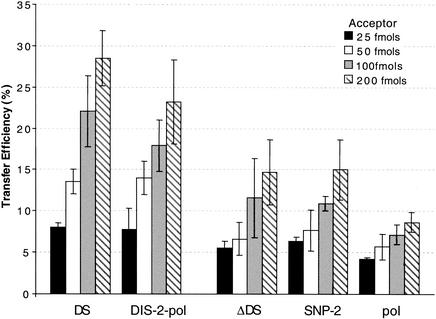
FIG. 4.
Effect of acceptor concentration on transfer efficiency. Transfer efficiency was measured for the dimerizing UTR and DIS-2-pol template pairs and the nondimerizing ΔDIS, SNP-2, and pol template sets as a function of increasing acceptor concentration. Donor and acceptor template pairs were allowed to predimerize prior to initiating the reaction. Reaction mixtures were incubated for 60 min and contained 25 fmols of donor and 25, 50, 100, or 200 fmols of the corresponding acceptor. Transfer efficiency was determined as described for Fig. 2. Values were averaged from a minimum of three independent experiments.
For all template pairs tested, a basal level of transfers was detectable even at an equimolar ratio of donor and acceptor (Fig. 4). For the ΔDIS, SNP-2, and pol templates, the transfer efficiencies ranged from 4 to 6%, while for the UTR and DIS-2-pol, the efficiencies were 7.5 to 8%. As the acceptor concentration was increased to two, four, and eight times the donor concentration, a concomitant increase in transfer efficiency was observed. Surprisingly, increased transfers were observed with both the DIS-containing and DIS-lacking template pairs. At sevenfold acceptor excess, the transfer efficiency reached 14 to 15% for the ΔDIS and SNP-2 templates and ∼9% for the pol templates. Under similar conditions, transfer efficiencies of the WT UTR and DIS-2-pol templates were about 28 and 23%, respectively, three times the efficiencies observed for equimolar donor and acceptor concentrations. Therefore, although transfer efficiencies for all of the template pairs were sensitive to acceptor concentration, WT UTR and DIS-2-pol templates showed larger increases in transfers in response to increasing acceptor concentrations.
The stimulatory effect of high acceptor concentrations on transfers with the UTR and DIS-2-pol templates prompted us to examine the effect of acceptor titration on the donor RNA dimerization. Using labeled donor and unlabeled acceptor RNA, dimerization assays were performed under template and buffer conditions identical to those used for the transfer assays. Analysis revealed an increase in the total dimer population as the total RNA concentration increased for the DIS-containing templates but not for the nondimerizing template pairs (data not shown). This was convincingly demonstrated with the pol and DIS-2-pol templates, with which, unlike the results seen with the UTR templates, differences in the donor and acceptor template sizes enabled the detection of donor homodimers and donor-acceptor heterodimers (data not shown). For the DIS-2-pol templates, the heterodimer population increased from 26% at equimolar concentrations of donor and acceptor (1×) to 46% at an acceptor concentration sevenfold higher (8×). Thus, the increased transfers observed with increasing acceptor concentration were effected through different mechanisms in the dimerizing and nondimerizing templates. With the nondimerizing templates, the increased transfers resulted from a simple acceptor concentration effect, while with the dimerizing templates, they resulted from increased donor-acceptor heterodimer formation.
Transfers are initiated through acceptor invasion behind the primer terminus.
Thus far, our study of the DIS-induced transfers had revealed that the DIS-induced donor-acceptor dimerization had a strong effect on transfer efficiency. However, sequence analysis of the transfer products showed that the peak of primer terminus switches occurred only after synthesis through the DIS hairpin (2). Our first impression from this was that the transfer peak could not be a consequence of dimerization. However, upon more thorough consideration, we realized that dimerization might promote an event occurring before the DIS that resulted in more efficient transfer beyond the DIS. We proposed that transfers involve extensive cDNA-acceptor interactions that are initiated prior to the region of primer terminus switch and that such interactions are efficiently promoted in the context of a dimerizing template (2). In such a model, the initial cDNA-acceptor contact would be made before primer extension through the DIS disrupted the donor-acceptor dimer. Branch migration of the cDNA-acceptor hybrid would progress through the DIS, eventually resulting in transfer of the primer terminus in the region downstream of the DIS.
To test this acceptor invasion model, we designed a series of blocking DNA oligomers to probe for acceptor-cDNA interaction sites (Fig. 5A). The oligomers each consisted of a sequence identical to that of a section of the donor template and therefore complementary to the nascent cDNA. If a transfer event were initiated through acceptor invasion at a specific region, then a DNA oligomer corresponding to that region would compete for the invasion site and thereby inhibit the transfer. The specific lengths and termini of the blocking oligomers were chosen such that the oligomers did not form highly stable primer dimers and secondary structures.
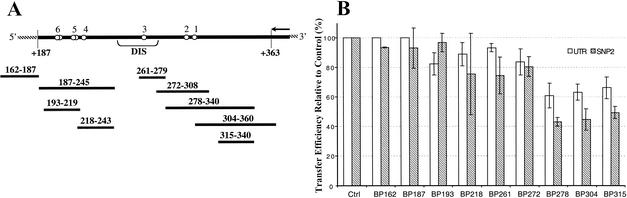
FIG. 5.
Blocking oligomers and their effect on transfer efficiency with the UTR and SNP-2 templates. (A) Schematic of the UTR donor template and the DNA oligomers designed to block invasion. Nucleotide positions within the genomic RNA to which the donor template corresponds are indicated and represent the region of homology shared between the donor and acceptor templates. Open circles on the donor indicate relative positions at which donor and acceptor differ in sequence by single- or double-nucleotide bases and are designated markers 1 to 6. Hatched lines on either end of the template represent plasmid-derived sequences. The 35-nt DIS hairpin site is indicated. The arrow at the 3′ end of the template represents the DNA primer and indicates the direction of synthesis. Blocking oligomers are designated by the template regions that they span and are aligned opposite their homologous regions on the template. BP162-187 falls outside the region of homology. (B) Transfer assays were performed for 60 min with the WT UTR and SNP-2 templates in the presence of 0.5 pmol of the blocking oligomer (see Materials and Methods for details). Transfer efficiency in the absence of blocking oligomers was taken as 100% (Ctrl). Transfer efficiency measured in the presence of each blocking oligomer was plotted as a percentage of the control reaction. Values were averaged from a minimum of three independent experiments.
The first set of DNA oligomers ranged in size from 57 to 63 nt and corresponded to the three major segments between markers within the region of homology (Fig. 5A). BP187-245 consisted of donor sequences 5′ to the DIS hairpin, while BP304-360 corresponded to the region of homology 3′ to marker 1. BP278-340 spanned the template segment 24 nt past the primer terminus to the start (3′ end) of the DIS hairpin. To reduce the possibility of the blocking oligomers interacting with the templates, and to prevent RTs from prebinding to the oligomers, they were added 1 to 2 min after initiation of reaction. Transfer assays were performed in the presence of the different blocking oligomers, and the transfer efficiencies were measured at 20 and 60 min after the initiation of reaction (Fig. 5B and data not shown). Compared to BP187, oligomers BP278 and BP304 inhibited transfers more efficiently, effecting 18 to 23% inhibition at 20 min and 37 to 40% at 60 min.
To define the invasion sites more precisely, we designed a second set of shorter blocking oligomers 19 to 37 nt in length (Fig. 5A). BP193-219 and BP218-243 had adjacent sequences spanning the region originally covered by BP187, while BP272-308 and BP315-340 spanned the region originally covered by BP278. BP261-279 comprised the 3′ stem sequence of the DIS hairpin. The five short oligomers together covered most of the homologous region within the template. The upper stem-loop region of the DIS hairpin was excluded as an oligomer binding site, primarily because of the potential of this sequence to fold into stable structures. Oligomer BP162-187 corresponded to a sequence in the acceptor template outside the region of homology and served as a control.
While all of the short oligomers, except for the control oligomer, inhibited transfers to some extent, they differed in relative effectiveness (Fig. 5B). BP261, which mapped to the DIS 3′ stem (which is part of the dimerization site), showed the least inhibition (5 to 10%). Between 10 and 25% inhibition was detectable with each of the oligomers BP193, BP218, and BP272, which map to the regions of homology 5′ to marker 4, between marker 4 and the DIS hairpin, and between markers 1 and 3, respectively. Among the five short oligomers, BP315 displayed the highest inhibition of transfers (between 26 and 40%). As shown, BP315 was a short sequence 3′ to marker 1, a region also common to BP278 and BP304 (Fig. 5A). Inhibition of transfer with BP315 was detectable at as early as 20 min after the initiation of reaction and was found at levels similar to those displayed by the longer oligomers, BP278 and BP304, at 60 min.
When the above results were considered in combination with the transfer distribution data (2), we recognized that while primer terminus transfer occurred predominantly in the region immediately after the DIS hairpin, a blocking oligomer corresponding to that region (BP218) showed only a minimal inhibition of transfers. Instead, blocking oligomers complementary to the 5′ end of the cDNA were more inhibitory. This implies that the invasion site need not be at or adjacent to the site of primer terminus transfer but could instead be at a distance.
Interestingly, the blocking oligomers were more effective in inhibiting transfers on the nondimerizing SNP-2 templates, although the overall inhibition profile remained the same as that observed with the WT UTR template (Fig. 5B). Oligomers BP278, BP304, and BP315 were the most inhibitory and blocked transfers by 50 to 60%, while oligomers BP218, BP261, and BP272 caused 20 to 25% inhibition. This suggests that the preferred invasion sites for transfers are identical in the WT UTR and SNP UTR templates. Most likely, the inability of the SNP templates to dimerize makes the acceptor invasion step less efficient, allowing the oligomers to compete more effectively with the acceptor for the interaction site on the cDNA. In contrast, the close proximity of the acceptor in the dimerizing templates facilitates invasion and therefore a higher efficiency of transfer and a lower inhibitory effect by the blocking oligomers.
Effects of increasing RNase H activity.
If invasion is an essential step in the transfer mechanism, then appropriate cleavage of the donor template is an important requirement. We therefore examined the effect of increases in RNase H activity on transfer efficiency (Fig. 6). To selectively increase donor template degradation with minimal effect on primer extension, transfer reaction mixtures were supplemented with increasing amounts of E. coli RNase H added after initiating the reaction (see Materials and Methods for details). Transfer efficiencies were compared between the WT UTR, SNP-2, and pol template sets (Fig. 6A). Supplementing the reactions with as little as 0.5 U of E. coli RNase H resulted in a 50 to 90% increase in transfers with the UTR and SNP-2 templates and a 150% increase in transfers with the pol templates. Further increasing E. coli RNase H concentrations did not cause substantial additional increases in transfer efficiency. At 2 U of E. coli RNase H, a general drop in transfer efficiency was observed, which most likely resulted from disruption of the acceptor-cDNA hybrids. For all three template sets, an increase in transfer efficiency was achieved through increasing RNase H activity supplied in trans. Interestingly, transfer efficiencies measured with the WT UTR templates were invariably higher than those supported by the SNP-2 and pol templates, suggesting that intrinsic features of the templates make a significant contribution to the facilitation of transfers in the UTR.
FIG. 6.
Effect of exogenous RNase H on transfer efficiency and invasion. (A) Transfer assays were performed with the WT UTR, SNP-2, and pol template sets in the presence of 0, 0.5, 1, or 2 U of E. coli RNase H (see Materials and Methods for details). Transfer efficiencies were averaged from a minimum of three independent experiments. (B) Transfer reaction mixtures with the WT UTR and SNP-2 template sets were supplemented with 0.5 U of E. coli RNase H and performed in the presence of 0.5 pmol of different blocking oligomers (see Materials and Methods for details). Transfer efficiency measured in the presence of each blocking oligomer was expressed as a percentage of the control reaction. Details are same as described for Fig. 5B.
To examine how the increased levels of RNase H might have been improving the efficiency of transfers, we reexamined the effect of the blocking oligomers in the presence of 0.5 U of E. coli RNase H (Fig. 6B). Results with WT UTR templates showed that the overall transfer inhibition profiles of the blocking oligomers remained unchanged, with BP278, BP304, and BP315 producing the most inhibition (compare Fig. 5B). The presence of exogenous RNase H caused a slight increase in the inhibition of transfer by the oligomers. In particular, the efficiency of the oligomers previously found to be the most effective, BP278, BP304, and BP315, was increased an additional 20%. Enhanced cleavages across the template did not significantly alter the preference for invasion sites. This suggests that specific regions within the template are intrinsically favorable sites for invasion.
Similar analysis of transfers with the SNP-2 template showed that, as with the WT UTR template, oligomers BP278, BP304, and BP315 were still the most inhibitory to transfers (Fig. 6B). In contrast to the WT template, enhancing degradation of the SNP template only marginally increased the inhibitory effect for these oligomers. Instead, increasing RNase H enhanced the inhibitory effect of oligomers BP187, BP261, and BP272, although they were still not as inhibitory as BP278, BP304, and BP315. Therefore, with the SNP-2 template, although invasion at additional sites was promoted with enhanced donor cleavage, the preferred invasion site still did not change. Preferred sites of invasion were therefore evident irrespective of dimerization.
Viral NC promotes DIS-related transfers.
The viral NC protein has been proposed to enhance DIS-induced dimerization by converting the kissing-loop complex, which involves the 6-nt loop sequence, to an extended, stable dimer involving the stem and loop sequences of the hairpin (40). We therefore examined the effect of the presence of NC on transfers with the DIS-containing UTR templates. In reactions with predimerized WT UTR templates, NC at a 200% coating level produced a 40 to 60% increase in transfer efficiency (data not shown). Interestingly, for reactions in which inhibiting predimerization of donor and acceptor caused a drop in transfers, addition of NC enhanced transfer efficiency to the levels observed under heat annealing-induced dimerization conditions (Fig. 7). NC most likely promoted DIS-induced donor-acceptor dimerization, thereby enhancing transfer efficiency. In contrast, NC had minimal effect on transfers with the pol templates. Incidentally, NC did not cause any noticeable change in the extension or pause profile under the various conditions tested for the UTR and pol templates. Similar results were observed with both the 55-aa and 72-aa forms of NC.
FIG. 7.
Effect of NC on strand transfer catalyzed by HIV-1 RT on the UTR template. The UTR acceptor RNA was mixed with the donor template during the heat-annealing step (+Ht) to promote donor-acceptor heterodimer formation or added to the reaction mixture at the start of synthesis (−Ht) to impede heterodimer formation. Transfer reactions were performed in the absence (−NC) or presence (+NC) of NC. Reactions were terminated at the 5 (lane 1)-, 15 (lane 2)-, 30 (lane 3)-, 45 (lane 4)-, and 60 (lane 5)-min time points, and products were resolved on 6% polyacrylamide gel under denaturing conditions. Full-length synthesis on the donor resulted in a 214-nt donor extension product (FL), while template switching and completion of synthesis on the acceptor generated a 250-nt transfer product (T).
To determine whether NC protein had an effect on transfer distribution, transfer products were isolated, sequenced, and analyzed (Fig. 8) as previously described (2). Since the donor and acceptor templates carry periodic nucleotide base variations, a template-switching event would result in a change in genetic markers in the cDNA, which identifies the region of primer terminus transfer. Transfers in the presence of NC (72-aa form) were assayed under the two different reaction conditions (see Materials and Methods for details). In the one case, heat annealing-induced donor-acceptor dimers were incubated with NC prior to initiation of the reaction. In the other case, following the annealing of primer and donor, acceptor template and NC were added to the reaction mixture and incubated for 10 min prior to initiation of the reaction (−HT+NC; Fig. 7).
FIG. 8.
Distribution of transfers within the 5′ UTR in the presence of NC. Template description is same as described for Fig. 5A. The region corresponding to the 35-nt DIS hairpin is indicated. (A) Transfer assays in the presence of 200% NC were performed with predimerized donor and acceptor (solid bars) or without predimerizing the templates (hatched bars). Transfer products were isolated, cloned, and sequenced to determine the distribution of crossover sites within the template. Values were averaged from two independent experiments with about 45 clones each and consisted of products resulting from single crossover events. Hatched and solid bars between markers represent average transfer frequencies within the marker segment and were calculated as follows: 100 × (number of transfers within a given marker segment/total number of transfer products analyzed). Corresponding numerical values are indicated above the bars. (B) The transfer frequencies presented in panel A were normalized for distance. Numerical values are indicated above the bars and were calculated by dividing the crossover frequency measured as described for panel A by the length of the respective marker segments and multiplying the final value by 10. Marker segments and segment lengths in nucleotides are highlighted by horizontal brackets.
For reactions in which the donor and acceptor templates were predimerized through heat annealing, the overall transfer distribution profile (Fig. 8A) was remarkably similar to that previously observed in the absence of NC (2). The preferred region for primer terminus switch, i.e., between markers 3 and 4, was preserved in the presence of NC. In fact, the 67% crossover within this 48-nt segment was slightly higher than that which Balakrishnan et al. had previously observed in the absence of NC (53% [2]). Transfers within the adjoining marker segments ranged between 8 and 15%. Interestingly, for reactions with NC in which the acceptor was not heat annealed with the donor, the profile of primer terminus transfer was altered, specifically in the segment between markers 2 and 4. In the shifted profile, each of the segments between markers 2 and 3 and markers 3 and 4 supported about 35% of the transfers. Transfer frequency in the remaining two segments remained largely unchanged. To normalize for the distance between the markers, the transfer frequency within each segment was corrected for the length of that segment (Fig. 8B). Under conditions of heat annealing, the segment between markers 3 and 4 supported a larger fraction of the primer terminus transfers, and NC did not significantly alter this. However, under conditions of NC-induced dimerization and acceptor folding, the resulting shift in the preferred region for primer terminus transfer broadened the transfer peak. One explanation for the altered transfer profile is that the acceptor assumes slightly different conformations under the different reaction conditions. Alternately, the NC-induced dimer linkage is different from that induced through heat annealing. An extended dimerization interaction proposed to be favored by NC should impede primer elongation because of the need for strand displacement synthesis. This might be the cause of the observed shift of the terminus transfer area closer to the expected site of invasion.
It was of interest then to determine whether the change in the transfer distribution profile was effected through a change in the invasion site or in the overall transfer mechanism. To address this, we reexamined the effect of the blocking oligomers under NC-induced template dimerization conditions. The profile of inhibition of transfers was essentially identical to that observed under the heat-annealing conditions (Fig. 5B), with the oligomers BP278, BP304, and BP315 effecting the most inhibition (data not shown). This indicated that the preferred region for acceptor invasion was not altered in the presence of NC. The data further confirmed that NC does not alter the basic transfer mechanism. The acceptor invasion step that initiates the transfer and the primer terminus switch which completes the transfer event are discrete steps in the transfer process and occur at distinct sites on the template. NC most likely stimulates transfers through its effects on one or more of these intermediate steps in the process.
DISCUSSION
Identification of recombination events associated with the genome dimerization sites in MLV suggested a role for template dimerization in promoting crossovers (34, 37, 38). Using RNA templates containing the HIV-1 leader sequence, Balakrishnan et al. recently showed that DIS-induced template dimerization enhances the efficiency of template switching by HIV-1 RT in vitro (2). The present study specifically examined the mechanism of template switching and the effect of template dimerization on the process.
Results from three separate experimental approaches demonstrated that it is the DIS-induced dimerization of the templates, and not the mere presence of this sequence, that enhances the efficiency of transfers in the HIV-1 UTR sequence. First, when the DIS was deleted, mutated, or introduced in a different template, we observed a direct correlation between the ability of the templates to dimerize and the efficiency of template switching. Second, inhibiting the donor-acceptor interaction was sufficient to reduce transfer efficiency, not only with the leader sequence containing the DIS but also with non-UTR sequences into which the DIS element was artificially introduced. Finally, increased donor-acceptor dimerization enhanced transfers.
The transfer distribution profile (Fig. 8) (2), taken together with the inhibition profile of the blocking oligomers (Fig. 5B), indicates that transfer involves an initial acceptor invasion step that is initiated at a site distant from the site of the primer terminus transfer. While transfer of the primer terminus occurred after synthesis through the DIS, the blocking oligomers that most effectively inhibited transfers were 50 to 60 nt upstream of the DIS. The preferred invasion sites were identical for the UTR and SNP-2 templates, indicating that its position is not determined by the DIS or dimerization. Additionally, using exogenous E. coli RNase H to increase the overall level of template degradation enhanced transfer efficiency without altering the preferred invasion site for both UTR and SNP-2 template sets. These combined observations led to the following conclusions. (i) Acceptor-cDNA interactions were initiated at preferred sites upstream of the dimerization site. (ii) The site of initiation of transfer and site of primer terminus transfer were separated by a significant distance (60 to 80 nt in the UTR templates) such that the overall transfer process occurred over an extended region.
We did not observe 100% inhibition of transfers with BP315 for two reasons. First, this sequence represents only 15% of the region of homology, and a portion of the invasion may be initiated at other sites. Notice that the other oligomers also caused some inhibition of transfers. Second, with the dimerizing templates, the DNA oligomers may be less effective in competing with the acceptor for the interaction site on the cDNA. Blocking oligomers were more effective with the nondimerizing SNP-2 templates. Invasion is therefore a key step in the transfer process. In dimerizing templates, the close proximity of the acceptor promotes effective invasion, thereby facilitating the transfer.
Studies show that excess RNase H stimulates transfers (12). Our findings corroborate these observations while suggesting that increased RNase H specifically enhances the invasion step. Analysis of phenotypically mixed virions shows that while reducing RT-polymerase activity was deleterious to viral replication for both HIV-1 and MLV, reducing RT-RNase H activity to 50% that in WT virus did not noticeably affect replication (4, 29). Limiting RNase H activity, however, caused a sharp decrease in template-switching frequency (4). The excess RNase H activity in some retroviruses, while not essential for normal replication (4, 29), might have been retained because of its positive effect on recombination and evolution of the virus. Most likely, extensive cleavage is required for dissociation of RNA fragments to facilitate acceptor invasion for transfer. The fact that limiting RNase H is more detrimental to minus strong-stop transfer than to the PPT primer biogenesis (29) further supports the notion that different aspects of retroviral replication and evolution require different levels of RNase H.
Results show that although NC stimulates transfer, it does not alter the transfer mechanism. When the UTR templates were predimerized, NC caused a modest increase in transfer efficiency without altering the transfer distribution. This stimulation can be explained by the ability of this chaperone protein to stimulate RT-RNase H (49, 70) and to promote acceptor-cDNA annealing (32, 66, 74), thereby promoting acceptor invasion and branch migration (Fig. 9, panels 2 and 3). For reactions in which acceptor was not heat annealed, NC stimulated an earlier transfer of the primer terminus. One possibility is that structures resulting from differences in heat-annealed versus NC-induced donor-acceptor dimerization influenced the terminus transfer. Alternately, acceptor folding under the different conditions may have been the significant factor. In the latter case, more effective promotion of the branch migration step by NC (Fig. 9, panel 3) would cause an earlier transfer of the primer terminus. This finding is comparable to that of the effects of NC observed by Negroni and Buc (42). They propose that folding and structure of the acceptor template can affect the site of primer terminus transfer.
FIG. 9.
Schematic of the proposed invasion-induced two-step transfer mechanism. Solid lines represent the dimerized donor (black) and acceptor (grey) templates; dotted lines represent the nascent cDNA. (Panel 1) Minus-strand synthesis is accompanied by cleavage of the donor RNA by RT RNase H activity. (Panel 2) Transfer is initiated through an invasion step in which the acceptor enters at gaps created within the cDNA-donor hybrid. Invasion is initiated before synthesis through the DIS separates the two templates. (Panel 3) While RT continues synthesis on the donor, the invasion propagates through branch migration, forming a three-strand intermediate. (Panel 4) The invasion eventually catches up with synthesis, and transfer is completed with the translocation of the primer terminus to the acceptor, the second step in the transfer process. In the UTR template, this second step occurs after synthesis through the DIS.
Based on the observed characteristics of the transfer reaction, we propose that template switching occurs through an active invasion mechanism (Fig. 9). While synthesis continues on the donor template, RNase H cleavages within the donor create gaps at which the acceptor can invade and initiate interactions with the nascent DNA (Fig. 9, panels 1 and 2). Acceptor invasion, the initial step in the transfer, is initiated before RT has synthesized through the DIS. Acceptor-cDNA interactions propagate through branch migration as cDNA synthesis and template cleavage continues on the donor template, creating a three-strand transfer intermediate (Fig. 9, panel 3). A significant length of the cDNA-acceptor hybrid is formed, and eventually the primer terminus, which has now extended past the DIS, also transfers (Fig. 9, panel 4). Three transient components in the transfer process are as follows: (i) acceptor invasion and initiation of the transfer, (ii) extension of this interaction through branch migration, and (iii) primer terminus switch, which completes the transfer.
The idea of an invasion-induced transfer mechanism is not new. Previous studies of template switching mechanisms have indicated the role of pausing and RNase H cuts in facilitating acceptor invasion (11, 12, 14, 31, 58, 63, 72). In this study, findings regarding the UTR and pol templates demonstrated that weak pause sites are sufficient to provide the required amount of cleavage for invasion and transfer. Template switching appears not to necessarily require intense pausing; rather, it depends on a site with characteristics that allow cleavage of the donor together with the ability of the acceptor to invade. Recent studies have proposed that transfers occur through a two-step mechanism in which the structure of the template, specifically the acceptor, is critical in the invasion (42, 58). Negroni and Buc recently suggested that transfer involves the formation of a complex containing both RNA strands and proceeds through an initial docking of the acceptor on the nascent DNA followed by strand invasion (42). The analysis by Roda et al. of transfer mechanisms in the context of a stable hairpin-containing template system led to the proposal of the two-step “Dock and Lock” model (58). Almost all of the transfers were initiated at the base of the hairpin, where extensive pause-induced cleavage of the donor template facilitated the Dock, or invasion, step. The Dock step, as previously described by Negroni and Buc (42), initiates the transfer. The point of transfer of the primer terminus, or the Lock step, occurred predominantly within the loop of the hairpin at a location at least 20 nt from the Dock site. In a smaller percentage of transfers, the Dock and Lock steps were completed at the base of the hairpin. In the UTR template, the primer terminus transfers to a location as far as 80 nt away from the invasion site. The transfer mechanism inferred from the UTR template system is consistent with the two-step model. We additionally conclude that the overall transfer process involves an extended region of homology, that invasion does not require strong pausing, and that invasion occurs at preferred sites in the template.
The two-step mechanism also explains the transfer profile observed by Wooley et al. (71), in which insertion of a homopolymeric region in the template increased the frequency of primer terminus switches in a template segment several nucleotides downstream of the site. Most likely, while pause-induced RNase H cleavage initiated the invasion step in the homopolymeric region, the subsequent primer terminus transfer step did not occur until synthesis was well past the homopolymer region. While the primer terminus transfer site defines the breakpoints in the recombinant genome, as demonstrated by the change in genetic markers, preferred recombination sites within the genome represent template regions with a favorable combination of sites for both invasion and primer terminus transfer.
The combined results of the present study and a previous study by Roda et al. (58) strongly suggest that retroviral recombination during minus-strand synthesis proceeds through an invasion-induced two-step mechanism. While the evidence indicates that the invasion site and the primer terminus transfer site can be separated by a large distance, factors that determine the size of the separation are not fully understood. Although we have some understanding of the factors that can facilitate the invasion step, we do not yet understand what causes the primer terminus to transfer at a specific site. Transient stalling of RT during synthesis, favorable structure of the acceptor template (42), and formation of an extended and stable hybrid are likely factors that affect the later terminus transfer. Such details are currently under investigation.
Acknowledgments
We thank the Genetics Institute for recombinant HIV-1 RT and Rob Gorelick for the HIV-1 NC (55-aa form). We are grateful to Jeff DeStefano for critical reading of the manuscript and to Yan Chen, Ricardo Roda, and Mark Hanson for helpful discussions.
This work was supported by NIH grant GM 49573 (to R.A.B. and P.J.F.).
REFERENCES
- 1.Allain, B., M. Lapadat-Tapolsky, C. Berlioz, and J. L. Darlix. 1994. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 13:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakrishnan, M., P. J. Fay, and R. A. Bambara. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482-36492. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, J. C., K. Bebenek, and T. A. Kunkel. 1992. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. USA 89:6919-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brincat, J. L., J. K. Pfeiffer, and A. Telesnitsky. 2002. RNase H activity is required for high-frequency repeat deletion during Moloney murine leukemia virus replication. J. Virol. 76:88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, D. S. 1997. Recombination in HIV: an important viral evolutionary strategy. Emerg. Infect. Dis. 3:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42:1-26. [DOI] [PubMed] [Google Scholar]
- 7.Darlix, J. L., A. Vincent, C. Gabus, H. de Rocquigny, and B. Roques. 1993. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV1 RNA. C. R. Acad. Sci. Ser. III 316:763-771. [PubMed] [Google Scholar]
- 8.Delviks, K. A., and V. K. Pathak. 1999. Effect of distance between homologous sequences and 3′ homology on the frequency of retroviral reverse transcriptase template switching. J. Virol. 73:7923-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Rocquigny, H., D. Ficheux, C. Gabus, M. C. Fournie-Zaluski, J. L. Darlix, and B. P. Roques. 1991. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 180:1010-1018. [DOI] [PubMed] [Google Scholar]
- 10.DeStefano, J. J. 1996. Interaction of human immunodeficiency virus nucleocapsid protein with a structure mimicking a replication intermediate. Effects on stability, reverse transcriptase binding, and strand transfer. J. Biol. Chem. 271:16350-16356. [PubMed] [Google Scholar]
- 11.DeStefano, J. J., R. A. Bambara, and P. J. Fay. 1994. The mechanism of human immunodeficiency virus reverse transcriptase-catalyzed strand transfer from internal regions of heteropolymeric RNA templates. J. Biol. Chem. 269:161-168. [PubMed] [Google Scholar]
- 12.DeStefano, J. J., L. M. Mallaber, L. Rodriguez-Rodriguez, P. J. Fay, and R. A. Bambara. 1992. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J. Virol. 66:6370-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeStefano, J. J., B. Roberts, and D. Shriner. 1997. The mechanism of retroviral recombination: the role of sequences proximal to the point of strand transfer. Arch. Virol. 142:1797-1812. [DOI] [PubMed] [Google Scholar]
- 14.Diaz, L., and J. J. DeStefano. 1996. Strand transfer is enhanced by mismatched nucleotides at the 3′ primer terminus: a possible link between HIV reverse transcriptase fidelity and recombination. Nucleic Acids Res. 24:3086-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz, R. S., E. C. Sabino, A. Mayer, J. W. Mosley, M. P. Busch, and The Transfusion Safety Study Group. 1995. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. J. Virol. 69:3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu, Z., Q. Gao, E. A. Faust, and M. A. Wainberg. 1995. Possible involvement of cell fusion and viral recombination in generation of human immunodeficiency virus variants that display dual resistance to AZT and 3TC. J. Gen. Virol. 76:2601-2605. [DOI] [PubMed] [Google Scholar]
- 19.Guo, J., L. E. Henderson, J. Bess, B. Kane, and J. G. Levin. 1997. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 71:5178-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison, G. P., M. S. Mayo, E. Hunter, and A. M. Lever. 1998. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res. 26:3433-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell, R. M., J. E. Fitzgibbon, M. Noe, Z. J. Ren, D. J. Gocke, T. A. Schwartzer, and D. T. Dubin. 1991. In vivo sequence variation of the human immunodeficiency virus type 1 env gene: evidence for recombination among variants found in a single individual. AIDS Res. Hum. Retrovir. 7:869-876. [DOI] [PubMed] [Google Scholar]
- 22.Hu, W. S., V. K. Pathak, and H. M. Temin. 1993. Role of reverse transcriptase in retroviral recombination, p. 251-274. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [PubMed]
- 23.Hu, W. S., and H. M. Temin. 1992. Effect of gamma radiation on retroviral recombination. J. Virol. 66:4457-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 26.Hwang, C. K., E. S. Svarovskaia, and V. K. Pathak. 2001. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc. Natl. Acad. Sci. USA 98:12209-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji, J. P., and L. A. Loeb. 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31:954-958. [DOI] [PubMed] [Google Scholar]
- 29.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kellam, P., and B. A. Larder. 1995. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J. Virol. 69:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, J. K., C. Palaniappan, W. Wu, P. J. Fay, and R. A. Bambara. 1997. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 272:16769-16777. [DOI] [PubMed] [Google Scholar]
- 32.Lapadat-Tapolsky, M., C. Pernelle, C. Borie, and J. L. Darlix. 1995. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 23:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464-13474. [DOI] [PubMed] [Google Scholar]
- 34.Lund, A. H., J. G. Mikkelsen, J. Schmidt, M. Duch, and F. S. Pedersen. 1999. The kissing-loop motif is a preferred site of 5′ leader recombination during replication of SL3-3 murine leukemia viruses in mice. J. Virol. 73:9614-9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, G. X., and J. Taylor. 1990. Template switching by reverse transcriptase during DNA synthesis. J. Virol. 64:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14:S31-S44. [PubMed] [Google Scholar]
- 37.Mikkelsen, J. G., A. H. Lund, M. Duch, and F. S. Pedersen. 2000. Mutations of the kissing-loop dimerization sequence influence the site specificity of murine leukemia virus recombination in vivo. J. Virol. 74:600-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikkelsen, J. G., A. H. Lund, K. D. Kristensen, M. Duch, M. S. Sorensen, P. Jorgensen, and F. S. Pedersen. 1996. A preferred region for recombinational patch repair in the 5′ untranslated region of primer binding site-impaired murine leukemia virus vectors. J. Virol. 70:1439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moutouh, L., J. Corbeil, and D. D. Richman. 1996. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc. Natl. Acad. Sci. USA 93:6106-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muriaux, D., H. De Rocquigny, B. P. Roques, and J. Paoletti. 1996. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 271:33686-33692. [DOI] [PubMed] [Google Scholar]
- 41.Muriaux, D., P. M. Girard, B. Bonnet-Mathoniere, and J. Paoletti. 1995. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J. Biol. Chem. 270:8209-8216. [DOI] [PubMed] [Google Scholar]
- 42.Negroni, M., and H. Buc. 2000. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. USA 97:6385-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negroni, M., and H. Buc. 1999. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J. Mol. Biol. 286:15-31. [DOI] [PubMed] [Google Scholar]
- 44.Paillart, J. C., R. Marquet, E. Skripkin, B. Ehresmann, and C. Ehresmann. 1994. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 269:27486-27493. [PubMed] [Google Scholar]
- 45.Palaniappan, C., M. Wisniewski, W. Wu, P. J. Fay, and R. A. Bambara. 1996. Misincorporation by HIV-1 reverse transcriptase promotes recombination via strand transfer synthesis. J. Biol. Chem. 271:22331-22338. [DOI] [PubMed] [Google Scholar]
- 46.Panganiban, A. T., and D. Fiore. 1988. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science 241:1064-1069. [DOI] [PubMed] [Google Scholar]
- 47.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc. Natl. Acad. Sci. USA 87:6024-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peeters, M., and P. M. Sharp. 2000. Genetic diversity of HIV-1: the moving target. AIDS 14:S129-S140. [PubMed] [Google Scholar]
- 49.Peliska, J. A., S. Balasubramanian, D. P. Giedroc, and S. J. Benkovic. 1994. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry 33:13817-13823. [DOI] [PubMed] [Google Scholar]
- 50.Peliska, J. A., and S. J. Benkovic. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112-1118. [DOI] [PubMed] [Google Scholar]
- 51.Pfeiffer, J. K., M. M. Georgiadis, and A. Telesnitsky. 2000. Structure-based Moloney murine leukemia virus reverse transcriptase mutants with altered intracellular direct-repeat deletion frequencies. J. Virol. 74:9629-9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfeiffer, J. K., R. S. Topping, N. H. Shin, and A. Telesnitsky. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 73:8441-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Preston, B. D., B. J. Poiesz, and L. A. Loeb. 1988. Fidelity of HIV-1 reverse transcriptase. Science 242:1168-1171. [DOI] [PubMed] [Google Scholar]
- 54.Quinones-Mateu, M. E., Y. Gao, S. C. Ball, A. J. Marozsan, A. Abraha, and E. J. Arts. 2002. In vitro intersubtype recombinants of human immunodeficiency virus type 1: comparison to recent and circulating in vivo recombinant forms. J. Virol. 76:9600-9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rein, A. 1994. Retroviral RNA packaging: a review. Arch. Virol. Suppl. 9:513-522. [DOI] [PubMed] [Google Scholar]
- 56.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 57.Robertson, D. L., B. H. Hahn, and P. M. Sharp. 1995. Recombination in AIDS viruses. J. Mol. Evol. 40:249-259. [DOI] [PubMed] [Google Scholar]
- 58.Roda, R. H., M. Balakrishnan, J. K. Kim, B. P. Roques, P. J. Fay, and R. A. Bambara. 2002. Strand transfer occurs in retroviruses by a pause initiated two-step mechanism. J. Biol. Chem. 277:46900-46911. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez-Rodriguez, L., Z. Tsuchihashi, G. M. Fuentes, R. A. Bambara, and P. J. Fay. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 270:15005-15011. [DOI] [PubMed] [Google Scholar]
- 60.Sharp, P. M., E. Bailes, D. L. Robertson, F. Gao, and B. H. Hahn. 1999. Origins and evolution of AIDS viruses. Biol. Bull. 196:338-342. [DOI] [PubMed] [Google Scholar]
- 61.Skripkin, E., J. C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 91:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stuhlmann, H., and P. Berg. 1992. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J. Virol. 66:2378-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Svarovskaia, E. S., K. A. Delviks, C. K. Hwang, and V. K. Pathak. 2000. Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J. Virol. 74:7171-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Telesnitsky, A., and S. P. Goff. 1993. Strong-stop strand transfer during reverse transcription, p. 49-83. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 65.Temin, H. M. 1993. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc. Natl. Acad. Sci. USA 90:6900-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsuchihashi, Z., and P. O. Brown. 1994. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 68:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vanin, E. F., M. Kaloss, C. Broscius, and A. W. Nienhuis. 1994. Characterization of replication-competent retroviruses from nonhuman primates with virus-induced T-cell lymphomas and observations regarding the mechanism of oncogenesis. J. Virol. 68:4241-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vartanian, J.-P., A. Meyerhans, B. Åsjö, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber, J., and F. Grosse. 1989. Fidelity of human immunodeficiency virus type I reverse transcriptase in copying natural DNA. Nucleic Acids Res. 17:1379-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wisniewski, M., Y. Chen, M. Balakrishnan, C. Palaniappan, B. P. Roques, P. J. Fay, and R. A. Bambara. 2002. Substrate requirements for secondary cleavage by HIV-1 reverse transcriptase RNase H. J. Biol. Chem. 277:28400-28410. [DOI] [PubMed] [Google Scholar]
- 71.Wooley, D. P., L. A. Bircher, and R. A. Smith. 1998. Retroviral recombination is nonrandom and sequence dependent. Virology 243:229-234. [DOI] [PubMed] [Google Scholar]
- 72.Wu, W., B. M. Blumberg, P. J. Fay, and R. A. Bambara. 1995. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J. Biol. Chem. 270:325-332. [DOI] [PubMed] [Google Scholar]
- 73.Xu, H., and J. D. Boeke. 1987. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84:8553-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.You, J. C., and C. S. McHenry. 1994. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 269:31491-31495. [PubMed] [Google Scholar]
- 75.Yu, H., A. E. Jetzt, Y. Ron, B. D. Preston, and J. P. Dougherty. 1998. The nature of human immunodeficiency virus type 1 strand transfers. J. Biol. Chem. 273:28384-28391. [DOI] [PubMed] [Google Scholar]
- 76.Yusa, K., M. F. Kavlick, P. Kosalaraksa, and H. Mitsuya. 1997. HIV-1 acquires resistance to two classes of antiviral drugs through homologous recombination. Antivir. Res. 36:179-189. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, J., L.-Y. Tang, T. Li, Y. Ma, and C. M. Sapp. 2000. Most retroviral recombinations occur during minus-strand DNA synthesis. J. Virol. 74:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and bio/technology. Kluwer Academic Publishers, Dordrecht, The Netherlands.