Abstract
The 2A proteinase (2Apro) of human rhinoviruses (HRVs) is a cysteine protease containing a structurally important zinc ion. In the viral polyprotein, the enzyme cleaves between the C terminus of VP1 and its own N terminus. 2Apro also processes the two isoforms of the cellular protein, eukaryotic initiation factor 4G (eIF4G). We have shown that mature HRV2 2Apro, when translated in vitro in rabbit reticulocyte lysates, efficiently cleaves eIF4GI, although the enzyme was not immediately active upon synthesis. Here, we examine the relationship between self-processing and eIF4GI cleavage. The onset of both reactions first occurred at least 10 min after initiation of protein synthesis. Furthermore, when self-processing was prevented by a specific mutation between VP1 and 2Apro, the VP1-2Apro precursor was essentially unable to cleave eIF4GI, implying that self-processing is a prerequisite for eIF4GI cleavage. 2Apro synthesized in the presence of a potent zinc chelator is inactive; however, upon addition of excess zinc, HRV2 2Apro rapidly gained activity. Finally, the presence of the zinc chelator in the culture medium can protect HeLa cells from HRV infection.
A crucial step in the life cycle of many viruses is the proteolytic processing of viral and cellular proteins (6). This is especially evident with certain picornaviruses which encode proteinases which not only process viral proteins but also carry out specific and controlled proteolysis of the eukaryotic initiation factor 4GI (eIF4GI) and its isoform 4GII (11, 20, 27). These proteins are components of the eIF4F complex, which is responsible for recruitment of capped mRNA to the ribosome (19). Cleavage of the eIF4G isoforms prevents recruitment of capped mRNAs, thus disabling host cell protein synthesis. Picornaviral mRNAs can, however, still be translated under these conditions, since protein synthesis initiates at a higher-ordered structure, termed the internal ribosome entry site, within the 5′ untranslated region (2).
Picornaviruses have evolved two different types of proteinases for cleavage of the eIF4G isoforms (23). Foot-and-mouth disease virus (FMDV) encodes a papain-like cysteine proteinase, the leader proteinase (Lpro), at the very N terminus of the polyprotein (10, 12). Lpro frees itself from the polyprotein by cleavage between its own C terminus and the adjacent protein, VP4 (26). Proteolysis of eIF4GI by Lpro has recently been shown to be enhanced by an apparently direct interaction of the C-terminal extension (CTE) of Lpro with the eIF4G isoforms (7).
In contrast, human rhinoviruses (HRVs) and enteroviruses (polioviruses and coxsackieviruses) encode a different cysteine proteinase, namely the 2A proteinase (2Apro), which is responsible for cleavage of the eIF4G isoforms (17, 23). This enzyme differs from the majority of cysteine proteinases by possessing a chymotrypsin fold as well as containing zinc (22, 25). The zinc ion, located about 20 Å from the active site, appears not to play a role in catalysis; instead, it has been proposed to play a structural role, possibly taking over the role of a disulfide bridge found at an equivalent position in extracellular chymotrypsin-like proteinases (22, 28). 2Apro is located toward the center of the viral polyprotein and processes itself from the growing polypeptide chain by cleavage between the C terminus of the preceding protein, VP1, and its own N terminus. Processing at the C terminus of 2Apro is carried out by the second viral proteinase, 3Cpro.
Despite much investigation, the mechanism of cleavage of eIF4G isoforms by human rhino- and enteroviral 2Apro is still not completely understood. Evidence for direct cleavage of eIF4GI by 2Apro from poliovirus, coxsackievirus B4, and HRV2 has been shown (3, 15, 16, 29). In contrast, cleavage of eIF4GI during poliovirus infection appears to occur indirectly via activation of cellular proteinases as well as by 2Apro (32, 33).
We have previously investigated in detail the relationship between FMDV Lpro self-processing and its ability to cleave eIF4GI. eIF4GI proteolysis was independent of whether the Lpro was expressed as a mature protein or whether it was expressed as a polyprotein with subsequent VP4/VP2 sequences. Indeed, inhibition of Lpro self-processing by mutation of the cleavage site between Lpro and VP4 did not inhibit eIF4GI processing (8). Finally, the onset of eIF4GI cleavage was rapid and occurred at low concentrations; proteolysis of eIF4GI could be observed even before mature Lpro was detectable by fluorography (8, 9).
HRV2 2Apro is not active immediately after synthesis.
In contrast to Lpro, when mature HRV2 2Apro was expressed in rabbit reticulocyte lysates (RRLs), cleavage of eIF4GI did not begin until at least 10 min after synthesis of 2Apro had been initiated (9). To investigate whether self-processing also did not commence until this time, we constructed a plasmid (pHRV2 VP1-2Apro) encoding the HRV2 VP1 and 2Apro (HRV2 nucleotides 2318 to 3586 cloned downstream of the encephalomyocarditis virus internal ribosome entry site in the plasmid pCITE), enabling us to examine HRV2 2Apro self-processing between the C terminus of VP1 and its own N terminus. Following linearization with BamHI, mRNA was transcribed in vitro as described previously (9) from pHRV2 VP1-2Apro and translated in vitro in RRLs. In vitro translation reactions (typical volume, 50 μl) contained 70% RRL (Promega), 20 μCi of [35S]methionine (1,000 Ci/mmol; Hartmann Analytic), 0.8 U of RNasin/μl, and unlabeled amino acids except methionine at 20 μM. After preincubation for 2 min at 30°C, translation was started by addition of mRNA to a final concentration of about 10 ng/μl. Samples were taken at different time points and placed on ice; unlabeled methionine and cysteine were then added to a 2 mM concentration (each) followed by Laemmli sample buffer. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and examined as described previously (8, 9) either by fluorography to detect the radiolabeled translation products (Fig. 1A) or by immunoblotting with an antibody against the N terminus of eIF4GI to monitor the status of the endogenous eIF4GI in the RRL (Fig. 1B).
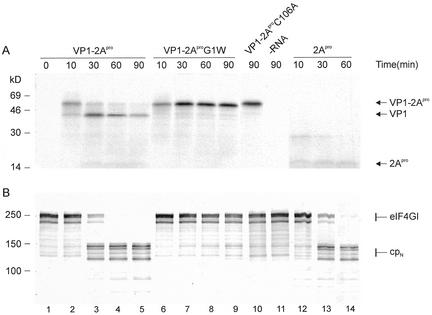
FIG. 1.
Kinetics of self-processing and eIF4GI cleavage by HRV2 2Apro and 2Apro G1W. RRLs were incubated with mRNAs (∼10 ng/μl) encoding HRV2 VP1-2Apro (lanes 1 to 5), VP1-2Apro G1W (lanes 6 to 9), or just 2Apro (lanes 12 to 14) or without RNA (lane 11). An mRNA encoding VP1-2AproC106A, an inactive 2Apro variant in which the active site cysteine is replaced by an alanine, was also translated (lane 10). Samples were taken at the designated time points after mRNA addition, and protein synthesis was terminated by addition of Laemmli sample buffer. Aliquots were then analyzed by PAGE. (A) Fluorogram of 17.5% acrylamide PAGE showing protein synthesis. The positions of the uncleaved precursor VP1-2Apro and the cleavage products VP1 and 2Apro are marked. The fluorogram was exposed for 17 h. (B) Immunoblot of 6% acrylamide PAGE showing the status of eIF4GI. The positions of the uncleaved eIF4GI and the N-terminal cleavage product (cpN) detected by the anti-eIF4GI antibody are marked. Protein standards in kDa are indicated in both panels.
The unprocessed precursor protein VP1-2Apro has a molecular mass of 49 kDa, while those of the processed products VP1 and 2Apro are 33 and 16 kDa, respectively. The 10-min time point (Fig. 1A, lane 2) shows more than 90% unprocessed VP1-2Apro, indicating that in contrast to FMDV Lpro, 2Apro is not immediately active upon synthesis. Complete cleavage is achieved between 60 and 90 min. The amounts of VP1 decrease over time, indicating nonspecific degradation of the protein in the lysate; this has been observed for certain proteins expressed in RRLs (21). The band corresponding to 2Apro is less intense than VP1, since it contains only two methionines, compared to the 9 methionines of VP1. To compare the effect of self-processing on eIF4GI cleavage, RNA from a plasmid encoding mature 2Apro without VP1 was also synthesized and translated (Fig. 1, lanes 12 to 14). The mature 2Apro in this construction is preceded by five additional amino acids from pCITE (MetAlaThrThrMet); therefore, the 2Apro has four methionines compared to the two present in 2Apro after processing from VP1. Thus, the intensity of the respective bands is about double.
The time course cleavage of eIF4GI is shown in Fig. 1B. Intact eIF4GI runs as a series of bands around 220 kDa due to microheterogeneity at the N terminus, while the N-terminal cleavage products of eIF4GI generated by 2Apro cleavage migrate at around 140 kDa. At the 10-min time point (Fig. 1B, lane 2), essentially no eIF4GI cleavage has occurred; once initiated, cleavage is rapid, being almost complete after 30 min. Similar rates of eIF4GI cleavage were observed in the absence of self-processing with 2Apro alone (lanes 12 to 14). Thus, independent of whether self-processing of HRV2 2Apro takes place or not, and in contrast to FMDV Lpro, 2Apro appears not to be immediately active upon synthesis but rather requires time to gain activity. In addition, the presence of the VP1 sequence appears to have no effect at the rate at which 2Apro gains activity.
The VP1-2Apro precursor cannot cleave eIF4GI.
To further investigate the relationship between the two cleavage reactions, we decided to inhibit self-processing by mutating the VP1-2Apro cleavage site. We demonstrated previously in a bacterial system (24) that replacement of the P1′ site amino acid glycine by a tryptophan completely prevents self-processing. Therefore, using standard methods of PCR mutagenesis, we constructed a variant of VP1-2Apro in which the first amino acid of 2Apro, glycine, was replaced by a tryptophan (pHRV2 VP1-2AproG1W). mRNA from this plasmid was then translated in vitro in RRLs, and the effect on eIF4GI cleavage was monitored (Fig. 1, lanes 6 to 9). As shown in Fig. 1A, only the unprocessed precursor protein is observed, confirming that 2Apro can no longer process itself from VP1. Interestingly, the failure to process VP1 from 2Apro also results in a lack of eIF4GI cleavage (Fig. 1B); even after 90 min of incubation, no significant eIF4GI cleavage by VP1-2AproG1W was seen. To rule out any effect of this mutation on the catalytic activity of 2Apro, we translated an mRNA encoding the 2AproG1W protein without VP1. This variant showed normal eIF4GI cleavage rates (data not shown). Thus, although it contains a potentially catalytically active enzyme, the uncleaved VP1-2Apro precursor has essentially almost no ability to process eIF4GI. This is in contrast to FMDV Lpro, which can still cleave eIF4GI even when self-processing is inhibited by mutations introduced at the cleavage site (8).
What are the molecular mechanisms behind these differences between 2Apro and Lpro? Examination of the three-dimensional structures of the two enzymes reveals a clear difference between the lengths and conformations of the termini at which processing occurs. Thus, Lpro has a C-terminal extension of 18 amino acids which projects well away from the main globular domain and lies about 30 to 40 Å from the active site. This suggests that the CTE can move away from the active site, leaving it available for other substrates. In contrast, the N terminus of HRV2 2Apro is closer to the active site, and only five amino acids of the N terminus project away from the globular domain; this suggests that the uncleaved VP1 protein blocks the enzyme's active site, thus preventing access to other substrates.
TPEN, a zinc chelator, prevents acquisition of an active conformation by HRV2 2Apro.
The delay in onset of 2Apro cleavage of both self-processing and eIF4GI cleavage suggested that the enzyme was initially synthesized in an inactive form. Since HRV 2Apro is known to contain zinc (25, 30), we decided to investigate whether the incorporation of the zinc ion into the enzyme was rate limiting by removal or addition of zinc to the lysate. We used the highly specific zinc chelator N,N,N′,N′-tetrakis-(2-pyridylmethyl)-ethylenediamine (TPEN) (Sigma). TPEN has a very high specificity for Zn2+ (pKa = 15.4) but low affinities for Ca2+ (pKa = 3) and Mg2+ (negligible) (4). The effects of zinc depletion on HRV2 2Apro activity were assayed by preincubating the RRL with different concentrations of TPEN (dissolved in ethanol at a stock concentration of 25 mM and diluted with water to the appropriate concentration immediately before use) before adding the VP1-2Apro mRNA. Figure 2A and B show that increasing concentrations of TPEN inhibit 2Apro self-processing and eIF4GI-processing ability. At 5 μM TPEN, 50% self-processing and eIF4GI cleavage are observed at 60 min, whereas the control shows 50% cleavage at 30 min and almost complete cleavage at 60 min (compare Fig. 2, lanes 5 and 6 with Fig. 1, lanes 3 and 4). In contrast, the addition of zinc in the absence of TPEN does not influence 2Apro activity in any way (Fig. 2, lanes 7 to 9). This suggests that the endogenous zinc concentration in the RRL is sufficient for 2Apro activity, despite the presence of the very high amounts of EGTA used to chelate the calcium ions required for micrococcal nuclease treatment. The translation machinery is not influenced by zinc depletion or higher zinc amounts, since concentrations up to 200 μM of TPEN and zinc, respectively, were tested without observable effects on protein synthesis (data not shown).
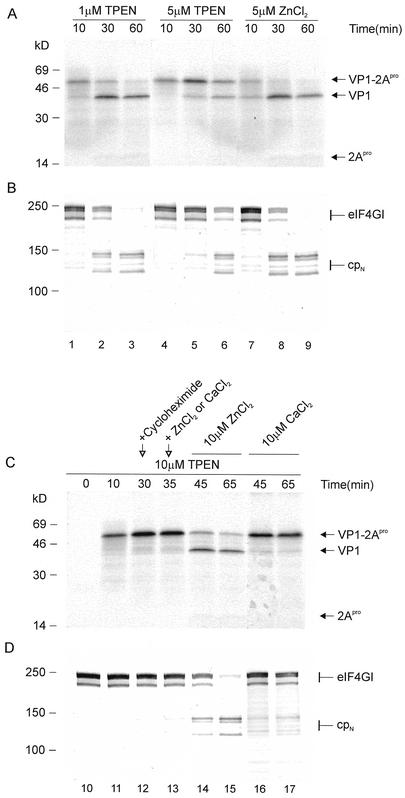
FIG. 2.
Effect of zinc concentration on HRV2 2Apro activity. (A and B) The mRNA VP1-2Apro (∼10 ng/μl) was used to program RRLs; the indicated concentrations of the zinc chelator TPEN or ZnCl2 were added to the translation mixture on ice prior to incubation. The HRV2 VP1-2Apro mRNA (∼10 ng/μl) was translated in RRLs in the presence of 10 μM TPEN. After 30 min, cycloheximide was added to a final concentration of 10 ng/μl to inhibit further protein synthesis. (C and D) After a further 5 min of incubation, ZnCl2 or CaCl2 was added to a final concentration of 10 μM, respectively. Aliquots were removed at the indicated time points, and protein synthesis (A and C) and eIF4GI state (B and D) were monitored as for Fig. 1.
Zinc-depleted HRV2 2Apro is rapidly activated by addition of zinc without requiring denaturation and renaturation.
Previous experiments (25, 30) have shown that HRV2 2Apro binds the zinc very tightly. The zinc cannot be extracted by 10 mM EDTA; removal of the zinc from HRV2 2Apro was achieved only after denaturation. The zinc-depleted denatured enzyme could be reactivated by renaturation in the presence of zinc. We were interested whether the de novo synthesized protease could be activated by addition of zinc without any special denaturation and renaturation steps. Therefore, it was necessary to completely inhibit HRV2 2Apro; this was achieved by translating the VP1-2Apro mRNA in the presence of 10 μM TPEN (Fig. 2C and D, lanes 11 and 12). After 30 min, cycloheximide was added to a concentration of 10 ng/μl to stop the synthesis of new 2Apro (Fig. 2, lane 12). At 35 min, we added either ZnCl2 to a 10 μM concentration to activate the 2Apro or CaCl2 as a control to the reaction mix and continued the incubation (Fig. 2, lanes 13 to 17). The presence of 10 μM TPEN inhibits 2Apro almost completely, since after 65 min without ZnCl2 (Fig. 2, lane 17) essentially no self-processing or eIF4GI cleavage is observed. In contrast, just 10 min after addition of 10 μM ZnCl2 (Fig. 2, lane 14), both VP1-2Apro and eIF4GI cleavage had reached about 50%. The absence of the previously observed 10-min delay indicates that the incorporation of zinc into de novo-synthesized VP1-2Apro is a very rapid step.
It is worth noting that neither the addition of TPEN nor that of zinc (both compounds were tested at 5 μM) affected Lpro activity in any way, showing that the effect is specific to HRV2 2Apro (data not shown).
TPEN can protect HeLa cells from the viral cytopathic effect.
Since the presence of TPEN can prevent the activation of de novo synthesized HRV2 2Apro, we investigated whether TPEN can be used to block HRV replication in cell culture. TPEN is able to permeate membranes and thus ideally suited for this purpose. To examine any possible differences between minor and major group HRVs, both HRV2 and HRV14 were used. HeLa cells were incubated with different concentrations of TPEN, and the cells were infected with HRV2 or HRV14 or incubated with medium alone. The viability of the cells was then assayed after 24 h using the Cell Titer 96 AQueous nonradioactive cell proliferation assay (Promega). Briefly, 106 cells were seeded into a 96-well plate 1 day prior to infection. Cells were infected using HRV serotypes 2 and 14 at 20 50% tissue culture infective doses/cell in the presence or absence of the indicated TPEN concentrations. Each experiment was repeated three times. Twenty-four hours after infection, cell viability was determined by adding the tetrazolium compound followed by incubation for 2 h at 37°C and subsequent measurement of absorption at 492 nm in a Labsystems Multiscan RC plate reader.
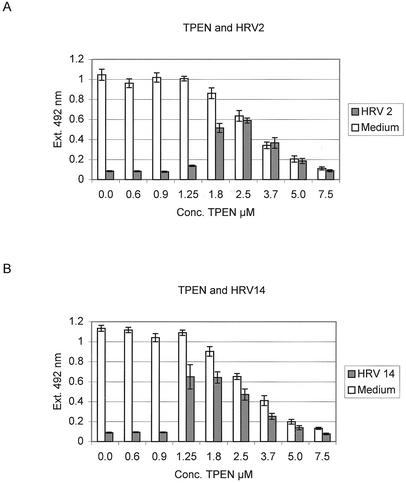
Figure 3 shows that uninfected HeLa cells remain viable in the presence of TPEN up to concentrations between 1.25 and 1.8 μM. Higher TPEN concentrations are increasingly cytotoxic. The cytotoxic effect of TPEN is not unexpected, since depletion of zinc in HeLa cells leads to activation of specific caspases which then induce apoptosis (5, 13, 18). The concentrations of TPEN at which apoptosis was induced were very similar to those reported previously (18).
FIG. 3.
Protection of HeLa cells by TPEN from HRV infection. HeLa cells were incubated in 96-well plates with the indicated TPEN concentrations and incubated with 20 50% tissue culture infective doses (open bars) of HRV2 (A) or HRV14 (B) or medium (closed bars) as a control. Twenty-four hours after infection, cell viability was determined. The mean and error bars were calculated from three separate experiments.
Despite the loss of cell viability due to the induction of apoptosis by TPEN, protection of cells from viral infection was observed at TPEN concentrations between 1.8 μM and 2.5 μM (Fig. 3). With HRV14 (Fig. 3B), the protection was more pronounced, with 60% of cells protected at a concentration of 1.25 μM, a concentration below that required for inducing apoptosis. Thus, the inhibition of HRV infection with a potent zinc chelator is feasible.
It has been known for many years that the addition of zinc itself inhibits HRV replication. The mechanism of this inhibition remains unclear, but it appears to involve binding of zinc to the part of the polyprotein containing the capsid precursors (14). In contrast, we have shown that inhibition of replication of two serotypes could be achieved by the chelation of zinc, brought about by the presence of TPEN. Although chelation of zinc by TPEN induces apoptosis, a concentration range was found which protected cells to a significant extent against infection by both HRV2 and HRV14. Notably, the concentration range of 1.5 to 2 μM is clearly lower than the 10 μM required for 2Apro inhibition in RRLs. This implies either that the zinc concentration is lower in HeLa cells than in RRLs or that TPEN is also affecting other cellular processes. Since TPEN has been shown to interfere with endocytosis (1), it seems possible that viral entry may also be affected.
The rapid induction of apoptosis by TPEN in HeLa cells would seem to preclude its use as a possible antiviral compound. However, other cell types, such as macrophages, have been shown to remain unaffected by TPEN concentrations as high as 150 μM (1). Thus, future work will be directed towards determining the TPEN sensitivities of cell lines capable of supporting HRV replication. An investigation of the usefulness of TPEN and related zinc chelators as inhibitors of HRV and human enterovirus replication can then be made with the cell lines most resistant to TPEN. Furthermore, it will be of interest to examine whether TPEN can also inhibit the replication of polio- and coxsackieviruses. Although the 2Apros of these viruses have not been directly shown to contain zinc, site-directed mutagenesis and amino acid alignments (31) strongly imply the ability of these proteinases to bind zinc and thus their sensitivity to TPEN during viral replication.
Acknowledgments
This work was supported by the Austrian Science Foundation (grants P-13367 and P-16189 to T.S.).
We thank D. Blaas, E. Gaudernak, J. Seipelt, and members of our laboratory for stimulating discussions.
REFERENCES
- 1.Aballay, A., M. N. Sarrouf, M. I. Colombo, P. D. Stahl, and L. S. Mayorga. 1995. Zn2+ depletion blocks endosome fusion. Biochem J. 312:919-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. R. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression, vol. 39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Bovee, M. L., B. J. Lamphear, R. E. Rhoads, and R. E. Lloyd. 1998. Direct cleavage of elF4G by poliovirus 2A protease is inefficient in vitro. Virology 245:241-249. [DOI] [PubMed] [Google Scholar]
- 4.Cherny, R. A., J. T. Legg, C. A. McLean, D. P. Fairlie, X. Huang, C. S. Atwood, K. Beyreuther, R. E. Tanzi, C. L. Masters, and A. I. Bush. 1999. Aqueous dissolution of Alzheimer's disease Abeta amyloid deposits by biometal depletion. J. Biol. Chem. 274:23223-23228. [DOI] [PubMed] [Google Scholar]
- 5.Chimienti, F., M. Seve, S. Richard, J. Mathieu, and A. Favier. 2001. Role of cellular zinc in programmed cell death: temporal relationship between zinc depletion, activation of caspases, and cleavage of Sp family transcription factors. Biochem. Pharmacol. 62:51-62. [DOI] [PubMed] [Google Scholar]
- 6.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases—functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foeger, N., W. Glaser, and T. Skern. 2002. Recognition of eIF4G isoforms by picornaviral proteinases. J. Biol. Chem. 277:44300-44309. [DOI] [PubMed] [Google Scholar]
- 8.Glaser, W., R. Cencic, and T. Skern. 2001. Foot-and-mouth disease leader proteinase: involvement of C-terminal residues in self-processing and cleavage of eIF4GI. J. Biol. Chem. 276:35473-35481. [DOI] [PubMed] [Google Scholar]
- 9.Glaser, W., and T. Skern. 2000. Extremely efficient cleavage of eIF4G by picornaviral proteinases L and 2A in vitro. FEBS Lett. 480:151-155. [DOI] [PubMed] [Google Scholar]
- 10.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarné, A., J. Tormo, K. Kirchweger, D. Pfistermueller, I. Fita, and T. Skern. 1998. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 17:7469-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, S., S. C. Chow, M. J. McCabe, Jr., and S. Orrenius. 1995. Lack of Ca2+ involvement in thymocyte apoptosis induced by chelation of intracellular Zn2+. Lab. Investig. 73:111-117. [PubMed] [Google Scholar]
- 14.Korant, B. D., and B. E. Butterworth. 1976. Inhibition by zinc of rhinovirus protein cleavage: interaction of zinc with capsid polypeptides. J. Virol. 18:298-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuechler, E., J. Seipelt, H.-D. Liebig, and W. Sommergruber. 2002. Picornavirus proteinase-mediated shutoff of host cell translation: direct cleavage of a cellular initiation factor, p. 301-311. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 16.Liebig, H.-D., E. Ziegler, R. Yan, K. Hartmuth, H. Klump, H. Kowalski, D. Blaas, W. Sommergruber, L. Frasel, B. Lamphear, R. Rhoads, E. Kuechler, and T. Skern. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4G and influence on translation. Biochemistry 32:7581-7588. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd, R. E., M. J. Grubman, and E. Ehrenfeld. 1988. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequencing. J. Virol. 62:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCabe, M. J., Jr., S. A. Jiang, and S. Orrenius. 1993. Chelation of intracellular zinc triggers apoptosis in mature thymocytes. Lab. Investig. 69:101-110. [PubMed] [Google Scholar]
- 19.Merrick, W. C., and J. W. B. Hershey. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, W. C. Merrick, and J. W. B. Hershey (ed.), Translational control of gene expression, vol. 39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Morley, S. J., P. S. Curtis, and V. M. Pain. 1997. eIF4G: translation's mystery factor begins to yield its secrets. RNA 3:1085-1104. [PMC free article] [PubMed] [Google Scholar]
- 21.Oberst, M. D., T. J. Gollan, M. Gupta, S. R. Peura, J. D. Zydlewski, P. Sudarsanan, and T. G. Lawson. 1993. The encephalomyocarditis virus 3C protease is rapidly degraded by an ATP-dependent proteolytic system in reticulocyte lysate. Virology 193:28-40. [DOI] [PubMed] [Google Scholar]
- 22.Petersen, J. F., M. M. Cherney, H. D. Liebig, T. Skern, E. Kuechler, and M. N. James. 1999. The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis. EMBO J. 18:5463-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skern, T., B. Hampoelz, E. Bergmann, A. Guarné, J. Petersen, I. Fita, and M. N. G. James. 2002. Structure and function of picornavirus proteinases, p. 199-212. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 24.Skern, T., W. Sommergruber, H. Auer, P. Volkmann, M. Zorn, H. D. Liebig, F. Fessl, D. Blaas, and E. Kuechler. 1991. Substrate requirements of a human rhinoviral 2A proteinase. Virology 181:46-54. [DOI] [PubMed] [Google Scholar]
- 25.Sommergruber, W., G. Casari, F. Fessl, J. Seipelt, and T. Skern. 1994. The 2A proteinase of human rhinovirus is a zinc containing enzyme. Virology 204:815-818. [DOI] [PubMed] [Google Scholar]
- 26.Strebel, K., and E. Beck. 1986. A second protease of foot-and mouth disease virus. J. Virol. 58:893-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukada, H., and D. M. Blow. 1985. Structure of alpha-chymotrypsin refined at 1.68 Å resolution. J. Mol. Biol. 184:703-711. [DOI] [PubMed] [Google Scholar]
- 29.Ventoso, I., S. E. MacMillan, J. W. Hershey, and L. Carrasco. 1998. Poliovirus 2A proteinase cleaves directly the eIF-4G subunit of eIF-4F complex. FEBS Lett. 435:79-83. [DOI] [PubMed] [Google Scholar]
- 30.Voss, T., R. Meyer, and W. Sommergruber. 1995. Spectroscopic characterization of rhinoviral protease 2A: Zn is essential for the structural integrity. Protein Sci. 4:2526-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, S. Y. F., and R. E. Lloyd. 1992. Characterization of the roles of conserved cysteine and histidine residues in poliovirus 2A-protease. Virology 186:725-735. [DOI] [PubMed] [Google Scholar]
- 32.Zamora, M., W. E. Marissen, and R. E. Lloyd. 2002. Multiple eIF4GI-specific protease activities present in uninfected and poliovirus-infected cells. J. Virol. 76:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zamora, M., W. W. Marissen, and R. E. Lloyd. 2002. Poliovirus-mediated shutoff of host translation: an indirect effect, p. 313-320. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.