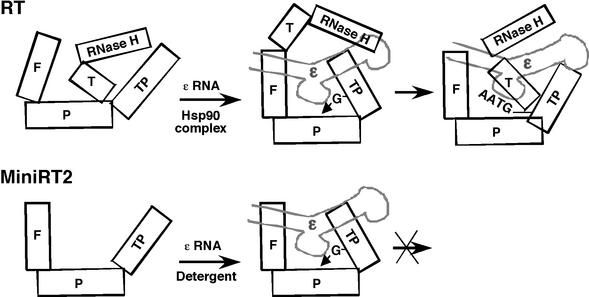
Abstract
The reverse transcriptase (RT) encoded by hepadnaviruses (hepatitis B viruses) is a multifunctional protein critical for several aspects of viral assembly and replication. Reverse transcription is triggered by the specific interaction between the RT and an RNA signal located on the viral pregenomic RNA, termed ɛ, and is initiated through a novel protein priming mechanism whereby the RT itself serves as a protein primer and ɛ serves as the obligatory template. Using the RT from duck hepatitis B virus as a model, we previously demonstrated that RT-ɛ interaction and protein priming require the assistance of a host cell chaperone complex, heat shock protein 90 (Hsp90) and its cochaperones, which associates with the RT and facilitates the folding of the RT into an active conformation. We now report that extensive truncation removing the entire C-terminal RNase H domain and part of the central RT domain could relieve this dependence on Hsp90 for RT folding such that the truncated RT variants could function in ɛ interaction and protein priming independently of Hsp90. The presence of certain nonionic or zwitterionic detergent was sufficient to establish and maintain the truncated RT proteins in an active, albeit labile, state. Furthermore, we were able to refold an RT truncation variant de novo after complete denaturation. In contrast, the full-length RT and also RT variants with less-extensive C-terminal truncations required Hsp90 for activation. Surprisingly, the presence of detergent plus some yet-to-be-identified cytoplasmic factor(s) led to a dramatic suppression of the RT activities. These results have important implications for RT folding and conformational maturation, Hsp90 chaperone function, and potential inhibition of RT functions by host cell factors.
Reverse transcription in hepadnaviruses (hepatitis B viruses [HBVs]) is carried out by a novel virally encoded reverse transcriptase (RT) (27, 30). The RT has the unique ability to initiate DNA synthesis de novo, using itself as a protein primer (18, 19, 35, 39, 43; for a review, see reference 9). This protein priming reaction requires the specific interaction between the RT and a short RNA signal, termed ɛ, located at the 5′ end of the viral pregenomic RNA (pgRNA; the template for reverse transcription) (22, 36). The product of protein priming is a three- to four-nucleotide DNA oligomer, representing the 5′ end of the viral minus-strand DNA, covalently attached to the RT via an invariant tyrosine residue located at its N-terminal domain (19, 21, 32, 34; for a review, see reference 11).
The ability of the hepadnavirus RT to carry out specific RNA recognition and protein priming is reflected in its structural organization, which displays both similarities to, as well as differences from, conventional RTs encoded by retroviruses and other retroelements (4, 9, 23). As mentioned above, the N-terminal domain (the so-called terminal protein [TP]) bears the invariant primer tyrosine residue; the TP domain is conserved among all hepadnaviruses but absent from any other known RTs. The central RT domain and the C-terminal RNase H domain share sequence homologies with conventional RTs. A highly variable spacer or tether domain appears to link the TP and RT domains. It is now well established that the TP and RT domains have to interact functionally to allow RT-ɛ interaction and protein priming (8, 17, 19, 22, 36).
We have previously shown that the RT from duck HBV (DHBV) associates with a host cell molecular chaperone complex composed of the heat shock protein 90 (Hsp90) and its cochaperones and requires the assistance of the Hsp90 complex in order to establish an active conformation competent in ɛ binding and protein priming (10, 13). More recently, we have succeeded in biochemically reconstituting a functional RT by using recombinant mini-DHBV RT proteins purified from bacteria and the Hsp90 chaperone complex, either as cell extract (8) or purified components (12). These mini-RT proteins bear both N- and C-terminal truncations and an internal deletion in the spacer region but contain the essential sequences from both the TP and RT domains that are required for ɛ binding and protein priming. By analogy with other chaperone substrates, the RT is thought to require Hsp90 assistance in order to establish a conformation active in ɛ binding and protein priming. However, the mechanism of this presumed RT conformational maturation remains unknown.
The dependence of hepadnavirus RT activation on Hsp90 is most likely a reflection of the novel structural organization of the RT and of the unique intramolecular and intermolecular interactions required for the distinct mechanism of hepadnavirus reverse transcription, as opposed to other conventional RTs that are not known to require Hsp90. For example, during the initiation of protein priming, the protein primer (i.e., the TP domain of the RT protein) and RNA template (i.e., the ɛ RNA) must interact with the RT active site in a way fundamentally different from that occurring during conventional reverse transcription, when a preexisting RNA (or DNA) primer, annealed to an RNA template, is elongated. In support of this notion, we have recently reported that a mini-DHBV RT protein with its entire C-terminal RNase H domain and part of the RT domain (the putative thumb subdomain) removed is still able to initiate protein priming but is defective in any DNA strand elongation, which is similar to conventional reverse transcription (38). We report here that the removal of these sequences in fact could relieve the dependence on Hsp90 for RT activation. Such truncated RT variants could be purified from bacteria or eukaryotic cells in an active form, without the need for in vitro reconstitution using the Hsp90 chaperone, which, as discussed above, is required for the full-length RT or longer RT variants. Instead, certain nonionic and zwitterionic detergents were able to facilitate the activation of these truncated mini-RT proteins into a conformation functional in ɛ binding and protein priming. These results have revealed the presence of autoinhibitory sequences in the RT C-terminal region that can block its folding into a conformation competent for ɛ binding and protein priming. They suggest that a major role of Hsp90 in facilitating RT conformational maturation is to counteract the suppressive effect of these autoinhibitory sequences. Unexpectedly, the combination of detergent and cellular extract led to a dramatic inhibition of RT activities.
MATERIALS AND METHODS
Plasmids and reagents.
pGST-MiniRT1 and pGST-MiniRT2 (for bacterial expression of glutathione (GSH) S-transferase [GST] or GST fusion proteins), as well as pHis-MiniRT1 and pHis-MiniRT2 (for expression of histidine-tagged fusion proteins), have been described before (8). pGST-MiniRT1/Pml was derived from pGST-MiniRT1 by the removal of the RT sequences C terminal to position 661 (at the PmlI restriction site). pEBG-MiniRT1 and pEBG-MiniRT2 (for expression of the GST-mini-RT fusion proteins in mammalian cells) have been described before (37, 38). All mini-RT proteins were tagged with the hemagglutinin (HA) epitope, as previously described (8). The monoclonal antibody against the HA epitope, HA.11 (clone 16B12), was purchased from BAbCO (Berkeley Antibody). Nonidet P-40 (NP-40) and its chemical equivalent, Igepal (CA-630), Triton X-100, CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, sodium dodecyl sulfate (SDS), and deoxycholate were purchased from Sigma and the nondetergent sulfobetaine, NDSB-195, from Calbiochem.
Protein purification.
GST-MiniRT1 and GST-MiniRT2 were expressed in Escherichia coli or 293T cells and purified by using GSH agarose beads as described previously (8, 38). When the RT proteins were purified from bacteria, 1% Triton X-100 was included in the lysis and wash buffers, whereas either 0.1 or 0.5% NP-40 (as indicated) was included in the buffers for protein purification from 293T cells. In some experiments (as indicated), additional washes with a detergent-free TNK buffer (20 mM Tris [pH 8.0], 15 mM NaCl, 20 mM KCl) were also performed before elution of the bound proteins. Additional specific modifications are indicated in individual figures.
His-MiniRT2 was purified by using the Ni+ affinity resin under nondenaturing conditions as previously described (8). To purify His-MiniRT2 under denaturing conditions, inclusion bodies were dissolved in lysis buffer containing 8 M urea and then purified by using the Ni+ affinity resin (Qiagen). To refold the purified, denatured His-MiniRT2, the denaturant (urea) was removed stepwise by dialysis into the refolding buffer (50 mM phosphate buffer [pH 8.0], 300 mM KCl, 2 mM EDTA, 5 mM reduced GSH, 0.5 mM oxidized GSH, 0.02% NaN3) containing a reduced amount of urea (3 M, 1 M, or none) in the presence or absence of detergent as indicated. In some experiments, l-arginine (1.0 M) was added to the refolding buffer and was decreased to 0.4 M via dialysis after RT refolding.
Recombinant human Hsp90β (7) and human Hsp70 (24) were expressed in Sf9 cells and purified as described previously. Human Hop (p60) (25), human p23 (29), and the yeast Hsp40 homolog Ydj1 (16) were expressed in bacteria and purified as previously described.
In vitro protein priming.
Approximately 10 ng of GST-MiniRT or His-MiniRT proteins, purified either from bacteria or 293T cells, were used in an in vitro protein priming reaction in a total volume of 10 μl as described previously (8, 10) by using [α-32P]dGTP as the radioactively labeled nucleotide precursor. In experiments with [α-32P]dATP as the labeled nucleotide precursor, unlabeled dGTP and TTP were also added, as described earlier (38). Various supplements to the protein priming reaction, including the rabbit reticulocyte lysate (nuclease treated; Promega), purified Hsp90 complex components (120 ng of Hsp90, 350 ng of Hsp70, 125 ng of Hop/p50, 1 μg of Ydj1, and 20 ng of p23), the ATP regenerating system (5 mM ATP, 10 mM creatine phosphate, and 50 μg of creatine phosphokinase/ml) were added to the protein priming reaction as described previously (8, 12). For the pretreatment of RT proteins with detergent, the purified RT proteins were first incubated with the indicated detergent at room temperature for 10 min, in a total volume of 2 μl, before being added to the protein priming reactions.
RESULTS
Dependence of protein priming activity of purified mini-DHBV RT protein on purification conditions.
We have recently shown that a mini-DHBV RT protein consisting of sequences from the TP and RT domains (see Fig. 5A for a schematic diagram) fused to GST, called GST-MiniRT2, can be stably expressed both in bacteria and 293T cells and readily purified (8, 12). Like the full-length RT and the longer MiniRT1 protein, protein priming activity of the purified MiniRT2 was found to require in vitro reconstitution with cell extract or purified components of the Hsp90 complex (12). However, we have noticed subsequently that under certain conditions, the MiniRT2 protein showed protein priming activity in vitro even without reconstitution. For example, when 0.5% of NP-40, a nonionic detergent, was used in the cell lysis and wash buffers, the MiniRT2 purified from 293T cells showed strong protein priming activity in vitro without the need of reconstitution (Fig. 1, lane 1). In contrast, the same mini-RT protein purified with buffers containing 0.1% NP-40 showed little activity and required in vitro reconstitution with the reticulcoyte lysate (Fig. 1, lanes 3 and 4) (12) or purified Hsp90 chaperone components (12). In fact, MiniRT2 purified with buffers containing 0.5% NP-40, without in vitro reconstitution, was nearly as active as that purified by using 0.1% NP-40 after in vitro reconstitution (Fig. 1, lanes 1 and 4). Another surprising result in Fig. 1 (described below) was the dramatic inhibition of RT activity when the reticulocyte lysate was added to MiniRT2 purified with 0.5% NP-40 (Fig. 1, lane 2); the protein priming activity under this condition was even threefold lower than the background activity (nonstimulated) obtained with the mini-RT protein purified with 0.1% NP-40 (Fig. 1, lane 3).
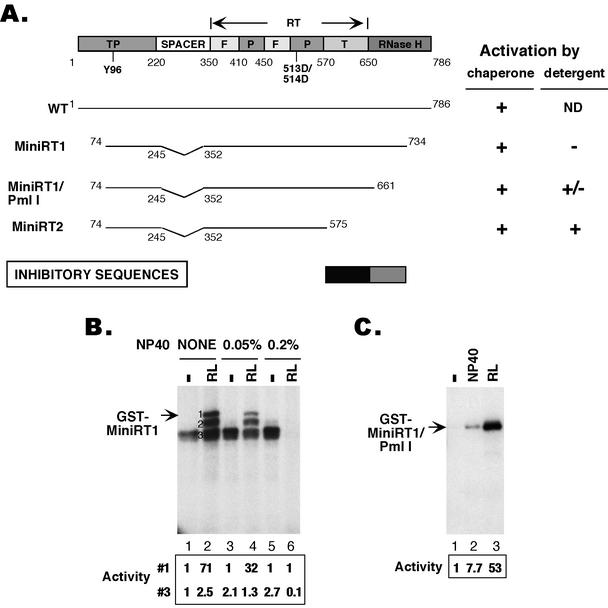
FIG. 5.
Mapping of RT sequences that auto-inhibited RT activity. (A) At the top is a schematic diagram of the DHBV RT domain structure (TP, spacer, RT, and RNase H). The primer tyrosine residue (Y96) and the double aspartate residues (D513/D514) at the RT active site are indicated. The RT domain is further divided into the “finger” (F), “palm” (P), and “thumb” (T) subdomains (with the approximate boundaries marked), based on alignment with the RT structure of the human immunodeficiency virus (5, 26). Summarized below the diagram are the protein priming activities of the RT proteins, following activation with the Hsp90 chaperone complex or detergent. The N- and C-terminal truncation points and the internal deletion in the spacer region are indicated. The RT sequences whose deletion led to chaperone-independent protein priming activity (autoinhibitory sequences) are denoted. ND, not tested. (B) GST-MiniRT1 was purified from bacteria and assayed for protein priming activity with or without treatment with NP40 or reticulocyte lysate (RL) as described in Fig. 1. The labeled GST-MiniRT1 is indicated. The numerals 1, 2, and 3 in lane 2 indicate the full-length MiniRT1 and its two degradation products, respectively. The protein priming activities of the intact MiniRT1 (species 1) and one of its degradation product (species 3), relative to those obtained in the absence of either detergent or the reticulocyte lysate (lane 1), are indicated at the bottom of the figure. (C) GST-MiniRT1/Pml was purified from bacteria and assayed for protein priming activity with or without treatment with NP-40 (0.2%) or reticulocyte lysate (RL) as described in Fig. 1, except that [α-32P]dATP (instead of [α-32P]dGTP) was used as the labeled nucleotide. The protein priming activities, relative to the activity obtained in the absence of either detergent or the reticulocyte lysate (lane 1), are given at the bottom of the figure.
FIG. 1.
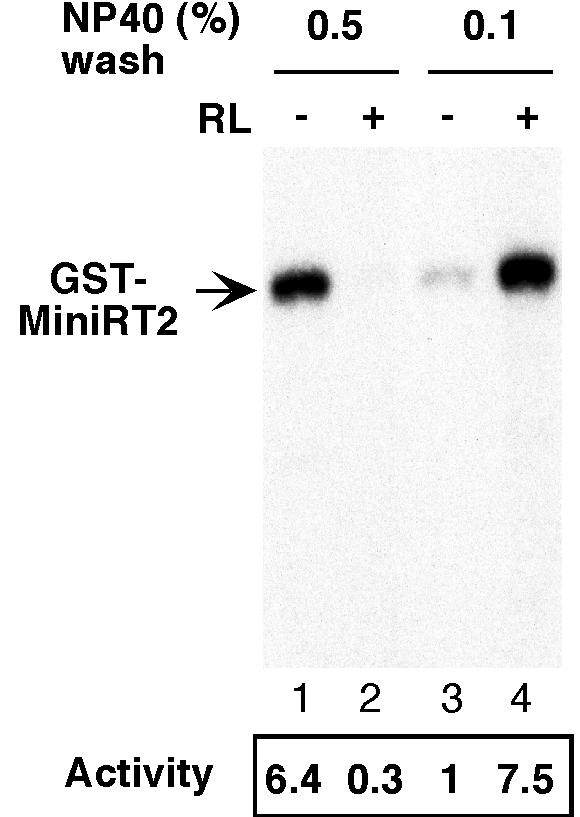
Protein priming activity of GST-MiniRT2 purified from 293T cells under low- and high-detergent conditions. Plasmid DNA expressing the GST-tagged MiniRT2 (pEBG-MiniRT2) was transfected into 293T cells. Transfected cells were lysed, and the GST-MiniRT2 was purified by using GSH affinity resins. Two similar purification conditions, lysis and washing buffer containing either 0.5% (lanes 1 and 2) or 0.1% (lanes 3 and 4) NP-40, were used. Proteins bound to the resins were eluted with GSH. The purified mini-RT protein was assayed for in vitro protein priming activity. All reactions were carried out in the presence of ɛ RNA and [α-32P]dGTP, with (lanes 2 and 4) or without (lanes 1 and 3) supplementation with the reticulocyte lysate (RL). The labeled GST-MiniRT2 is indicated. The labeled mini-RT bands were quantified by phosphorimaging, and the protein priming activities under the different conditions, relative to the activity of the mini-RT purified with 0.1% NP-40 assayed in the absence of the reticulocyte lysate (lane 3), are given at the bottom of the figure.
Since no detergent was added to the elution buffer, we suspected that residual amounts of NP-40 were carried over to the purified mini-RT preparation from the wash buffers and may have affected the protein priming activity of the MiniRT2 protein purified under high- versus low-detergent conditions. We estimated that the concentration of NP-40 in the purified RT (the eluate) could be up to 0.1% when 0.5% NP-40 was included in the wash buffer. Initially, we tested this possibility by sequentially washing the resin-bound mini-RT protein with low (0.1%), then high (0.5%), and again low (0.1%) concentrations of NP-40 before eluting the mini-RT. Strikingly, by simply washing the bound mini-RT protein, which had been initially purified with 0.1% NP-40, with 0.5% NP-40 before elution, we could activate the mini-RT protein such that it became active in protein priming without the need for in vitro reconstitution with cellular factors (Fig. 2A, lane 3 versus lane 1), as if the RT had been purified under conditions of a high detergent concentration (Fig. 1, lane 1). Furthermore, subsequent washing with a low concentration (0.1%) of NP-40 reversed the activation of the mini-RT protein such that it again required in vitro reconstitution with cellular factors (Fig. 2A, lanes 5 and 6), just like the mini-RT protein purified with a low detergent concentration (Fig. 2A, lanes 1 and 2, and Fig. 1, lanes 3 and 4).
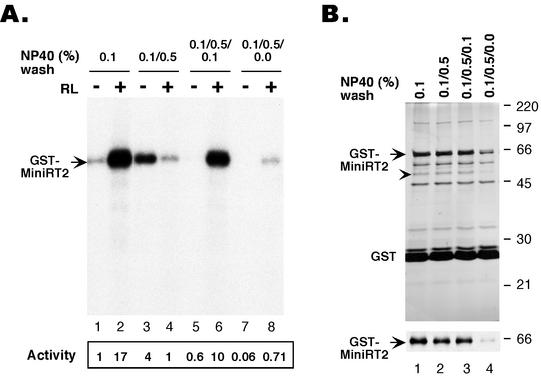
FIG. 2.
Effect of washing conditions on the purification and activity of GST-MiniRT2. GST-MiniRT2 was expressed in 293T cells and purified by using GSH affinity resins as described in Fig. 1, except that 0.1% NP-40 was used initially in the lysis and washing buffers (A, lanes 1 and 2; B, lane 1) and this was followed by additional rounds of washing with 0.5% NP-40 (A, lanes 3 and 4; B, lane 2), 0.5% and then 0.1% NP-40 (A, lanes 5 and 6; B, lane 3), or 0.5% NP-40 and then detergent-free washes (A, lanes 7 and 8; B, lane 4), before elution of the bound proteins with GSH. (A) The mini-RT proteins, purified under the different conditions, were assayed for in vitro protein priming activity as described in Fig. 1. The various results are as described in the legend to Fig. 1. (B) The purified mini-RT proteins were resolved by SDS-polyacrylamide gel electrophoresis and were detected by either silver staining (top) or Western blotting (bottom) with the anti-HA monoclonal antibody. GST-MiniRT2 is indicated, as is GST. The arrowhead indicates a factor (identity not yet known) that appeared to be associated with the RT, the amount of which was proportional to that of the purified RT. Other nonidentified bands seemed to represent nonspecifically bound proteins whose levels did not vary with that of the RT.
Since the stimulating effect of washing with higher concentrations of detergent was reversible by a subsequent wash with low concentrations of detergent, the detergent effect on the RT activity was probably not due to any potential effect on the yield or purity of the purified mini-RT protein. This was confirmed when we determined the yield and the purity of the purified mini-RT protein under different detergent conditions. As shown in Fig. 2B, the different detergent conditions did not affect the yield of the mini-RT protein purified, as determined by either protein staining or Western blot analysis (Fig. 2B, lanes 1 to 3). Moreover, although the mini-RT protein was only partially purified, the pattern of copurifying (or contaminating) cellular proteins did not change with the different detergent conditions (Fig. 2B, lanes 1 to 3).
When the resin-bound mini-RT proteins were washed finally with a detergent-free buffer to remove residual amounts of detergent, we found that most of the mini-RT protein was lost (probably as a result of aggregation), so that the amount of eluted mini-RT protein was decreased by 5- to 10-fold compared to the eluates in the absence of the detergent-free washes (Fig. 2B, lane 4). The mini-RT protein thus eluted, like that purified under low-detergent concentration, also required in vitro reconstitution to gain protein priming activity (Fig. 2A, lanes 7 and 8).
In summary, the inclusion of relatively high concentrations of the nonionic detergent, NP-40, in the purification process was able to activate MiniRT2, in a reversible manner. The low- and high-detergent conditions did not affect either the yield or the purity of the mini-RT. Only when the detergent was completely removed did the mini-RT appear to aggregate and was it poorly recovered. These results suggest that residual amounts of NP-40, carried over from the purification process, could stimulate the RT activity when present above a certain threshhold concentration. In addition, at concentrations too low to stimulate RT activity, the detergent could nevertheless help to maintain the RT in a soluble state.
Direct activation of MiniRT2 by nonionic and zwitterionic detergents.
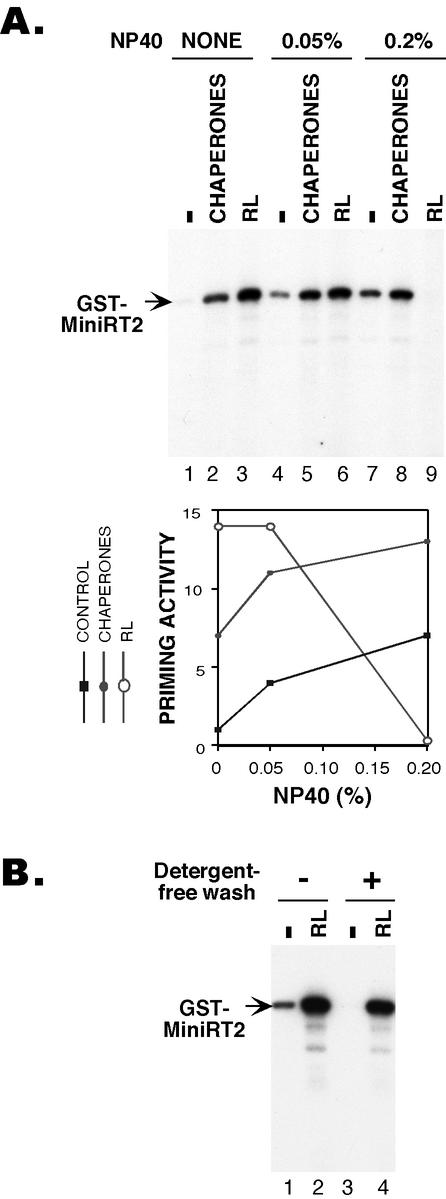
Although the results described above suggested that nonionic detergents may be able to stimulate RT activity, it remained possible that some unknown factor(s) from the mammalian expression host copurified with the mini-RT and affected the RT activity under the different detergent conditions (although silver staining did not reveal any differences in the quality or quantity of the copurifying cellular factors [Fig. 2B, lanes 1 to 3]). To test whether the detergent alone, in the absence of any eukaryotic host factor (e.g., Hsp90), could directly activate the RT, we purified GST-MiniRT2 from bacteria. We then added NP-40 at different concentrations to the purified RT before we conducted the priming reactions. Indeed, the addition of NP-40 could directly stimulate the RT activity in vitro, in a dose-dependent manner (Fig. 3A, lanes 1, 4, and 7). At the optimal concentration, NP-40 activated the RT as efficiently as the Hsp90 complex (Fig. 3A, lanes 2 and 7). When the two were added together, the stimulatory effect of the detergent and the chaperone on the RT were additive (Fig. 3A, lanes 5 and 8).
FIG. 3.
Direct in vitro activation of GST-MiniRT2 by detergent. (A) GST-MiniRT2 was purified from bacteria. As indicated, the RT was either mock treated (lanes 1 to 3) or pretreated with 0.05% (lanes 4 to 6) or 0.2% (lanes 7 to 8) NP-40 (NP-40) before the protein priming reaction. Chaperones (Hsp90, Hsp70, p60/Hop, Ydj1, and p23 supplemented with an ATP regenerating system; see Materials and Methods) (lanes 2, 5, and 8) or reticulocyte lysate (RL; lanes 3, 6, and 9) was then added together with the reaction buffer, the ɛ RNA, andnucleotide substrates to initiate protein priming. In the graph, the 32P-labeled RT bands were quantified by using phosphorimaging, and protein priming activities at various conditions were expressed relative to the control reaction (lane 1, no detergent, chaperones or reticulocyte lysate). (B) GST-MiniRT2 was purified from bacteria as in panel A, except for the reactions shown in lanes 3 and 4, for which an additional detergent-free washing step was included before elution of the RT in order to remove residual amounts of detergent carryover from the detergent-containing washing buffer. The RT was then assayed for in vitro protein priming activity, with (lanes 2 and 4) or without (lanes 1 and 3) reconstitution with the reticulocyte lysate (RL).
Occasionally, a low protein priming activity could be detected with the MiniRT2 purified from bacteria without any stimulation with detergents or Hsp90 (Fig. 3B, lane 1). We suspected that this was probably due to the carryover of residual amounts (up to 0.05% in the eluate) of Triton X-100, a nonionic detergent used in the purification process, as was the case with NP-40 when the mini-RT was purified from 293T cells. In support of this proposition, when additional washing steps with a detergent-free wash buffer before elution of the RT was performed to remove the residual amount of detergent, the “background” (without chaperone reconstitution in vitro) priming activity was essentially eliminated (Fig. 3B, lane 3). Interestingly, the yield of the purified RT after the detergent-free wash was not decreased in this case and the RT activity after in vitro reconstitution with the cellular extract was the same regardless of the washing conditions (Fig. 3B, lanes 2 and 4, and data not shown). This finding was in contrast to what was observed when the RT was purified from the mammalian cells shown above; in that case, the yield of RT purification was dramatically decreased by the detergent-free wash (Fig. 2B, lane 4). Since the RT purified from bacteria was associated with the bacterial chaperones, GroEL and DnaK, these bacterial chaperones most likely helped to prevent the aggregation and the resultant loss of the RT in the absence of detergents, as we suggested earlier (8).
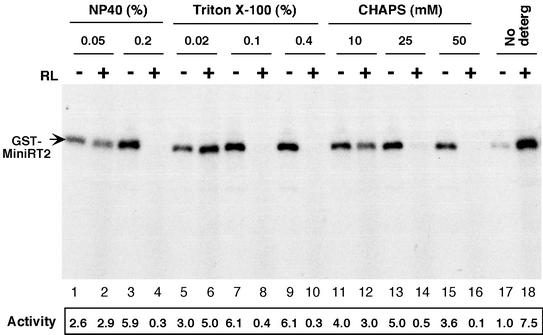
The results presented in Fig. 3B further suggested that not only NP-40 but also Triton X-100 and perhaps other detergents might have a similar stimulatory effect on RT activity. To test this possibility, we purified MiniRT2 from either bacteria or 293T cells and added various nonionic (NP-40 and Triton X-100) and zwitterionic (CHAPS) detergents to the purified RT before testing its protein priming activity (Fig. 4 and data not shown). The results showed that, as anticipated, not only NP-40 but also Triton X-100 and CHAPS could stimulate RT activity in a dose-dependent manner. Furthermore, like NP-40, the other detergents (when used at higher concentrations) also induced a dramatic inhibition of RT activity when added together with the reticulocyte lysate. On the other hand, anionic detergents, such as deoxycholate and SDS, never activated RT at any of the concentrations tested and inhibited RT at high concentrations when these strong detergents most likely denatured the RT (data not shown).
FIG. 4.
Activation of MiniRT2 by nonionic and zwitterionic detergents. GST-MiniRT2 was purified from bacteria. Prior to being used in the protein priming reaction, GST-MiniRT2 was pretreated with the indicated detergents as described in Fig. 3. The mini-RT protein was then assayed for protein priming activity (as described in Fig. 1) in the presence (even-numbered lanes) or absence (odd-numbered lanes) of reticulocyte lysate (RL). The protein priming activities, relative to the activity obtained in the absence of either detergent or the reticulocyte lysate (lane 17), are given at the bottom of the figure.
Mapping of RT truncation boundary that conferred Hsp90-indepedent activation.
In contrast to MiniRT2, another mini-RT protein, MiniRT1, which has a less extensive C-terminal deletion removing only part of the RNase H domain, could never be activated by any of the detergents tested and always required the Hsp90 chaperone for activity (Fig. 5B, species 1) (8, 12). Strikingly, a degradation product from MiniRT1 (labeled species 3), which was approximately the same size as MiniRT2 and probably shared a similar C-terminal deletion to MiniRT2 due to degradation (38), could nevertheless be activated by detergent in the same reaction (Fig. 5B, lanes 1, 3, and 5). Also, as noted before for MiniRT2 (Fig. 3B), this degradation product of MiniRT1 showed significant priming activity without the addition of exogenous detergent (Fig. 5B, lane 1) due to the carryover of detergent from the purification process. In addition, another degradation product (labeled species 2, which was apparently larger than species 3) seemed also to be weakly activated by detergent (Fig. 5B). This suggested that RT proteins with longer C-terminal sequences than MiniRT2 could be activated by detergent. It is also interesting that the degradation products (Fig. 5B, species 2 and 3) of MiniRT1 appeared to have a higher specific activity upon activation than the intact MiniRT1 (Fig. 5B, species 1), since these degradation products were less abundant (although their exact concentrations varied) (8; data not shown) than the intact protein and yet they showed an equal or higher activity (Fig. 5B, lane 2).
To further define the C-terminal RT sequences that had to be removed to allow RT activation independent of Hsp90, we constructed MiniRT1/Pml, which was truncated at amino acid 661, approximately halfway between the C termini of MiniRT1 (at position 734) and MiniRT2 (at position 575). This mini-RT protein could indeed be weakly activated by the detergent NP-40 (Fig. 5C). However, whereas MiniRT2 could be activated by detergent almost as efficiently as by the Hsp90 complex or reticulocyte lysate, activation of MiniRT1/Pml by detergent was only ca. 10% as efficient as that by reticulocyte lysate.
In summary, removal of RT C-terminal sequences (amino acids 575 to 734) allowed the truncated RT to fold into an active conformation competent in protein priming. Instead of requiring the Hsp90 chaperone complex for their conformational maturation, the truncated RT proteins could apparently adopt an active conformation with the assistance of mild detergent. These results suggest that autoinhibitory sequences may be present in the RNase H and the RT domains that can block the folding of the full-length RT protein into a protein-priming active state and that this inhibitory effect can be counteracted by the Hsp90 chaperone complex. The strong activation of MiniRT2 (and the shorter of the two MiniRT1 degradation products, species 3) compared to the rather weak activation of MiniRT1/Pml (and the longer MiniRT1 degradation product, species 2) (Fig. 3 and 5) indicates the presence of a major inhibitory sequence from amino acids 575 to 661 in the C-terminal portion of the RT domain, whereas the absolute requirement of Hsp90 for MiniRT1 activation and the inability of MiniRT1 to be activated by detergent compared to the weak detergent activation observed for MiniRT1/Pml (and the longer MiniRT1 degradation product, species 2) (Fig. 5) (8, 12) suggests the existence of a secondary inhibitory sequence from amino acids 661 to 734 in the RNase H domain.
In vitro refolding of denatured MiniRT2.
Although the results presented above strongly suggested that removal of some RT sequences from its C terminus could relieve its dependence on Hsp90 for the establishment of a protein priming-competent conformation, two potential caveats could complicate the interpretation of the results. One was the fact that the mini-RT proteins were all fused to GST, which might have affected the folding of the fusion proteins. The other concern was that the mini-RT proteins were only partially purified; in particular, two bacterial chaperones, DnaK and GroEL, were associated with the RT proteins purified from bacteria and might have affected the folding of the RT. To address these issues, we purified mini-RT proteins by using the six-histidine tag (8) instead of the GST tag. First, we purified the His-tagged MiniRT2 protein (His-MiniRT2) under native conditions (in the presence of either NP-40 or Triton X-100) and found that it was active in protein priming without the need for in vitro reconstitution with chaperone proteins (data not shown), a finding similar to that described above with GST-MiniRT2. This result indicated that the GST fusion was not necessary for the folding of the mini-RT in the absence of Hsp90. However, the purified His-tagged mini-RT was also associated with DnaK and GroEL under these purification conditions, as reported previously (1, 8). To exclude a role of the bacterial chaperone proteins in RT folding, we then purified the His-tagged mini-RT proteins under denaturing conditions from inclusion bodies. Under these conditions, His-MiniRT2 was purified to >95% homogeneity (Fig. 6, lane 1) and was essentially free of any significant contamination of bacterial proteins, including DnaK and GroEL. We then attempted to refold the denatured mini-RT by gradual removal of the denaturant (urea) with or without the presence of detergent. Indeed, we succeeded in refolding His-MiniRT2 by using NP-40 in the refolding buffer. The refolded His-MiniRT2 was active in protein priming without the need for in vitro reconstitution (Fig. 6, lane 2). In addition, we also found that we could refold His-MiniRT2 into a soluble state (but inactive in protein priming) with a detergent-free refolding buffer containing l-arginine (Fig. 6, lane 3). This partially refolded MiniRT2 could then be activated to a protein priming competent conformation by simply incubating it with detergent at the time of the priming assay (Fig. 6, lane 4).
FIG. 6.
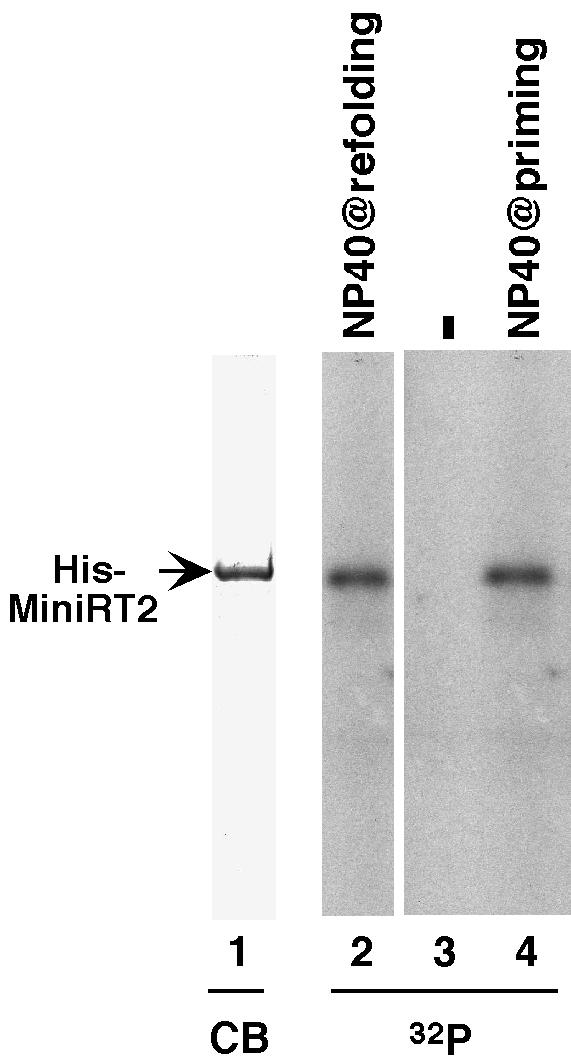
De novo refolding of MiniRT2. His-MiniRT2 was purified from bacteria under denaturing conditions, resolved by SDS-polyacrylamide gel electrophoresis, and detected by Coomassie blue (CB) staining (lane 1). Denatured His-MiniRT2 was refolded in the absence (lanes 3 and 4) or presence (lane 2) of 0.1% NP-40 before being assayed for protein priming activity. In the reaction shown in lane 4, the refolded mini-RT was pretreated with 0.2% NP-40 right before protein priming (as described in Fig. 3). 32P denotes the labeling of His-MiniRT2 as a result of protein priming in the presence of [α-32P]dGTP (lanes 2 to 4).
In summary, MiniRT2, without the GST fusion, could fold into an active conformation in the absence of Hsp90. Furthermore, highly purified, denatured MiniRT2 could productively refold de novo with the assistance of detergent and in the absence of any eukaryotic or prokaryotic chaperone proteins. These results confirmed that removal of the C-terminal RT sequences could indeed relieve the requirement for cellular chaperones in RT folding and activation.
RT inhibition by cell lysate in the presence of detergent.
As was clear from the results presented above, the various detergents could also lead to dramatic (10- to 40-fold) inhibition of RT activity when added together with the reticulocyte lysate (Fig. 1, lane 2; Fig. 3A, lane 9; Fig. 4, lanes 4, 8, 10, 14, and 16; and Fig. 5B, lane 6). This inhibitory effect of the detergent, in the presence of the cell lysate, was also dose dependent, like its stimulatory effect on MiniRT2 in the absence of the lysate. Furthermore, whereas the detergents were able to activate only MiniRT2 but not MiniRT1, they showed the inhibitory effect (in the presence of the reticulocyte lysate) on both MiniRT1 and MiniRT2 (Fig. 5B) regardless of whether the RT proteins were fused to GST or the histidine tag (data not shown). In contrast to the cell lysate, the addition of the purified Hsp90 chaperone components plus the detergents never inhibited RT activity (Fig. 3A and data not shown), indicating that these chaperone components were not responsible for mediating the inhibitory effects of the lysate in the presence of the detergent.
DISCUSSION
Previous work has shown that the hepadnavirus RT requires the host cell chaperone, the Hsp90 complex, in order to establish and maintain an active conformation functional in ɛ binding and protein priming (8, 10, 12, 13). We have demonstrated here that the removal of the C-terminal RNase H domain and part of the RT domain could relieve the dependence of RT folding on Hsp90. Instead, the truncated RT proteins could fold (refold) into a functional state in the absence of any cellular chaperone proteins. We propose that “autoinhibitory” sequences are present in the RNase H domain and part of the RT domain that can block, in the absence of Hsp90 assistance, the folding of RT into a conformation competent in ɛ binding. A major role of Hsp90 in RT folding, therefore, appears to counteract the effect of these inhibitory sequences so as to facilitate productive RT folding. It is, however, important to note that these results by no means exclude any additional role that the Hsp90 complex potentially plays in the subsequent steps of viral reverse transcription after ɛ binding and the initiation of protein priming, as suggested by its incorporation into the nucleocapsids (13).
Although dispensable for ɛ binding and protein priming in vitro, the C-terminal sequences removed in MiniRT2 are essential for other aspects of RT functions, including pgRNA packaging, minus-strand DNA elongation, degradation of the pgRNA template after minus-strand DNA synthesis, and plus-strand DNA synthesis (9, 27). In the absence of Hsp90 assistance, these sequences may prevent RT folding by interfering with the domain-domain interactions between the TP and the N-terminal portion of the RT domain required for ɛ binding and protein priming. The removal of these self-inhibitory sequences could then allow the interaction between the TP and RT domains to occur in the truncated mini-RT proteins without Hsp90 assistance. This suggestion is also consistent with the fact that C-terminally truncated RT proteins can be much more readily expressed than the full-length RT in bacteria as reported previously by us and others (1, 8), presumably because the C-terminal sequences interfere with RT folding in bacteria. Our results indicate that the inhibitory sequences seem to be bipartite; a major inhibitory sequence lies within the C-terminal region of the RT domain (amino acids 575 to 661, the putative thumb subdomain) (20, 26) and a secondary inhibitory sequence in the RNase H domain (amino acids 661 to 734).
The thumb subdomain forms part of the polymerase active site and is responsible for template-primer interactions in other known RTs and polymerases in general (3, 28). However, we have shown that these sequences are dispensable for the initiation of protein priming, i.e., the covalent linkage of the 5′-terminal nucleotide of the viral minus-strand DNA to the RT (38; the present study), indicating that the interaction between the hepadnavirus RT and its template (the ɛ RNA) and primer (the TP domain of the RT itself) during the initiation of protein priming is fundamentally different from other polymerase template-primer interactions. On the other hand, we have shown that the thumb subdomain is indispensable for any subsequent DNA strand elongation. Thus, immediately after the initiation of protein priming, the RT must undergo some dramatic conformational change so that the thumb subdomain can interact with the other subdomains (the so-called finger and palm, Fig. 5A and 7) of the RT domain in order to establish a more conventional polymerase structure to carry out DNA elongation. Without Hsp90 assistance, the thumb subdomain may prematurely associate with the rest of the RT domain and preclude the TP-RT domain interactions necessary for ɛ binding and the initiation of protein priming. Similarly, the RNase H domain, which has to interact with the RT domain during the later stages of viral DNA synthesis, may also exert its inhibitory effect on protein priming by interfering with TP-RT domain interactions necessary for initiating protein priming due to its premature interaction with the RT domain. The role of Hsp90 would thus be to prevent these premature intramolecular interactions so as to allow an ordered transition of the different RT conformations that are required for the different stages of reverse transcription and viral replication.
FIG. 7.
Working model for Hsp90-dependent and -independent folding of the DHBV RT and MiniRT2. The domains and subdomains of the RT are depicted as blocks. TP, TP domain; F, P, and T, the “finger,” “palm,” and “thumb” subdomains of the RT domain, respectively. The ɛ RNA is depicted as a stem-loop structure, with its internal bulge (the template for protein priming) facing the palm subdomain. In the case of the full-length RT, the thumb subdomain (and the RNase H domain) may prematurely interact with the palm subdomain and preclude the TP from accessing the RT active site to establish a conformation competent for ɛ binding and initiation of protein priming. The Hsp90 chaperone complex is proposed to counteract this inhibitory effect of the thumb subdomain and RNase H domain by preventing these inappropriate interactions and facilitating the productive interactions between the TP and the RT domains. In the case of MiniRT2, the removal of the thumb subdomain and RNase H domain allows the mini-RT to fold independently of Hsp90, so long as some mild detergent is present to facilitate folding. On the other hand, after the initiation of protein priming (the covalent linkage of the dGMP residue to the TP domain), the thumb subdomain has to access the RT active site (and the TP has to exit) in order to facilitate the subsequent DNA extension (leading to the synthesis of the nascent DNA oligomer, 5′-GTAA-3′). Lacking the thumb subdomain, MiniRT2 is thus unable to carry out any DNA elongation. See the text for details.
The proposed role for Hsp90 in preventing nonproductive intramolecular interactions of its substrates is consistent with several observations in the literature. In the case of steroid receptors, which normally require Hsp90 to establish a hormone binding-competent conformation, the isolated hormone-binding domain seems nevertheless able to fold into a hormone-binding competent state independently of the Hsp90 complex (2, 31, 41). Recently, a short segment of the estrogen receptor, ca. 45 residues long and lying adjacent to the hormone-binding domain, has indeed been suggested to play an inhibitory role in receptor function by restraining the hormone binding domain in an inactive conformation (15). These results suggest that the inhibition-relieving role proposed here for Hsp90 in RT folding could be a more generalized function of Hsp90 in chaperoning its various substrates.
The precise mechanism of detergent-assisted activation of the truncated RT proteins folding is not yet known. However, the results presented here can exclude the following trivial explanations. First, the role of detergent is not simply to prevent RT aggregation or loss to the surface. The differential effect of detergent on the activity of the RT proteins with different truncations, even in the same reaction, clearly indicates a specific effect on RT activity (and, by inference, RT folding) (Fig. 5). In addition, at concentrations below certain threshhold levels, the detergents could still help to prevent RT aggregation but failed to activate the RT (Fig. 2), a finding similar to the effect of the bacterial DnaK and/or GroEL chaperones, which also can prevent RT aggregation but cannot activate RT function (Fig. 3B) (8). Second, it is also unlikely that the role of detergent was to suppress an unknown inhibitor of RT functions that copurified with the RT proteins. The detergents showed the same effect on the RT proteins whether they were purified from bacteria or mammalian cells. Although the RT proteins purified under native conditions from either source were not completely pure, it is highly unlikely that the same inhibitor of RT function, which can be relieved by the detergent, was copurified from both the prokaryotic and eukaryotic cells. Furthermore, we could purify the truncated RT proteins under denaturing conditions to near homogeneity and refold these pure RT proteins de novo into an active form with detergent assistance.
Nonionic and zwitterionic detergents have been widely used as “artificial chaperones” to facilitate protein folding in vitro (6, 42), although the precise mechanism of detergent-assisted protein folding in general is still largely unknown. We noticed that, for the detergents we have tested, concentrations above their respective critical micelle concentrations seemed to be required to facilitate MiniRT2 folding. However, the critical micelle concentration is known to be affected by many factors (6, 42), and we do not yet have direct evidence that detergent micelles indeed were formed under our assay conditions. We have also found that some detergent analogs that cannot form micelles, the so-called nondetergent sulfobetaines (33), can also facilitate MiniRT2 folding, suggesting that micelle formation may not be necessarily required for the detergents to assist MiniRT2 folding. We also believe that the role of Hsp90 in MiniRT2 activation, in contrast to its role in activation of the full-length RT or MiniRT1, was most likely similar to that of the detergents, acting largely as a so-called passive chaperone (14, 40, 42). In support of this, we found that Hsp90 activation of MiniRT2, in contrast to that of MiniRT1 or the full-length RT (10, 13), was ATP independent (X. Wang and J. Hu, unpublished results). Interestingly, the activated state of RT remains unstable whether RT activation is induced by Hsp90 (8, 10, 13) or detergent (Fig. 2). Further studies on RT activation by detergents as well as chaperone proteins may shed new light on the mechanism of action of both artificial and cellular chaperones. Regardless of the exact mechanism of activation of the truncated mini-RT proteins, the ability to purify an active RT protein without the need of the host cell chaperones may in practice finally make feasible high-resolution structural studies on the RT, e.g., through X-ray crystallography.
Remarkably, the combination of detergent and reticulocyte lysate led to a dramatic inhibition of protein priming activity. An intriguing interpretation of this result is that the detergents may induce the formation of a factor(s) in the cell lysate that can inhibit RT activity. Future work aimed at elucidating the mechanism of RT inhibition and identifying the putative cellular inhibitors may suggest novel ways of inhibiting RT function for specific anti-HBV treatments.
Acknowledgments
We thank David Toft for generously providing the purified chaperone proteins. We thank David Toft, Christoph Seeger, and Bill Mason for a critical reading of the manuscript.
This work was supported by Public Health Service grants R01 AI43453 (to J.H.) and R01 DK53893 (to H.-C.G.) from the National Institutes of Health.
REFERENCES
- 1.Beck, J., and M. Nassal. 2001. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 75:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bledsoe, R., V. Montana, T. Stanley, C. Delves, C. Apolito, D. McKee, T. Consler, D. Parks, E. Stewart, T. Willson, M. Lambert, J. Moore, K. Pearce, and H. Xu. 2002. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93-105. [DOI] [PubMed] [Google Scholar]
- 3.Brautigam, C. A., and T. A. Steitz. 1998. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 8:54-63. [DOI] [PubMed] [Google Scholar]
- 4.Chang, L. J., R. C. Hirsch, D. Ganem, and H. E. Varmus. 1990. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J. Virol. 64:5553-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, K., X. Xiong, H. Yang, C. E. Westland, C. S. Gibbs, S. G. Sarafianos, and E. Arnold. 2001. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 75:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garavito, R. M., and S. Ferguson-Miller. 2001. Detergents as tools in membrane biochemistry. J. Biol. Chem. 276:32403-32406. [DOI] [PubMed] [Google Scholar]
- 7.Grenert, J., W. Sullivan, P. Fadden, T. Haystead, J. Clark, E. Mimnaugh, H. Krutzsch, H.-J. ochel, T. Schulte, E. Sausiville, L. Neckers, and D. Toft. 1997. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J. Biol. Chem. 272:23843-23850. [DOI] [PubMed] [Google Scholar]
- 8.Hu, J., and D. Anselmo. 2000. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 74:11447-11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, J., and C. Seeger. 1996. Expression and characterization of hepadnavirus reverse transcriptases. Methods Enzymol. 275:195-208. [DOI] [PubMed] [Google Scholar]
- 10.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu, J., and C. Seeger. 1997. RNA signals that control DNA replication in hepadnaviruses. Semin. Virol. 8:205-211. [Google Scholar]
- 12.Hu, J., D. Toft, D. Anselmo, and X. Wang. 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakob, U., and J. Buchner. 1994. Assisting spontaniety: the role of Hsp90 and small Hsps as molecular chaperones. Trends Biochem. Sci. 19:205-211. [DOI] [PubMed] [Google Scholar]
- 15.Koide, A., S. Abbatiello, L. Rothgery, and S. Koide. 2002. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc. Natl. Acad. Sci. USA 99:1253-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosano, H., B. Stensgard, M. C. Charlesworth, N. McMahon, and D. Toft. 1998. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J. Biol. Chem. 273:32973-32979. [DOI] [PubMed] [Google Scholar]
- 17.Lanford, R. E., Y. H. Kim, H. Lee, L. Notvall, and B. Beames. 1999. Mapping of the hepatitis B virus reverse transcriptase TP and RT domains by transcomplementation for nucleotide priming and by protein-protein interaction. J. Virol. 73:1885-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanford, R. E., L. Notvall, and B. Beames. 1995. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J. Virol. 69:4431-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanford, R. E., L. Notvall, H. Lee, and B. Beames. 1997. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J. Virol. 71:2996-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, X., Z. H. Yuan, L. Wu, J. P. Ding, and Y. M. Wen. 2001. A single amino acid in the reverse transcriptase domain of hepatitis B virus affects virus replication efficiency. J. Virol. 75:11827-11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nassal, M., and A. Rieger. 1996. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 70:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollack, J. R., and D. Ganem. 1994. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 68:5579-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radziwill, G., W. Tucker, and H. Schaller. 1990. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 64:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumacher, R. J., W. J. Hansen, B. C. Freeman, E. Alnemri, G. Litwack, and D. O. Toft. 1996. Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry 35:14889-14898. [DOI] [PubMed] [Google Scholar]
- 25.Schumacher, R. J., R. Hurst, W. P. Sullivan, N. J. McMahon, D. O. Toft, and R. L. Matts. 1994. ATP-dependent chaperoning activity of reticulocyte lysate. J. Biol. Chem. 269:9493-9499. [PubMed] [Google Scholar]
- 26.Seeger, C., E. H. Leber, L. K. Wiens, and J. Hu. 1996. Mutagenesis of a hepatitis B virus reverse transcriptase yields temperature-sensitive virus. Virology 222:430-439. [DOI] [PubMed] [Google Scholar]
- 27.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steitz, T. A. 1998. A mechanism for all polymerases. Nature 391:231-232. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan, W., B. Stensgard, G. Caucutt, B. Bartha, N. McMahon, E. Alnemri, G. Litwack, and D. Toft. 1997. Nucleotides and two functional states of hsp90. J. Biol. Chem. 272:8007-8012. [DOI] [PubMed] [Google Scholar]
- 30.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 31.Tanenbaum, D. M., Y. Wang, S. P. Williams, and P. B. Sigler. 1998. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc. Natl. Acad. Sci. USA 95:5998-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavis, J. E., S. Perri, and D. Ganem. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vuillard, L., T. Rabilloud, and M. E. Goldberg. 1998. Interactions of non-detergent sulfobetaines with early folding intermediates facilitate in vitro protein renaturation. Eur. J. Biochem. 256:128-135. [DOI] [PubMed] [Google Scholar]
- 34.Wang, G. H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, G. H., and C. Seeger. 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663-670. [DOI] [PubMed] [Google Scholar]
- 36.Wang, G. H., F. Zoulim, E. H. Leber, J. Kitson, and C. Seeger. 1994. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J. Virol. 68:8437-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, X., N. Grammatikakis, and J. Hu. 2002. Role of p50/CDC37 in hepadnavirus assembly and replication. J. Biol. Chem. 277:24361-24367. [DOI] [PubMed] [Google Scholar]
- 38.Wang, X., and J. Hu. 2002. Distinct requirement for two stages of protein-primed initiation of reverse transcription in hepadnaviruses. J. Virol. 76:5857-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber, M., V. Bronsema, H. Bartos, A. Bosserhoff, R. Bartenschlager, and H. Schaller. 1994. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 68:2994-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiech, H., J. Buchner, R. Zimmermann, and U. Jacob. 1992. Hsp90 chaperones protein folding in vitro. Nature 358:169-170. [DOI] [PubMed] [Google Scholar]
- 41.Williams, S. P., and P. B. Sigler. 1998. Atomic structure of progesterone complexed with its receptor. Nature 393:392-396. [DOI] [PubMed] [Google Scholar]
- 42.Zardeneta, G., and P. M. Horowitz. 1994. Detergent, liposome, and micelle-assisted protein refolding. Anal. Biochem. 223:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Zoulim, F., and C. Seeger. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]