Abstract
Human respiratory syncytial virus (HRSV) and bovine RSV (BRSV) infect human beings and cattle in a species-specific manner. We have here analyzed the contribution of RSV envelope proteins to species-specific entry into cells. In contrast to permanent cell lines, primary cells of human or bovine origin, including differentiated respiratory epithelia, peripheral blood lymphocytes, and macrophages, showed a pronounced species-specific permissivity for HRSV and BRSV infection, respectively. Recombinant BRSV deletion mutants lacking either the small hydrophobic (SH) protein gene or both SH and the attachment glycoprotein (G) gene retained their specificity for bovine cells, whereas corresponding mutants carrying the HRSV F gene specifically infected human cells. To further narrow the responsible region of F, two reciprocal chimeric F constructs were assembled from BRSV and HRSV F1 and F2 subunits. The specificity of recombinant RSV carrying only the chimeric F proteins strictly correlated with the origin of the membrane-distal F2 domain. A contribution of G to the specificity of entry could be excluded after reintroduction of BRSV or HRSV G. Virus with F1 and G from BRSV and with only F2 from HRSV specifically infected human cells, whereas virus expressing F1 and G from HRSV and F2 from BRSV specifically infected bovine cells. The introduction of G enhanced the infectivities of both chimeric viruses to equal degrees. Thus, the role of the nominal attachment protein G is confined to facilitating infection in a non-species-specific manner, most probably by binding to cell surface glycosaminoglycans. The identification of the F2 subunit as the determinant of RSV host cell specificity facilitates identification of virus receptors and should allow for development of reagents specifically interfering with RSV entry.
Human respiratory syncytial virus (HRSV) and bovine RSV (BRSV) are closely related members of the Pneumovirus genus within the Paramyxoviridae family, causing lower respiratory tract disease in humans and cattle, respectively. Although HRSV is one of the most important respiratory pathogens in childhood, causing bronchiolitis and pneumonia (10, 16, 21), effective prophylactic or therapeutic tools are not available. As clinical features after infection are very similar for both viruses, the natural disease caused by BRSV is an important animal model for all aspects of HRSV infection (6).
RSVs encode three envelope glycoproteins, a small hydrophobic (SH) protein of unknown function, a glycoprotein (G) known as attachment protein, and a fusion (F) protein. The RSV F protein is structurally similar to F proteins from other Paramyxoviridae with respect to the location of hydrophobic domains, heptad repeats, and cysteine residues, as well as to proteolytic activation resulting in the exposition of a hydrophobic fusion peptide (7). The inactive precursor F0 is cleaved by the endoprotease furin into an N-terminal F2 subunit and a C-terminal, membrane-anchored F1 subunit carrying the fusion peptide. A peculiarity of RSV F is cleavage at two neighboring multibasic cleavage motifs, resulting in release of a peptide, pep27 (11, 37, 38), whose function is currently under investigation. In contrast to F, the RSV G protein has no sequence or structural similarity to Paramyxovirinae attachment proteins H (hemagglutinin) and HN (hemagglutinin-neuraminidase). Specific receptors for RSVs are not known; however, cell surface glycosaminoglycans (GAGs), and in particular heparan sulfates, have been shown to be important for infection. Both G and F are able to bind GAGs, with G contributing to the majority of virus binding to cell surface GAGs (3, 8, 9, 12, 13, 19, 24, 33).
Surprisingly, however, and in contrast to the case for virtually all members of the Paramyxovirinae subfamily, neither G nor SH is required for RSV infectivity, as suggested first by the isolation of an RSV mutant (cp52) lacking part of these genes (20). This was confirmed subsequently by a series of studies using recombinant HRSV and BRSV SH and/or G deletion mutants (5, 17, 19, 30, 32, 36). In addition, in cells transfected with the RSV F gene alone, formation of multinucleated syncytia is observed, although coexpression with G enhances fusion activity (reference 26 and unpublished observations). Vesicular stomatitis virus pseudotypes (18) and artificial RSV RNAs complemented with F alone (34) are infectious, but the presence of G greatly enhanced passage efficiency (34). Accordingly, the RSV F protein must combine activities in membrane fusion and cell attachment and is sufficient to mediate infection of cells with virus. Intriguingly, recombinant HRSV having F as the sole membrane protein was shown to retain considerable cell binding and fusion activity even in the absence or after blocking of GAGs, suggesting a GAG-independent mechanism of entry and the presence of specific cell surface receptors (33).
To more closely determine the contribution of RSV envelope proteins in host cell entry, we here exploited significant differences in the host tropisms of BRSV and HRSV in vitro. In striking contrast to the case for permanent cell lines of bovine or human origin, which are well infected by both HRSV and BRSV, we noticed a very pronounced species-specific infection of primary hematopoietic cells, such as peripheral blood lymphocytes (PBLs) and macrophages, as well as of differentiated respiratory epithelial cells. By employing recombinant and chimeric RSVs generated by reverse genetics techniques, we could dissect the functions of F and G in entry. The results identify the F2 subunit as the sole factor responsible for species-specific entry of RSV, whereas F1 and G do not contribute. The role of the RSV attachment protein G in receptor binding and entry appears to be limited to initially binding GAGs in a non-species-specific manner and thereby facilitating access of F2 to specific receptors. This is similar to the situation with many enveloped DNA viruses or positive-strand RNA viruses, in which GAGs serve as initial binding partners, followed by recruitment of specific entry receptors.
MATERIALS AND METHODS
Cells and virus.
Human or bovine peripheral blood mononuclear cells were isolated as described previously (30). Buffy coats of healthy adult donors or heparinized blood from cows was used for Ficoll (Lymphoflot; Biotest AG, Dreieich, Germany) gradient centrifugation at 1,500 rpm in a Heraeus Varifuge. PBLs were separated from monocytes by adherence to plastic for 1 h. Adherent macrophages were separated from B cells by overnight incubation and rigorous washing three times. Remaining adherent macrophages were trypsinized, and primary cultures of macrophages or PBLs were grown in suspension in RPMI (Gibco) supplemented with 10% fetal calf serum (FCS) at 37°C and 5% CO2. As determined by fluorescence-activated cell sorting (FACS) analysis with monoclonal CD3 and CD14 antibodies (Serotec), the respective primary cells were enriched to more than 90%. Prior to virus infection, PBLs and macrophages were activated by incubation for 16 h with phytohemagglutinin (PHA) (5 μg/ml) (Sigma) and lipopolysaccharide (10 μg/ml) (Sigma), respectively. BJAB, HL60, and Jurkat cell lines were maintained in RPMI (Gibco) with 10% FCS, and A549, HEp2, and MDBK cell lines were maintained in Dulbecco modified Eagle medium (DMEM) (Gibco) with 5% FCS. Differentiated human respiratory epithelial cells grown on membranes under ambient airflow (1) were a generous gift of R. Bals, Marburg, Germany.
HRSV strain Long and recombinant BRSVs were grown on MDBK cells as described previously (30). For preparation of virus stocks, 80% confluent MDBK cell layers were infected at a multiplicity of infection (MOI) of 0.1 in DMEM in the absence of FCS. The inoculum was removed after 1 h, and cells were incubated in DMEM supplemented with 2.5% FCS at 37°C in a 5% CO2 atmosphere. Upon development of extensive cytopathic effects, virus was released by freezing and thawing, followed by centrifugation at 3,500 rpm for 20 min at 4°C. Infectious virus titers were determined on Vero cells by end point dilution and counting of infected-cell foci stained for indirect immunofluorescence with RSV F-specific monoclonal antibody F56 (kindly provided by J. A. Melero) or RSV-F (Serotec).
Infection assays and immunostaining.
For infection of cell lines (A549, HEp2, MDBK, BJAB, Jurkat, and HL60) or primary, activated PBLs and macrophages, cells were suspended in virus-containing medium without FCS, incubated for 1 h, and collected by centrifugation. A549, HEp2, and MDBK cells were resuspended in DMEM with 2.5% FCS and seeded in 12-well plates at a concentration of 2.5 × 105 cells/ml. BJAB cells, Jurkat cells, HL60 cells, PBLs, and macrophages were seeded in six-well plates at a concentration of 106 cells/ml in RPMI with 2.5% FCS. At 2 days postinfection (dpi), virus foci were visualized in adherent cells after fixation in 80% acetone by indirect immunostaining with monoclonal antibody RSV-F. Infection of hematopoietic cell lines, primary T cells, and macrophages was monitored by FACS analysis of 2 × 105 cells after fixation with 3% paraformaldehyde at room temperature for 5 min. After washing with FACS buffer (phosphate-buffered saline containing 0.4% FCS and 0.02% NaN3), cell surface staining of viral envelope proteins for 30 min on ice was performed with RSV-F antibody and an isotype-specific antibody control provided by Serotec. After washing, cells were incubated for 30 min with fluorescein isothiocyanate-labeled anti-mouse antibody (Dianova, Hamburg, Germany), followed by washing and FACS analysis. For infection of human respiratory epithelial cells, membrane inlays with monolayers of approximately 107 cells were incubated with 1 ml of medium containing viruses resulting in an MOI of 0.1. Inocula were removed after 1 h, and cells were grown for 3 days in DMEM. Cells were fixed with 3% paraformaldehyde and stained with RSV-F (Serotec).
Construction of chimeric F genes.
Two different chimeric RSV F genes were assembled in the pTM1 plasmid (25) by using an overlapping PCR technique. For cloning of the chimeric F1b2h gene, the F2-coding region of the HRSV (strain Long) F gene corresponding to amino acids 1 to 136 of the F protein was amplified from the pTM1-hF plasmid (39) by using oligonucleotides (MWG Biotech, Ebersberg, Germany) hF-S (5′-TTTCCATGGAGTTGCCAATCCTCAAAGC) and hF2-AS (5′-TAGCAAGAATCCTAAAAATCTTCTTTTCCTTTTCTTGCTTAATGTTAC). In a separate PCR, the F1-coding region from the BRSV (strain ATue51908) F gene corresponding to amino acids 137 to 574 of the F protein was amplified from the pTM1-bF plasmid by using oligonucleotides bF1-S (5′-GCAAGAAAAGGAAAAGATTTTTAGGATTCTTGCTAGGTATTGG) and bF-AS (5′-TTCGGATCCTCATTTACTAAAGGAAAGATTGTTG). The two PCR products were gel purified with a Qiagen gel extraction kit, mixed at a molar ratio of 1:1, and heated for 2 min at 95°C for denaturation. The mixture was then incubated at 60°C for 30 s to allow the two fragments to anneal to each other. Hybridization was mediated by overlapping complementary sequences that were introduced into the PCR fragments by 5′ overhangs of the oligonucleotides hF2-AS and bF1-S. A complete double-stranded DNA hybrid was obtained after incubating the mixture with Pfu polymerase (Promega) at 72°C for 3 min. The chimeric gene was subsequently amplified by PCR with oligonucleotides hF-S and bF-AS, which contained NcoI and BamHI restriction sites (underlined above), respectively. Taking advantage of these restriction sites, the PCR product was ligated into the pTM1 vector to obtain the pTM1-F1h2b plasmid. The same overlapping PCR technique was used for the construction of pTM1-F1b2 h. The plasmid was generated by using two overlapping PCR fragments that were amplified from pTM1-bF and pTM1-hF by using oligonucleotides bF-S (5′-AATCCATGGCGACAACAACCATGAGGATGATC) and bF2-AS (5′-TAACAAAAAACCAAGAAATCTCCTTTTTCTCTTCTTGCCCATTAGCCC), or hF1-S (5′-CAAGAAGAGAAAAAGGAGATTTCTTGGTTTTTTGTTAGGTGTTGGATC) and hF-AS (5′-TTTCTCGAGTTTTATTCAGTTACTAAATGC), respectively. The hybrid gene was cloned into the pTM1 plasmid, taking advantage of the unique NcoI and XhoI restriction sites (underlined) carried by the oligonucleotides bF-S and hF-AS, respectively. The total open reading frames (ORFs) of all chimeric genes were sequenced to rule out any amplification errors.
Generation of recombinant BRSV.
All BRSV cDNA constructs were based on the cDNA clone pBRSV ΔO, which lacks the SH, G, and F genes (30) and is derived from full-length cDNA of strain Atue51908 (4). F and G genes were introduced into pBRSV ΔO as described previously. First, introduction of the F genes from BRSV and HRSV resulted in pBRSV ΔSHΔG/Fb and pBRSV ΔSHΔG/Fh, respectively. Then, the G genes of BRSV and HRSV were inserted, giving rise to pBRSV ΔSH/GbFb and pBRSV ΔSH/GhFh, respectively (30). For generation of viruses containing chimeric F genes (pBRSV ΔSH/F1h2b and pBRSV ΔSH/F1b2h, respectively), pBRSV ΔO was digested with XhoI and treated with Klenow polymerase to fill in the 5′ overhang, followed by insertion of the complete ORFs of chimeric fusion proteins F1b2h and F1h2b (see above), which had been excised from plasmids pTM-F1/2 and pTM-F3/4 with NcoI (1 nucleotide upstream of the start codon) and StuI (21 nucleotides downstream of stop codon) and treated with Klenow polymerase.
To generate recombinant BRSVs expressing BRSV G along with the chimeric F protein F1b2h, pTIT BRSV-G (4) was used as the template for generation of a cDNA fragment spanning a sequence from 3 nucleotides upstream of the BRSV G translation start codon to 11 nucleotides downstream of the BRSV F start codon by PCR with primers Gb-XhoI (5′-CACCCGCTCGAGACCATGTCCAACCATAC) and Fb-NcoI (5′-GGCTGTTGTCGCCATGG*TTATTT GCCCC), which also introduced an NcoI site by a C-to-G mutation at the nucleotide indicated by the asterisk. This PCR fragment, designated Gb-stop/start, was digested with XhoI and NcoI (restriction sites are underlined) and ligated to the F1b2h cDNA fragment generated by NcoI and XhoI digestion (the XhoI cleavage site is 12 nucleotides 3′ of the stop codon). Recleavage with XhoI and insertion into XhoI-digested pBRSV ΔO gave rise to pBRSVΔSH/Gb-F1b2h.
For generation of a recombinant BRSV expressing the HRSV G protein and the chimeric F protein F1h2b, the same strategy was used. A DNA fragment generated by PCR on pTIT HRSV-G (30) with primers GhXhoI (5′-GGGCCGCTCGAGAACATGTCCAAAAACAAG) (the start codon is in boldface) and GhstopClaI (5′-GCAGCCATCGATCTACTGGCGTGTTGTGTTG) (the stop codon is in boldface) was digested with XhoI and ClaI (restriction sites are underlined). To fuse the authentic BRSV G/F intergenic region to the HRSV G cDNA, a PCR was done with pBRSV ΔSH/GbFb by using primers GbstopClaI (5′-ACCTCCATCGATACCTCCATATAATATCAATT) and Fbstop (5′-GACTAACGACGCGTTTTTATATAACTATCAAC), amplifying DNA starting 6 nucleotides downstream of the BRSV G stop codon and ending with the transcription stop signal of BRSV F. After ClaI digestion, this PCR product was ligated to the ClaI site (underlined) of the HRSV G PCR fragment. After removal of the BRSV-F part by NcoI digestion, the remaining fragment was ligated to the ORF of F1h2b that was released from pTM-F3/4 by NcoI and XhoI digestion. The fragment containing the Gh-coding region, the BRSV G/F intergenic region, and the F1h2b ORF was inserted into XhoI-digested pBRSV ΔO, giving rise to BRSVΔSH/GhF1 h2b.
All recombinant BRSVs were recovered from cDNA as described before (4) in BSR T7/5 cells expressing T7 RNA polymerase and transfected with 10 μg of CaPO4-precipitated cDNA of the respective rBRSV full-length cDNA together with a set of four support plasmids (4 μg of pTit-N, 4 μg of pTit-P, 2 μg of pTit-L, and 2 μg of pTit-M2) expressing the BRSV N-, P-, L-, and M2 proteins from a T7 promoter. The transfection medium was replaced after 4 h with BHK-21 medium (Gibco) with 5% FCS. After 4 days, cells were split 1:3 and maintained in BHK-21 medium with 2.5% FCS until a cytopathic effect was detectable.
RESULTS
Similar permissivities of cell lines for HRSV and BRSV.
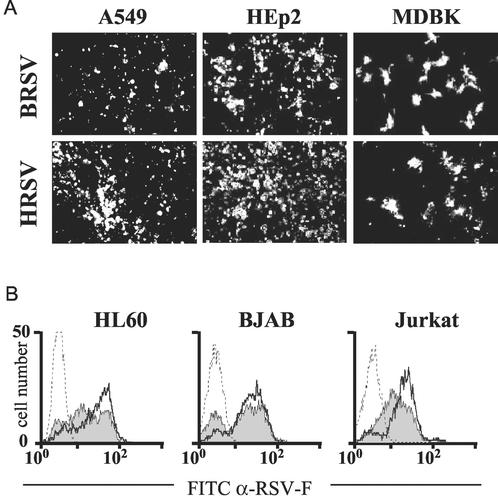
To assess the possibility of reproducing species-specific differences of HRSV and BRSV infection in vitro, permanent cell lines were first screened by infection with BRSV and HRSV at an MOI of 0.2. At 2 dpi, infected cells were identified by immunostaining with an antibody recognizing F protein. As indicated already by the appearance of extensive cytopathic effects, the human cell lines HEp-2 and A549 were well permissive for both HRSV and BRSV (Fig. 1A). A lower degree of infection was observed with the bovine MDBK cell line for both BRSV and HRSV. Since we previously noticed that cell lines of hematopoietic origin also are permissive for infection with HRSV or BRSV (30), human T-cell-derived Jurkat, B-cell-derived BJAB, and macrophage-derived HL60 cell lines were also included. As revealed by FACS analysis, all of these cell lines were equally permissive for both HRSV and BRSV. After 4 days of infection at an MOI of 0.1, more than 80% of the cells stained positive for RSV F protein (Fig. 1B).
FIG. 1.
Nonspecific infection of permanent cell lines by HRSV and BRSV. (A) Human cell lines A549 and HEp2 and the bovine cell line MDBK are permissive for both HRSV and BRSV. Infection was visualized by immunostaining of RSV F. (B) Infection or mock infection (dotted lines) of human hematopoietic cell lines BJAB, HL60, and Jurkat with BRSV (filled curves) or HRSV (heavy lines). Surface expression of RSV F was determined by FACS at 4 dpi. Results from a single representative experiment are shown. FITC, fluorescein isothiocyanate; α-RSV-F, anti-RSV-F.
Primary cells are infected in a species-specific manner.
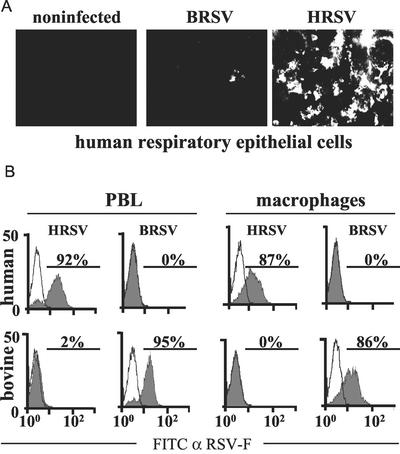
As the permanent hematopoietic or airway-derived cell lines were not suitable to analyze species-specific differences of HRSV and BRSV infection, we also sampled primary cells. Monolayers of differentiated primary human respiratory epithelial cells grown under ambient airflow (kindly provided by R. Bals) revealed highly specific infection of HRSV. After 3 days of infection at an MOI of 0.1 more than 50% of the cells were infected as revealed by immunostaining. In contrast, only a few small clusters of infected cells were observed in cultures incubated with BRSV, indicating that entry of BRSV is highly restricted (Fig. 2A). Furthermore, ongoing HRSV infection led to formation of large syncytia at 5 dpi, while this was never observed after BRSV infection (data not shown).
FIG. 2.
Species-specific infection of primary cells by HRSV and BRSV. (A) Monolayers of differentiated human respiratory epithelial cells were infected with the indicated viruses. Infection was visualized after 4 days by indirect immunostaining with an RSV F-specific antibody. (B) PBLs or macrophages of the indicated origin were infected with BRSV or HRSV as indicated (filled curves) or mock infected (open curves). The percentage of infected cells was determined by surface staining of RSV F with a monoclonal antibody and FACS after 6 days of infection. Results from one representative experiment of three are shown. FITC, fluorescein isothiocyanate; α-RSV-F, anti-RSV-F.
Interestingly, a similarly pronounced species-specific entry was observed with primary hematopoietic cells. Freshly isolated human and bovine T cells or macrophages were stimulated with PHA and lipopolysaccharide, respectively, and were infected with HRSV and BRSV at an MOI of 0.1. As revealed by immunostaining and FACS analysis at 6 dpi, more than 90% of human PBLs and more than 85% of human macrophages stained positive after incubation with HRSV, whereas BRSV infection was not detectable (Fig. 2B). In contrast, more than 90% of bovine PBLs and more than 85% of bovine macrophages revealed infection with BRSV, whereas only minor populations of up to 2% of cells stained positive after incubation with HRSV (Fig. 2B). This indicated that in PBLs and macrophages virus entry is highly restricted to the homotypic virus, although species-specific differences in replication were not yet excluded.
Species-specific infection is directed by the F protein.
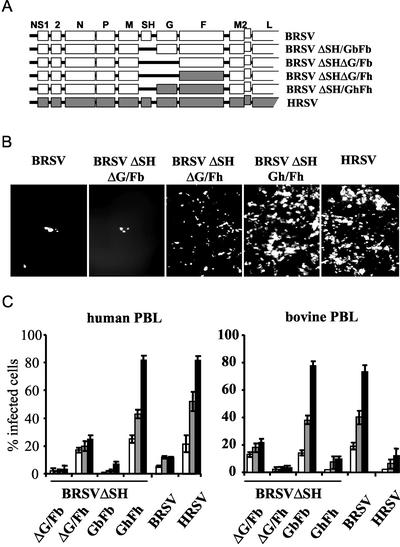
To verify that the observed species-specific permissivity of primary cells is determined by the viral surface glycoproteins, we made use of BRSV cDNA constructs lacking the SH gene and which allowed further deletion or swapping of individual F and G genes (Fig. 3A). First, a BRSV in which the G and F genes were exchanged with the HRSV counterparts, BRSV ΔSH/GhFh, was generated and used for infection of human respiratory epithelial cells at an MOI of 0.1. In striking contrast to BRSV, which infected only a few cells, the chimeric BRSV ΔSH/GhFh was able to infect cells as efficiently as HRSV and caused extensive viral spreading indistinguishable from that with HRSV (Fig. 3B). Thus, the origin of G and F, rather than a potential restriction of virus gene expression and replication in heterotypic host cells, is responsible for the observed phenotype. Corresponding results were obtained in stimulated PBLs and macrophages (see below).
FIG. 3.
Species-specific infection of primary cells is determined by the RSV F protein. (A) Organization of recombinant BRSV-derived viruses. BRSV-derived ORFs are shown by open boxes; ORFs from HRSV are shown by filled boxes. (B) Monolayers of differentiated human respiratory epithelial cells were infected with the indicated viruses and immunostained at 3 dpi for F expression. (C) PBLs of human or bovine origin were infected with recombinant viruses as indicated. The percentage of infected cells was determined by FACS analysis after 3 (white bars), 5 (grey bars), or 8 (filled bars) dpi. Experiments were performed with blood cells from four individual donors. Error bars indicate standard deviations.
As recombinant RSV lacking SH and G genes is infective, the F protein must be able to mediate attachment to the cell surface and entry into target cells. To determine the contribution of F to the observed species-specific infection of primary cells, we analyzed the behavior of recombinant BRSV expressing either BRSV F or HRSV F as the only surface protein (rBRSV ΔSHΔG/Fb, and rBRSV ΔSHΔG/Fh, respectively) (30). Compared to BRSV or BRSV lacking SH and G (rBRSV ΔSHΔG/Fb), the recombinant virus expressing only HRSV F infected human epithelial cells effectively (Fig. 3B). Thus, HRSV F is necessary and sufficient to mediate infection and, moreover, to determine the species specificity of infection.
Although it was able to specifically infect human airway cells, the virus expressing HRSV F as the sole surface protein was less effective in infection than HRSV. However, a recombinant BRSV expressing both HRSV F and G was indistinguishable from HRSV in the specificity and efficiency of infection (Fig. 3B, rBRSV ΔSH/GhFh). Also, extensive viral spread and syncytium formation indistinguishable from that with HRSV was observed with this recombinant (not shown), confirming an important contribution of G to the efficiency of F-mediated infection.
To more quantitatively assess the contribution of F and G to the specificity and efficiency of infection, freshly isolated human and bovine PBLs stimulated with PHA were infected with wild-type (wt) RSVs and the recombinant viruses at an MOI of 0.1. Infection was monitored by FACS analysis at 3, 5, and 8 dpi. Successful infection of human or bovine PBLs correlated with the origin of the F gene (Fig. 3C). wt HRSV and the recombinant BRSV carrying HRSV G and F (rBRSV ΔSH/GhFh) infected virtually equal numbers of cells at all time points, reaching approximately 80% of cells at 8 dpi. This applied also to infection of bovine PBLs with wt BRSV and BRSV ΔSH/GbFb. Moreover, in the absence of G, F-mediated infection was also species specific. Recombinant BRSV expressing HRSV F as the only surface protein (rBRSV ΔSHΔG/Fh) infected a significant population of human PBLs (20 to 30%), whereas it was not able to infect bovine PBLs (<5%). In contrast, virus expressing only BRSV F (rBRSV ΔSHΔG/Fb) infected only bovine PBLs effectively (20%), whereas it could not effectively enter human PBLs (<5%), as shown in Fig. 3C. Thus, F alone is able to determine the specificity of entry. As estimated from these experiments, the presence of homotypic (with respect to F) G protein enhances the efficiency of F-mediated entry by approximately fourfold. In addition, these results show that neither the specificity nor the efficiency of entry is affected in the absence of SH, supporting previous reports challenging a role of HRSV SH protein in virus entry (32).
F2 subunit is responsible for species-specific infection.
To further identify regions or domains of F important for species-specific entry, we generated chimeric F constructs in which the two F subunits, F1 and F2, were derived from different viruses. Both combinations, namely, (i) a protein in which subunit F1 was derived from BRSV and subunit F2 was derived from HRSV F (F1b2h) and (ii) the counterpart, F1h2b, were successfully expressed from cDNA. Expression levels and cell surface transport were comparable for both constructs (not shown). The cDNAs were then used to generate recombinant BRSV having the chimeric F1b2h or F1h2b protein as the only surface proteins (BRSV ΔSHΔG/F1b2h and BRSV ΔSHΔG/F1h2b) (Fig. 4A). Both viruses were readily recovered from cDNA in BSR T7/5 cells by using the standard reverse genetics protocol, suggesting reasonable functionality of the chimeric F proteins.
FIG. 4.
The F2 subunit of F determines species specificity of RSV infection. (A) Schematic representation of viruses expressing chimeric F proteins composed of F1 (amino acids 1 to 136) and F2 (amino acids 137 to 574) subunits from HRSV (black bars) and BRSV (white bars). (B) Human and bovine PBLs were infected as indicated, and the percentage of infected cells was determined by FACS after 3, 5, and 8 dpi (white, grey, and filled bars, respectively). Experiments were done with cells from three individual donors. Error bars indicate standard deviations.
To determine the effect of the two F subunits on the specificity of infection, PHA-stimulated human or bovine PBLs were infected at an MOI of 0.1 in parallel with SH and G deletion mutants carrying either the authentic HRSV or BRSV F gene or the chimeric F gene as indicated (Fig. 4A). After immunostaining at 3, 5, and 8 dpi, infected cells were identified by FACS analysis. In this setting, mutants expressing the authentic HRSV F protein or the chimeric protein F1b2h (with an HRSV-derived F2 domain) specifically infected human PBLs. In contrast, viruses carrying the authentic BRSV F or F1h2b led to infection of bovine PBLs but not of human PBLs (Fig. 4B). Thus, solely the N-terminal F2 unit of F contains a domain responsible for mediating species specificity. Compared to the authentic F proteins, the chimeras were approximately 50% less efficient in mediating infection, which might indicate minor paucities in protein functions. However, non-species-specific infection, i.e., infection by viruses with the heterotypic F2 subunit, was less than 5% in all cases.
G protein is not involved in the specificity of RSV entry.
To exclude a possible contribution of G protein to the specificity of RSV infection, we generated recombinant BRSV expressing G proteins along with the above-described chimeric F proteins. One virus was constructed to encode HRSV-derived G and F1h2b (rBRSV ΔSH/GhF1h2b). Thus, of all surface protein components, only the F2 subunit is of BRSV origin. Similarly, in the other virus, rBRSV ΔSH/GbF1b2h, all surface protein components except for the HRSV F2 subunit are derived from BRSV (Fig. 5A). Human or bovine PBLs were infected with these viruses in parallel with viruses carrying only the chimeric F proteins to directly measure the effect of G on infection. After immunostaining, infected cells were quantified by FACS analysis at 3, 5, and 8 dpi.
FIG. 5.
RSV G protein has no influence on species specificity. (A) Organization of recombinant BRSV expressing chimeric F genes and, in addition, HRSV (black bars) or BRSV (open bars) G genes. (B) PBLs of the indicated origin were infected as indicated, and the percentage of infected cells was determined by FACS analysis after 3 (open bars), 5 (grey bars), or 8 (filled bars) dpi. Individual experiments were performed with cells isolated from three individual donors. Error bars indicate standard deviations.
As before, the chimeric F proteins directed infection according to the origin of F2. The chimeric F1b2h in the absence of G led to infection of approximately 20% of human PBLs, whereas fewer than 5% of bovine PBLs were found to be infected (Fig. 5B). The additional presence of BRSV G led to markedly enhanced infection of human PBLs, to more than 60% at 8 dpi. However, it did not enhance infection of bovine PBLs (<5%). Similarly, the infectivity of F1h2b for bovine PBLs was increased by the presence of HRSV G by a factor of 3, whereas human PBL infection remained at background levels of <5%. Comparable values were obtained with human and bovine macrophages (not shown). Thus, in this setting, G was able to greatly support infection of heterotypic (with respect to G) target cells but not infection of homotypic cells. Accordingly, G merely has a role in enhancing the F2-directed infectivity but does not have any influence on the species specificity.
DISCUSSION
The previous finding that RSV F proteins can initiate virus infection in the absence of the attachment protein G (and SH) is in striking contrast to the situation with most Paramyxovirinae. There, the glycoproteins H and HN are essential not only as the receptor binding proteins but also as factors required for triggering conformational changes in the F protein that finally lead to fusion of viral and cell membranes (7). Similarly, F and H complexes rather than the single proteins are required for other functions of Paramyxovirinae surface proteins, such as surface contact-mediated inhibition of T-cell proliferation by Morbillivirus members. As we have demonstrated recently, RSV F displayed alone on a presenter cell surface is able to inhibit the proliferation of T cells by contact, again distinguishing RSV F from Paramyxovirinae F proteins. Interestingly, HRSV and BRSV F proteins showed clear species-specific activity in inhibiting T cells from bovine or human origin, respectively, suggesting species-specific binding partners for this function. In the present work we have addressed the species specificity of RSV entry into target cells and again found the F protein to be solely responsible. We further could demonstrate that the F2 subunit of F is the determinant of species specificity, and we provide evidence that the RSV attachment protein G supports entry in a non-species-specific way.
In contrast to the situation in vivo, HRSV and BRSV readily infect cell lines from different species, including human, bovine, or hamster cells. This applies not only to laboratory strains but also to clinical HRSV isolates (not shown). Human cell lines of hematopoietic origin were also found to be highly permissive for both viruses, suggesting the presence of common or unspecific receptor structures on the surfaces of these cells allowing for virus entry. We therefore first searched for conditions under which a more pronounced species-specific infection by HRSV and BRSV in vitro is possible. In addition to differentiated respiratory epithelial cells, primary hematopoietic cells were found to be very suitable, with the advantage of easy handling and quantification. In fact, hematopoietic cells are also natural targets in vivo, as shown by isolation of infected macrophages ex vivo. Species-specific differences in infection were most pronounced in PBLs at MOIs of between 0.1 and 1 with a very low background of heterotypic infection. With regard to the intrinsic much slower replication of BRSV compared to HRSV, as well as to the described differential activity of HRSV and BRSV in escaping host-specific interferon responses (2, 29), the availability of recombinant (BRSV) virus constructs with identical genetic backbones was crucial. In order to facilitate cloning, a BRSV construct lacking SH was used throughout the study. In fact, virtually no difference in entry and spread was observed between the SH deletion mutant and wt BRSV. After exchange of F and G, the BRSV-derived virus lacking SH was identical in all aspects with HRSV. This confirmed previous work suggesting that SH does not support RSV entry and spread in vitro (32).
The replacement of BRSV surface protein genes F and G with the HRSV counterparts allowed efficient entry and spread of the chimeric virus in human respiratory epithelia cells, human PBLs, and human macrophages. Further deletion of G from BRSV expressing either BRSV F or HRSV F markedly reduced entry efficiency and spread. This was also expected, as the presence of G has been shown to considerably increase the infectivity of RSVs (19, 32) and RSV-like particles (34) and the cell fusion activity of F (14, 26, 32). Mostly, this has been attributed to binding of G to cell surface GAGs. However, whereas the efficiency of entry was reduced, the species specificity of infection was retained in the absence of G, identifying F as the responsible factor.
The availability of in vitro cell culture systems allowing the discrimination of virus entry mediated by HRSV and BRSV F led us to further define the structures of F involved. As for all Paramyxoviridae fusion proteins, RSV F is synthesized as an inactive precursor (F0) which is cleaved by host proteases into two disulfide-linked subunits, the membrane-anchored F1 subunit and the N-terminal F2 subunit. As the amino acid sequences of the mature HRSV and BRSV F proteins are highly similar, with an identity of 83% (35), and, in particular, as the structural elements important for membrane fusion are conserved, we presumed that chimeric F proteins, in which F1 and F2 were derived from different viruses, would be functional in processing and membrane fusion. As first observed after transient expression in BSR T7/5 cells, the two chimeric proteins, composed of HRSV F1 and BRSV F2 and vice versa, were transported to the cell surface. Recovery of recombinant BRSVs carrying the chimeric F genes also was readily achieved, indicating successful cooperation of the heterologous F1 and F2 domains. Both viruses grew to titers which were similar to those observed for recombinant BRSVs expressing the genuine BRSV or the genuine HRSV F protein on the viral envelope.
Infection of PBLs revealed a clear correlation of permissivity with the origin of the F2 subunit. Although it did so somewhat less efficiently than virus expressing the authentic HRSV F, the chimeric virus having only the F2 subunit of HRSV F infected human PBLs but not bovine PBLs. Similarly, exchange of the HRSV F2 domain with that of BRSV F resulted in a switch of tropism from human to bovine PBLs. Notably, the background infection was not higher than with the heterotypic authentic F proteins, suggesting that the F1 domain does not contribute at all to the specificity of infection. These results strongly suggest the presence of a specific cell receptor binding site in the F2 subunit of HRSV and BRSV F protein.
RSV G is the nominal virus attachment protein and contributes considerably to RSV cell surface binding (19, 33), and it enhances the infectivity of RSV (14, 26, 32). In our experiments, the presence of G led to a three- to fourfold increase of infected PBLs after 8 dpi (Fig. 3C). It should be noted that, in contrast to the F proteins, the G proteins of HRSV and BRSV are highly dissimilar in sequence. Also, within the HRSV species, G is variable and is well known for its contribution to HRSV antigenic variation (23, 27). Differential binding of G to bovine and human cells, and hence a possible contribution to the specificity of infection, could therefore not be excluded. To assess this, the G proteins of HRSV and BRSV were expressed with the chimeric F proteins from recombinant virus, such that only the F2 subunit originated from one virus species and both F1 and G originated from the other. If HRSV G, for example, bound to human cells more efficiently than to bovine cells, a shift in the specificity of infection, or in other words, an increase of heterotypic (with respect to F2) infection, would be expected. Strikingly, G did augment the infectivity of recombinant viruses, again by a factor of approximately 3, but with a species specificity exclusively correlating to the origin of F2. As an increase of heterotypic (with respect to F2) infection was not observed by G “fitting” to the host cell, G is not able to contribute in a specific manner to the specificity of RSV entry.
The nature of specific RSV receptor structures bound by the F2 subunit is not yet known. So far, GAGs, and in particular heparin sulfate and chondroitin sulfate B (8, 9, 12, 13, 19, 22), have been identified as being important for initiating RSV infection. As suggested by experiments utilizing CHO cell lines defective in GAG synthesis, RSV G binds GAGs with high affinity, accounting for 50% of all virus binding to cells (33). Also the F proteins of HRSV and BRSV can bind to heparan sulfate (8, 19) accounting for approximately 25% of all virus binding. Most interestingly, however, the remaining 25% of the entire HRSV cell binding activity could be attributed to GAG-independent binding of HRSV F (33). Heparan sulfate proteoglycans on the cell surface can be used as initial attachment receptors by a wide range of viruses, such as herpesviruses, picornaviruses, alphaviruses, and flaviviruses (for a review see reference 31). It appears probable that, similar to the case for these viruses, cellular GAGs may serve as initial binding partners for RSV G, and maybe also for F, thereby facilitating access of the RSV F2 subunit to so-far-unknown specific receptors required for membrane fusion and virus entry.
It appears unlikely that the species specificity of RSV infection observed in our experiments is determined by cell surface GAGs binding to F2, although GAGs may provide specific configurations that may be used as ligands for virus attachment. Species-specific binding to GAGs was expected for the HRSV and BRSV G proteins, as these are highly dissimilar. However, in the experiments in which G proteins were expressed in combination with the chimeric F1/F2 proteins, this was not the case. Compared to viruses expressing solely the chimeric F1/F2 proteins, the enhancement of infection by HRSV and BRSV G was equal, suggesting equal binding of HRSV and BRSV G to GAGs. Although potential GAG binding sites have been identified in the F2 subunit, the similar behavior of G may indicate that GAGs do not contribute to the host tropism in our in vitro system. Similar to GAGs, however, the specific receptors responsible for F2-directed infection of cells must be expressed on a broad variety of cells, as indicated by the identical results obtained for respiratory epithelial cells, T cells, and macrophages. Notably, all of these cell types represent natural targets in vivo.
The finding that neither F1 nor the attachment protein G, but only the F2 subunit of the fusion protein, is responsible for the specificity of HRSV and BRSV entry underscores the unique position of the Pneumovirinae within the Paramyxoviridae family. Unlike the attachment proteins H and HN of the Paramyxoviridae, the RSV G is an accessory protein not involved in specific receptor binding. Specific receptor binding is achieved by the RSV F protein. This makes RSV F similar to the spike proteins of viruses which are required for attachment to specific receptors and membrane fusion, such as human immunodeficiency virus type 1 Env or influenza virus hemagglutinin. Also in these cases, the receptor binding sites map to the N-terminal subunits of the proteins, gp120 and HA1, respectively (15, 28). The identification of F2 as the binding partner of specific RSV entry receptors should facilitate the identification of such receptors and provide the basis for the development of tools specifically interfering with RSV infection.
Acknowledgments
This work was supported by the European Commission (EC 5th FP-RSV Vac QLK2-CT-1999-00443) and the Deutsche Forschungsgemeinschaft (SFB 455-A3 to K.C. and He/1168/11-3 to G.H.).
We thank S. Weber and S. Hake, Veterinary School Munich, for providing bovine blood; R. Bals for providing cultured human respiratory epithelial cells; and Stefan Finke and Birgit Bossert for critical reading of the manuscript.
REFERENCES
- 1.Bals, R., W. Xiao, N. Sang, D. J. Weiner, R. L. Meegalla, and J. M. Wilson. 1999. Transduction of well-differentiated airway epithelium by recombinant adeno-associated virus is limited by vector entry. J. Virol. 73:6085-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossert, B., and K. K. Conzelmann. 2002. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 76:4287-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourgeois, C., J. B. Bour, K. Lidholt, C. Gauthray, and P. Pothier. 1998. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J. Virol. 72:7221-7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins, P. L., K. McIntosh, and R. M. Chanock. 1996. Respiratory syncytial virus, p. 1313-1352. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnik, T. P. Monath, and S. E. Straus (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 7.Dutch, R. E., T. S. Jardetzky, and R. A. Lamb. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci. Rep. 20:597-612. [DOI] [PubMed] [Google Scholar]
- 8.Feldman, S. A., S. Audet, and J. A. Beeler. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glezen, P., and F. W. Denny. 1973. Epidemiology of acute lower respiratory disease in children. N. Engl. J. Med. 288:498-505. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Reyes, L., M. B. Ruiz-Arguello, B. Garcia-Barreno, L. Calder, J. A. Lopez, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 98:9859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallak, L. K., P. L. Collins, W. Knudson, and M. E. Peeples. 2000. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271:264-275. [DOI] [PubMed] [Google Scholar]
- 13.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heminway, B. R., Y. Yu, Y. Tanaka, K. G. Perrine, E. Gustafson, J. M. Bernstein, and M. S. Galinski. 1994. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology 200:801-805. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman, T. L., and R. W. Doms. 1999. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 16:57-65. [DOI] [PubMed] [Google Scholar]
- 16.Holberg, C. J., A. L. Wright, F. D. Martinez, C. G. Ray, L. M. Taussig, and M. D. Lebowitz. 1991. Risk factors for respiratory syncytial virus-associated lower respiratory illnesses in the first year of life. Am. J. Epidemiol. 133:1135-1151. [DOI] [PubMed] [Google Scholar]
- 17.Jin, H., H. Zhou, X. Cheng, R. Tang, M. Munoz, and N. Nguyen. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210-218. [DOI] [PubMed] [Google Scholar]
- 18.Kahn, J. S., M. J. Schnell, L. Buonocore, and J. K. Rose. 1999. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology 254:81-91. [DOI] [PubMed] [Google Scholar]
- 19.Karger, A., U. Schmidt, and U. J. Buchholz. 2001. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J. Gen. Virol. 82:631-640. [DOI] [PubMed] [Google Scholar]
- 20.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krilov, L. R. 2001. Respiratory syncytial virus: update on infection, treatment, and prevention. Curr. Infect. Dis. Rep. 3:242-246. [DOI] [PubMed] [Google Scholar]
- 22.Krusat, T., and H. J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 23.Martinez, I., J. Dopazo, and J. A. Melero. 1997. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J. Gen. Virol. 78:2419-2429. [DOI] [PubMed] [Google Scholar]
- 24.Martinez, I., and J. A. Melero. 2000. Binding of human respiratory syncytial virus to cells: implication of sulfated cell surface proteoglycans. J. Gen. Virol. 81:2715-2722. [DOI] [PubMed] [Google Scholar]
- 25.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 26.Pastey, M. K., and S. K. Samal. 1997. Analysis of bovine respiratory syncytial virus envelope glycoproteins in cell fusion. J. Gen. Virol. 78:1885-1889. [DOI] [PubMed] [Google Scholar]
- 27.Prozzi, D., K. Walravens, J. P. Langedijk, F. Daus, J. A. Kramps, and J. J. Letesson. 1997. Antigenic and molecular analyses of the variability of bovine respiratory syncytial virus G glycoprotein. J. Gen. Virol. 78:359-366. [DOI] [PubMed] [Google Scholar]
- 28.Rogers, G. N., J. C. Paulson, R. S. Daniels, J. J. Skehel, I. A. Wilson, and D. C. Wiley. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76-78. [DOI] [PubMed] [Google Scholar]
- 29.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlender, J., G. Walliser, J. Fricke, and K. K. Conzelmann. 2002. Respiratory syncytial virus fusion protein mediates inhibition of mitogen-induced T-cell proliferation by contact. J. Virol. 76:1163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider-Schaulies, J. 2000. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 81:1413-1429. [DOI] [PubMed] [Google Scholar]
- 32.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Techaarpornkul, S., P. L. Collins, and M. E. Peeples. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294:296-304. [DOI] [PubMed] [Google Scholar]
- 34.Teng, M. N., and P. L. Collins. 1998. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J. Virol. 72:5707-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walravens, K., R. Kettmann, A. Collard, P. Coppe, and A. Burny. 1990. Sequence comparison between the fusion protein of human and bovine respiratory syncytial viruses. J. Gen. Virol. 71:3009-3014. [DOI] [PubMed] [Google Scholar]
- 36.Whitehead, S. S., A. Bukreyev, M. N. Teng, C. Y. Firestone, M. St Claire, W. R. Elkins, P. L. Collins, and B. R. Murphy. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J. Biol. Chem. 276:31642-31650. [DOI] [PubMed] [Google Scholar]
- 38.Zimmer, G., K. K. Conzelmann, and G. Herrler. 2002. Cleavage at the furin consensus sequence RAR/KR(109) and presence of the intervening peptide of the respiratory syncytial virus fusion protein are dispensable for virus replication in cell culture. J. Virol. 76:9218-9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmer, G., I. Trotz, and G. Herrler. 2001. N-glycans of F protein differentially affect fusion activity of human respiratory syncytial virus. J. Virol. 75:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]