Abstract
Herpes simplex virus thymidine kinase is important for reactivation of virus from its latent state and is a target for the antiviral drug acyclovir. Most acyclovir-resistant isolates have mutations in the thymidine kinase gene; however, how these mutations confer clinically relevant resistance is unclear. Reactivation from explanted mouse ganglia was previously observed with a patient-derived drug-resistant isolate carrying a single guanine insertion within a run of guanines in the thymidine kinase gene. Despite this mutation, low levels of active enzyme were synthesized following an unusual ribosomal frameshift. Here we report that a virus, generated from a pretherapy isolate from the same patient, engineered to lack thymidine kinase activity, was competent for reactivation. This suggested that the clinical isolate contains alleles of other genes that permit reactivation in the absence of thymidine kinase. Therefore, to establish whether thymidine kinase synthesized via a ribosomal frameshift was sufficient for reactivation under conditions where reactivation requires this enzyme, we introduced the mutation into the well-characterized strain KOS. This mutant virus reactivated from latency, albeit less efficiently than KOS. Plaque autoradiography revealed three phenotypes of reactivating viruses: uniformly low thymidine kinase activity, mixed high and low activity, and uniformly high activity. We generated a recombinant thymidine kinase-null virus from a reactivating virus expressing uniformly low activity. This virus did not reactivate, confirming that mutations in other genes that would influence reactivation had not arisen. Therefore, in strains that require thymidine kinase for reactivation from latency, low levels of enzyme synthesized via a ribosomal frameshift can suffice.
Clinically relevant resistance to antiviral drugs requires both evasion of antiviral activity and retention of pathogenicity. Acyclovir (ACV) and related nucleoside analogs are the predominantly prescribed drugs, used prophylactically and therapeutically, for the treatment of herpes simplex virus (HSV) infections. Resistance to antiviral chemotherapy is rare in the immunocompetent, where infections are usually self-limiting (4, 20, 30). However treatment of immunocompromised patients, where HSV infection is a frequent cause of serious morbidity and even mortality, is often hindered by the emergence of ACV-resistant (ACVr) viruses, estimated to occur in ∼5% of those treated (4, 11). These clinical ACVr isolates most often contain mutations in the viral thymidine kinase gene (tk), which encodes the protein that activates the drug (details of drug action and resistance are discussed in reference 6). Several TK phenotypes of clinical ACVr isolates have been reported; these include altered substrate specificity, low but measurable activity (TKL), and lack of measurable activity (TK−) (13, 28). The last phenotype presents something of a paradox because, although TK is not required for propagation of laboratory strains in cell culture, it has been shown to be crucial for pathogenesis in mouse models of HSV infection, in particular, for reactivation from explanted sensory ganglia (8, 10).
The most common ACVr mutation found in clinical isolates is an insertion of a single guanine (G) into a run of 7 guanines (G string) (2, 13, 18, 24, 28-30). One such mutant, 615.9, was one of a series of isolates taken from a bone marrow transplant patient suffering from severe esophagitis, despite antiviral therapy (25). Despite the single-G insertion into the G string (G7+1G), which would be predicted to result in a truncated polypeptide without TK activity, 615.9 expressed low levels of active TK via a net +1 ribosomal frameshift (18). Moreover, 615.9 reactivated from explanted mouse ganglia, and plaque autoradiography of the reactivating virus revealed a uniformly TKL phenotype. This observation provoked the hypothesis that 615.9 made enough TK to cause disease but not enough to activate effective levels of ACV. The mechanism of the ribosomal frameshift on the G string is unusual, in that its efficiency correlates with the ability of the mRNA sequence G8AG to form inter- or intramolecular structures through non-Watson-Crick base pairing; furthermore, this sequence was functional in the absence of canonical simulators of frameshifting (17). (Ribosomal frameshifting has been reviewed previously [1, 12]).
An alternative explanation for reactivation of a virus with a G7+1G mutation was suggested following the observation that a virus with this mutation could reactivate from mouse ganglia as a mixed population of TK+ (high TK activity) and TK− viruses (27, 28). It was suggested that the increased error rate during replication of long iterated sequences resulted in the emergence of a TK+ population that could complement the reactivation defect of the TK− virus. A third explanation for reactivation of G7+1G mutants stems from our report describing a clinical isolate that could reactivate from latency in mouse ganglia in the absence of any TK activity (16). This observation suggests that allelic variation within the viral genome may sometimes reduce the importance of TK for pathogenesis.
Herein we report studies on the contribution of the low levels of TK synthesized via frameshifting on the G7+1G sequence to reactivation in a mouse model of latency. We first asked if reactivation of the clinical isolate from which mutant 615.9 was derived required TK activity. It did not. We therefore engineered the G7+1G mutation into the laboratory strain KOS, which requires TK for reactivation; this permitted comparisons to other mutant viruses based on KOS. This virus was tested for its ability to replicate in and reactivate from mouse trigeminal ganglia following corneal inoculation of mice. This virus reactivated, indicating that the low levels of TK synthesized via frameshifting were sufficient to support reactivation from latency.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) and TK− human osteosarcoma (143B) cells (American Type Culture Collection, Manassas, Va.) were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum at 37°C and 5% CO2. The viruses used in this study were wild-type HSV-1 strain KOS and KOS mutant tkLTRZ1 (9), together with clinical HSV-1 strains 294.1 and 615.9 (25) (Fig. 1). Viruses 294 and 615 were derived from the same bone marrow transplant patient before and after the commencement of antiviral therapy and have been confirmed to be genetically related by comparison of restriction endonuclease digestion patterns (25). Virus 294.1 is a plaque isolate of 294 that was isolated prior to the onset of antiviral therapy and is ACV sensitive (ACVs). Virus 615.9 is an ACVr plaque isolate of 615 that was isolated 28 days after 294, during treatment with ACV. 615.9 carries the G7+1G mutation (18).
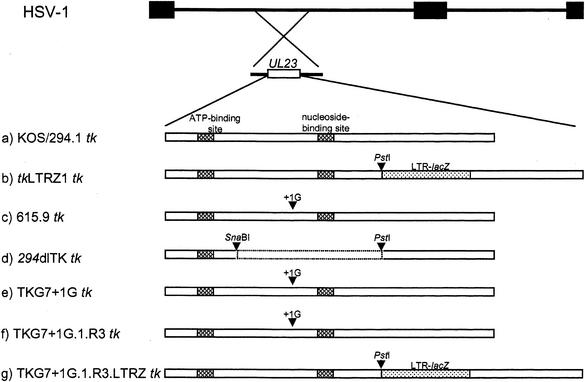
FIG. 1.
Structure of the tk genes of viruses used in this study. The top two lines represent the HSV genome and the location of the tk gene (UL23). Below are schematic diagrams of the tk genes of the viruses used in this study. a, KOS and 294.1 tk (cross-hatched boxes, functional domains of TK); b, tkLTRZ1 (tk with LTR-lacZ inserted into the PstI site [dotted box]); c, 615.9 (a clinical isolate with the G7-to-G7+1G mutation [arrow, site of the mutation]); d, 294dlTK (tk with a deletion between the SnaBI and PstI sites [dotted lines], generated from 294.1); e, TKG7+1G (tk with a single G inserted into the G string of KOS tk); f, TKG7+1G.1.R3 (plaque isolate of reactivated TKG7+1G that reactivated as a uniformly TKL population); g, TKG7+1G.1.R3.LTRZ (tk with LTR-lacZ inserted into the PstI site replacing the tk gene of TKG7+1G.1.R3).
Plasmids.
p294ΔTK was made by digesting pBH15 (which contains the tk gene from virus 294.1 [18]) with SnaBI and PstI, blunt ending with T4 DNA polymerase, and then recircularizing. Plasmid pAG5 (14) contains the BamHI P fragment of HSV-1 KOS cloned into pBluescript SK(+) (Promega, Madison, Wis.) such that tk is in the same orientation as the T7 promoter. Plasmid pAG6.TKG7+1G was made by introducing the G7+1G mutation into pAG5 with the QuikChange kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions by using two complementary oligonucleotides (Integrated DNA Technologies, Coralville, Iowa); however only the forward primer is listed here: CTGGCTCCTCATATCGGGGGGGGAGGCTGGGAGCTC. The presence of the expected mutations in both p294ΔTK and pAG6.TKG7+1G was confirmed by sequencing (the entire BamHI P fragment of pAG6.TKG7+1G was sequenced). Plasmid ptkLTRZ1 contains an insertion into tk of lacZ downstream of the Moloney murine leukemia virus long terminal repeat (LTR) (9).
Construction of recombinant viruses.
Virus 294dlTK (Fig. 1) was generated following cotransfection of p294ΔTK with infectious DNA derived from 294.1, as previously described (16). This procedure involved one round of purification under 120 μM ACV, followed by three rounds of plaque purification in the absence of the drug. Two independent isolates were generated from separate transfections, and the presence of the mutations was confirmed by sequencing and Southern blot hybridization.
Virus TKG7+1G (Fig. 1) was made following cotransfection of pTKG7+1G and infectious DNA from mutant virus tkLTRZ1, which is TK− due to an insertion of lacZ driven by a strong promoter into tk, as described previously (14). Briefly, recombinant viruses were isolated following limiting dilution in 96-well dishes; wells were screened for a single white plaque following staining for β-galactosidase activity with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and counterstaining with neutral red, as described previously (15). Two independent isolates were generated from separate transfections, and the presence of the mutation was confirmed by sequencing. Virus TKG7+1G.1.R3.LTRZ (Fig. 1) was generated in the same manner, except that infectious DNA from TKG7+1G.1.R3 was cotransfected with ptkLTRZ1 DNA, and the wells were screened for blue plaques.
Plaque autoradiography.
Plaque autoradiography was performed as described previously (14). Briefly, ∼120 PFU of each virus were inoculated onto a 60-mm-diameter dish that had been seeded 24 h earlier with 8 × 105 143B cells and then overlaid with medium containing 0.75% carboxymethylcellulose and 2% newborn calf serum. Cells were incubated at 37°C, and, once plaques had reached a reasonable size, the overlay was replaced with an overlay containing 2.6 μCi of [3H]thymidine ([3H]methyl; 64.5 Ci/mmol; Moravek, Brea, Calif.)/ml in medium for 14 h. The cells were then stained with 2% crystal violet in 10% ethanol, washed, and air dried. The base of each dish was cut away from the rim and exposed to a tritium phosphor storage screen (Fuji Medical Systems, Stamford, Conn.), which was then scanned with a phosphorimager (Bio-Rad, Hercules, Calif.).
Assays of acute and latent infections in mice.
Male 8-week-old randomly bred CD-1 (Charles River Laboratories, Indianapolis, Ind.) or CD-1-derived Hsd:ICR (Harlan Sprague-Dawley, Kingston, N.Y.) mice (similar results were obtained with both suppliers) were infected via the cornea as described previously (7, 22). Acute viral replication was monitored by assaying virus in the tear film at 1, 2, and 3 days postinfection (p.i.) and in excised and homogenized trigeminal ganglia at day 3 p.i. (23). Reactivation from latency was measured by enzymatically dissociating ganglia excised at 30 days p.i. and culturing on Vero cells. Cells were screened for 10 days for cytopathic effect. If none was observed, the cultures were replated and then screened for a further 4 days (22).
RESULTS
Reactivation of 294dlTK from latency.
It was previously reported that virus reactivated from approximately 25% of trigeminal ganglia latently infected with the clinical isolate 615.9 and that the viruses that reactivated had a TKL phenotype (18). However, a loss of dependence on TK for reactivation has been shown for a different clinical isolate (16). The latter observation called into question the importance of the TKL phenotype in the reactivation of 615.9. To address this, we introduced a deletion into tk of 294.1, an ACVs virus that was isolated from the same patient as 615.9 prior to the onset of antiviral therapy (25). Two independently isolated viral mutants were generated, 294dlTKA and 294dlTKB, and their ability to reactivate from mouse latency was tested following inoculation onto scarified cornea with either 2 × 106 or 2 × 107 PFU of virus. 294dlTK replicated efficiently on the eye during the first 24 h; however eye titers were reduced relative to those for 294.1 at day 3 p.i. (Table 1). It is interesting that at 3 days p.i. we detected lower titers of 615.9 than of 294dlTK in the eye, although titers in the ganglia were similar (Table 1). Although laboratory strains that lack TK activity do not reactivate from latency in mouse ganglia (3, 8, 10, 14) (see below), 294dlTKA and 294dlTKB both reactivated, even at the lower inoculum of 2 × 106 PFU (Table 1). These data indicate that this clinical isolate does not have an absolute requirement for TK for reactivation.
TABLE 1.
Acute replication and reactivation from latency of 294.1-derived viruses
| Virus | Inoculum (PFU) | Log mean titer ± SEa at:
|
Ganglia with reactivating virus/total ganglia | ||
|---|---|---|---|---|---|
| Day 1 p.i. in eye | Day 3 p.i. in eye | Day 3 p.i. in TGb | |||
| 294.1 | 2 × 105 | 5.4 ± 0.2 (3) | 4.6 ± 0.5 (3) | 4.3 ± 0.4 (4) | NDc |
| 615.9 | 2 × 106 | 5.1 ± 0.1 (3) | 1.1 ± 0.6 (3) | 0.9 ± 0.3 (4) | 5/10 |
| 294dlTKA | 2 × 106 | 6.1 ± 0.2 (3) | 3.2 ± 0.7 (3) | 0.1 ± 0.1 (4) | 5/10 |
| 294dlTKA | 7 × 107 | 6.1 ± 0 (3) | 2.2 ± 1.1 (3) | 0.2 ± 0.3 (4) | 7/10 |
| 294dlTKB | 2 × 106 | 6.5 ± 0.1 (3) | 3.4 ± 0.2 (3) | 0.8 ± 0.1 (4) | 6/10 |
| 294dlTKB | 7 × 107 | 6.3 ± 0.1 (3) | 3.3 ± 0.3 (3) | 1.2 ± 0.4 (4) | 7/8 |
The number of samples titrated for each group is shown in parentheses. Values were calculated by averaging the logs of the titers. When no plaques were detected, titers were assigned a value of 1.
TG, trigeminal ganglia.
ND, not done. Since a corneal inoculation of 2 × 105 PFU of 294.1 was 100% fatal to mice, KOS was used as a positive control for reactivation (virus in six of six ganglia reactivated).
Construction of recombinant virus TKG7+1G.
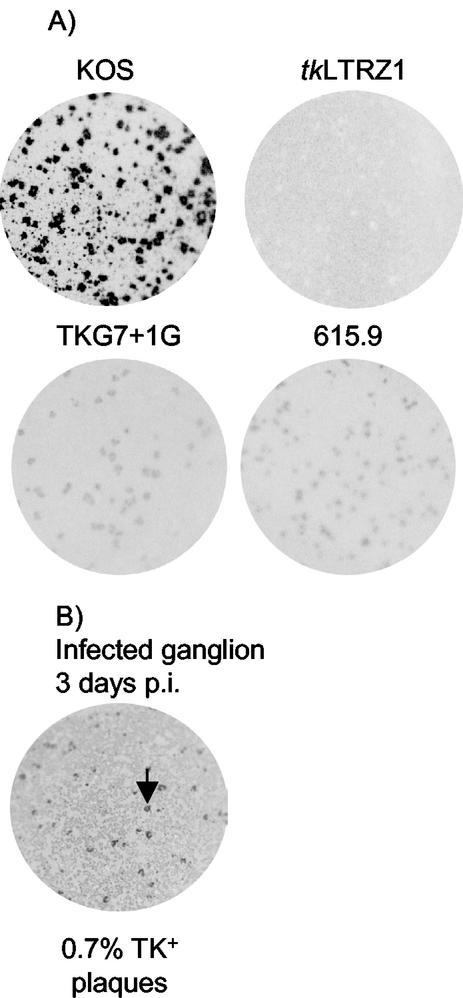
The reactivation of virus 294.1 (a genetically related pretherapy counterpart of 615.9) from latency in the absence of functional TK makes the contribution of the TKL phenotype to reactivation of 615.9 unclear. Therefore to ask whether the TKL phenotype due to ribosomal frameshifting could support reactivation, we chose to engineer the G insertion into the tk gene of the laboratory strain KOS. We considered KOS appropriate because it has been shown to be absolutely dependent on TK for reactivation from latency (3, 8, 19). To enable blue/white screening, thereby avoiding the use of ACV, we used a recombinant virus made from KOS, tkLTRZ1 (Fig. 1), which is TK− due to an insertion into tk of lacZ driven by the Moloney murine leukemia virus LTR (9). tkLTRZ1 does not reactivate from ganglia in the mouse model of latency (3, 19). Crucially for this study, using a TK-negative mutant as a starting point for isolation of recombinants eliminated a source of contaminating TK+ virus. TKG7+1G was generated by screening for white recombinants (which lacked the LTR-lacZ insertion; Fig. 1) and was confirmed to have a TKL phenotype qualitatively similar to that of 615.9 by plaque autoradiography (Fig. 2A). TK activity was also measured by quantitative plaque autoradiography and was shown to be ∼0.5% of KOS TK activity (data not shown).
FIG. 2.
Plaque autoradiography. (A) Images of plates infected with the indicated viruses and labeled with [3H]thymidine to show TK activity associated with each plaque. (B) Image of plate showing plaque autoradiography of a virus amplified from a ganglion excised at 3 days p.i. Approximately 0.7% of the virus had a TK+ phenotype. Arrow, example of a plaque with the TK+ phenotype.
Acute replication of recombinant viruses in mice.
To assess the ability of TKG7+1G to replicate in mice, 7 × 107 PFU of mutant viruses or 2 × 106 PFU of KOS were inoculated onto scarified mouse cornea. The tear films of infected mice were sampled at 1, 2, and 3 days p.i., and infected ganglia were harvested at 3 days p.i. TKG7+1G.1 and TKG7+1G.2 exhibited declines in titer over the first 3 days of infection similar to that exhibited by tkLTRZ1 (Table 2); however, greater levels of replication were observed in ganglia infected with TKG7+1G.1 and TKG7+1G.2 than in those infected with tkLTRZ1. Virus isolated from the homogenized ganglia of a mouse infected with TKG7+1G.1 was amplified on Vero cells, which would not offer any selective advantage for TK+ viruses. Plaque autoradiography revealed that this virus contained ∼0.7% TK+ virus among the majority TKL virus (Fig. 2B). However, despite the examination of more than 500 plaques from each of the TKG7+1G stocks used to inoculate the mice, a TK+ plaque was not observed (<0.1%). It is not possible to say whether the TK+ virus occurred as a reversion during acute replication or was always present as an “ultralow” population within the inoculum; either way, a TK+ virus is expected to have a strong selective advantage over TKL virus in ganglia.
TABLE 2.
Acute replication and reactivation from latency of engineered KOS mutants
| Virus | Inoculum (PFU) | Log mean titer ± SEa at:
|
Ganglia with reactivating virus/total ganglia | |||
|---|---|---|---|---|---|---|
| Day 1 p.i. in eye | Day 2 p.i. in eye | Day 3 p.i. in eye | Day 3 p.i. in TGb | |||
| KOS | 2 × 106 | 5.1 ± 0.5 (4) | 5.2 ± 0.2 (4) | 4.2 ± 0.4 (4) | 4.4 ± 0.4 (2) | 30/30 |
| tkLTRZ1 | 7 × 107 | 5.9 ± 0.4 (4) | 4.2 ± 0.2 (4) | 2.3 ± 0.1 (4) | 0.5 ± 0.5 (2) | 0/34 |
| TKG7 + 1G.1 | 7 × 107 | 5.5 ± 0.3 (2) | 4.3 ± 0.2 (2) | 2.6 ± 0 (2) | 1.6 ± 0.5 (2) | 9/20 |
| TKG7 + 1G.2 | 7 × 107 | 5.5 ± 0.1 (3) | 4.3 ± 0.1 (4) | 2.8 ± 0.1 (4) | 1.8 ± 1.8 (2) | 2/15 |
| TKG7 + 1G.1.R3 | 7 × 107 | NDc | ND | ND | ND | 3/14 |
| TKG7 + 1G.1.R3.LTRZ | 7 × 107 | 5.7 ± 0.2 (2) | 5.1 ± 0.1 (2) | 2.8 ± 0.1 (2) | 0 (1) | 0/16 |
The number of samples titrated for each group is shown in parentheses. Values were calculated by averaging the logs of the titers. When no plaques were detected titers were assigned a value of 1.
TG, trigeminal ganglia. Each sample contains the pooled ganglia from one mouse.
ND, not done.
Reactivation of TKG7+1G from latency.
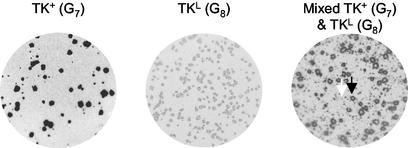
To establish whether TKG7+1G was able to reactivate from latency despite the frameshift mutation within tk, ganglia were harvested from latently infected mice, enzymatically dissociated, and plated onto Vero cells that were monitored for the appearance of plaques. Nine of 20 ganglia latently infected with TKG7+1G.1 and 2 of 15 infected with TKG7+1G.2 yielded virus, compared to all of those infected with KOS and none of those infected with tkLTRZ1 (Table 2). Four of the samples with reactivating virus were amplified on Vero cells and analyzed by plaque autoradiography (Fig. 3). Two of the four appeared to be mixed TK+ and TKL, and one appeared to be uniformly TK+. However the fourth sample appeared to be uniformly TKL. The tk genes from the uniformly TK+ and uniformly TKL stocks were sequenced and were shown to contain G7 and G7+1G sequences, respectively.
FIG. 3.
Plaque autoradiography of viruses isolated following reactivation of TKG7+1G. In the image of the mixed population, one example of a TK+ plaque and one of a TKL plaque are shown (black and white arrows, respectively).
Cloning and study of reactivated TKL virus.
We considered two possible explanations for reactivation of a uniformly TKL population following infection with TKG7+1G. First, low levels of TK were sufficient to support reactivation of this virus. Alternatively, it was possible that the reactivated virus had acquired a second mutation that rendered the mutant able to reactivate independently of TK activity. To address this, we removed the capacity of the reactivated virus to synthesize active TK. To this end, we recombined the LTR-lacZ (from plasmid ptkLTRZ1, the same plasmid cotransfected with KOS to make tkLTRZ1 [9]) into a virus that was cloned by limiting dilution from the reactivated TKL stock (TKG7+1G.1.R3), giving TKG7+1G.1.R3.LTRZ (Fig. 1). The viruses (7 × 107 PFU each) were inoculated onto scarified mouse cornea, and trigeminal ganglia were harvested 30 days p.i. and plated onto Vero cells, which were monitored for the appearance of cytopathic effect for 14 days (Table 2). Virus from approximately 20% (3 of 14) of the ganglia from mice that were infected with TKG7+1G.1.R3 reactivated, a frequency similar to that observed with TKG7+1G. However, TKG7+1G.1.R3.LTRZ did not reactivate from latency (0 of 16 ganglia). We take this to indicate that this virus had not acquired any mutations that could confer the ability to reactivate in the absence of TK. We therefore conclude that the low level of TK, synthesized as the result of a ribosomal frameshift, was sufficient to support reactivation in explanted mouse ganglia.
DISCUSSION
We are interested in mechanisms that permit HSV to evade antiviral chemotherapy yet retain the ability to be pathogenic. We have previously reported on pathogenic ACVr isolate 615.9, which synthesizes low levels of active TK (TKL) via frameshifting despite a G7+1G mutation in tk (18). Herein we demonstrate that a genetically related pretherapy isolate from the same patient, engineered to lack TK activity (294dlTK), can reactivate from explanted mouse ganglia. Therefore to investigate whether the TKL phenotype was sufficient to permit reactivation from latency, we engineered the G7+1G mutation into the laboratory strain KOS (TKG7+1G). This virus reactivated, and we observed several phenotypes of reactivating viruses, including a uniformly TKL population. This observation is consistent with the idea that the ∼1% of wild-type TK activity synthesized despite the G7+1G mutation is sufficient for reactivation. We discuss below the reactivation of 294dlTK, the phenotypes of TKG7+1G, and how different mechanisms may compensate for loss of TK in the pathogenesis of HSV.
Reactivation of a clinical isolate independently of TK activity.
The introduction of a large deletion into the tk gene of 294.1 (294dlTK) did not abolish the ability of this virus to reactivate from latency (Table 1). In contrast, laboratory strains that have been engineered to lack TK activity do not reactivate from latency (8, 10). However, this laboratory has previously reported that an ACVr clinical isolate (GGdlTK) reactivated from latency despite the same engineered mutation in tk (16). It was suggested that there were alleles present in GGdlTK, but not in KOS, that could overcome the requirement for TK for reactivation; this also most readily explains the reactivation of 294dlTK. It is possible that these alleles result in the increased activity of other viral nucleotide metabolism enzymes (e.g., dUTPase and ribonucleotide reductase). This would be consistent with the observation that supraphysiological concentrations of thymidine can overcome the lack of viral TK activity (31). Experiments to test this hypothesis are under way.
GGdlTK reactivated from approximately 30% of infected ganglia following inoculation with 7 × 107 PFU (16), and no reactivation was observed following an inoculum of 2 × 106 PFU (S.-H. Chen and D. M. Coen, unpublished observations). However, 615.9 and 294dlTK both reactivated (with efficiencies of ∼50%) following inoculation with 2 × 106 PFU (Table 1). These data suggest that the alleles that may be supporting reactivation of 294dlTK do so more effectively than those supporting reactivation of GGdlTK. There is an interesting difference between the clinical histories of the GG and 294.1 isolates. The GG virus was isolated from a bone marrow transplant patient following a prophylactic course of intravenous ACV, and thus its TK independence could have been selected during prophylaxis (16). In contrast, 294.1 was isolated prior to any reported ACV therapy; indeed, the patient was not given ACV prophylaxis because she was HSV seronegative (25). Although we cannot completely discount the possibility that this patient had undergone ACV therapy that was not reported, our observations raise the interesting possibility that there are viruses circulating within the population that do not absolutely require TK activity for pathogenesis. Presumably, given that the simplest route to ACVr is removal of TK activity, such viruses would be significantly more disposed to clinically significant ACV resistance. There have been reports of ACVr viruses isolated from patients in which TK activity was not detected. However, as has been reviewed, assays to detect TK activity can be insufficiently sensitive, leaving the possibility that these viruses possessed sufficient TK activity to permit reactivation but not enough to be detected (5). We believe that the best way to confirm the ability of a clinical isolate to reactivate in the absence of any TK activity is to generate a recombinant virus that has been engineered to unambiguously lack TK activity.
G7+1G mutation associated with TKL phenotype in strain KOS.
We engineered the G7+1G mutation into strain KOS to give virus TKG7+1G and observed that the mutation conferred a TKL phenotype (Fig. 2). Quantitative plaque autoradiography demonstrated that TKG7+1G had TK activity roughly similar to that of 615.9 (∼0.5 and ∼1% of wild-type activity, respectively [data not shown]). Therefore, the G7+1G genotype has now been associated with the TKL phenotype in two HSV-1 strains. However, several reports have described HSV-1 and HSV-2 strains with this mutation as TK− (2, 13, 25, 26, 28). We believe that the reason for the description of viruses with this mutation as TK− is most likely that the level of activity is below the sensitivity of the particular assays used in the cited reports. Indeed, the TK activity of TKG7+1G, which was lower than that of 615.9, may have been undetectable in plaque autoradiography assays previously utilized by this laboratory (3, 16). The improved assay is sensitive to as little as 0.5% of wild-type TK activity (14). Moreover, TK activity of viruses carrying the G7+1G mutation is consistent with the finding that the sequence G8AG in an mRNA is sufficient to cause ∼1% ribosomal frameshifting in vitro (17).
Reversion of TKG7+1G to TK+ observed during acute replication in the mouse.
Despite the inefficient replication in the eyes of TKG7+1G-infected mice by day 3 p.i., viral titers above levels typically associated with TK− mutants were detected in infected ganglia (Table 2). Virus from a TKG7+1G-infected ganglion was analyzed by plaque autoradiography, and ∼0.7% was shown to be TK+ (Fig. 2B). This was at least a sevenfold-higher frequency than was observed for the TKG7+1G or 615.9 stock (data not shown). The appearance of a TK+ population following infection of mice with an HSV-2 mutant that had the G7+1G mutation in tk has previously been reported (27). These authors suggested that the TK+ population was related to the inability of the HSV DNA polymerase to accurately replicate the extended homopolymeric sequence; indeed there have been several reports demonstrating a relationship between the length of a homopolymeric sequence and DNA replication fidelity (reviewed in reference 21). The data presented in this paper provide further evidence that replication of the extended G string is error prone and can result in a TK+ population that is selected for in mouse ganglia. Regardless, our results indicate that such reversion of a TKL mutant is not required for reactivation from latency.
TKG7+1G reactivated from latency.
It had previously been shown that a virus with only ∼5% of wild-type TK activity efficiently reactivated from mouse latency (3). That a uniformly TKL reactivation was observed implies that only ∼0.5% of wild-type TK activity is required to support reactivation from latency, albeit with reduced efficiency. This is further evidence that HSV-1 expresses much more TK activity than is required for ganglionic functions, at least in the mouse.
It is interesting to consider the source of the TK+ viruses present following reactivation from the other ganglia. One possibility is that they were a consequence of TK+ virus that appeared during acute replication (Fig. 2B). However, it is also possible that the mixed population occurred as a consequence of a reversion to TK+ that occurred during reactivation of a hitherto uniformly TKL population. Indeed, the observation of mixed-phenotype reactivations following a laboratory infection may mimic what is sometimes seen in the clinic, where isolates have been shown to have heterogeneous TK phenotypes (16, 25, 28).
Contribution of the different mechanisms of ACVr to pathogenesis.
The results that we report in this paper illustrate three different mechanisms that can be exploited by HSV to compensate for a mutation that would otherwise inactivate TK and prevent reactivation from latency: alleles in loci other than tk, ribosomal frameshifting to express low levels of TK, and replication errors that create subpopulations of TK+ virus. As the tk mutation that is compensated is the most common one found in ACVr clinical isolates, we suggest that these mechanisms may allow HSV to evade ACV therapy yet cause human disease. It also seems quite possible that these mechanisms could act in concert to increase the pathogenicity of ACVr isolates. Although we did not see any evidence for increased reactivation or acute ganglionic replication of 615.9, which exhibits frameshifting, compared with the genetically related deletion mutant 294dlTK, which exhibits only compensating alleles (Table 1), this may be due to the relatively strong compensating alleles that permit reactivation in the absence of TK. It will be interesting to see if a GG strain G7+1G mutant is more pathogenic than GGdlTK, which reactivates less efficiently than does 294dlTK.
Is rescue of functional TK by frameshifting an example of a more general mechanism?
It is tempting to speculate that HSV has evolved mechanisms that exploit the machinery involved in translation and replication to compensate for mutations in tk. Indeed, it is possible that they may represent mechanisms that are ubiquitous in biology to permit some (rather than zero) synthesis of a protein, despite a mutation that would otherwise result in loss of function. We are unaware of any other ribosomal frameshift events that have been shown to rescue a mutant's function; however this could be because most studies of translational recoding have concentrated on programmed frameshifting. As the frameshift on the G string is relatively inefficient, it may be that many other examples simply go unnoticed. There has been a report of an inefficient “out-of-frame” net +1 recoding event in lacZ that shifted ribosomes back into the wild-type reading frame (32); could this be an example of a bacterial translational compensation mechanism? Investigations of other mutations that confer clinically relevant ACVr on HSV may reveal other interesting mechanisms that permit these viruses to evade chemotherapy and retain pathogenicity.
Acknowledgments
We thank Jean Pesola and Angela Pearson for assistance with the animal experiments.
This work was supported by grants PO1 NS35138, RO1 AI26126, and T32 AI07245 from the National Institutes of Health.
REFERENCES
- 1.Baranov, P. V., R. F. Gesteland, and J. F. Atkins. 2002. Recoding: translational bifurcations in gene expression. Gene 286:187-201. [DOI] [PubMed] [Google Scholar]
- 2.Chatis, P. A., and C. S. Crumpacker. 1991. Analysis of the thymidine kinase gene from clinically isolated acyclovir-resistant herpes simplex viruses. Virology 180:793-797. [DOI] [PubMed] [Google Scholar]
- 3.Chen, S. H., W. J. Cook, K. L. Grove, and D. M. Coen. 1998. Human thymidine kinase can functionally replace herpes simplex virus type 1 thymidine kinase for viral replication in mouse sensory ganglia and reactivation from latency upon explant. J. Virol. 72:6710-6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christophers, J., J. Clayton, J. Craske, R. Ward, P. Collins, M. Trowbridge, and G. Darby. 1998. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob. Agents Chemother. 42:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coen, D. M. 1994. Acyclovir-resistant, pathogenic herpesviruses. Trends Microbiol. 2:481-485. [DOI] [PubMed] [Google Scholar]
- 6.Coen, D. M. 1996. Herpes simplex virus and varicella zoster virus: nucleosides and foscarnet. Mechanisms, p. 81-102. In D. Richman (ed.), Antiviral drug resistance. Wiley, Chichester, England.
- 7.Coen, D. M., A. F. Irmiere, J. G. Jacobson, and K. M. Kerns. 1989. Low levels of herpes simplex virus thymidine-thymidylate kinase are not limiting for sensitivity to certain antiviral drugs or for latency in a mouse model. Virology 168:221-231. [DOI] [PubMed] [Google Scholar]
- 8.Coen, D. M., M. Kosz-Vnenchak, J. G. Jacobson, D. A. Leib, C. L. Bogard, P. A. Schaffer, K. L. Tyler, and D. M. Knipe. 1989. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc. Natl. Acad. Sci. USA 86:4736-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davar, G., M. F. Kramer, D. Garber, A. L. Roca, J. K. Andersen, W. Bebrin, D. M. Coen, M. Kosz-Vnenchak, D. M. Knipe, X. O. Breakefield, and O. Isacson. 1994. Comparative efficacy of expression of genes delivered to mouse sensory neurons with herpes virus vectors. J. Comp. Neurol. 339:3-11. [DOI] [PubMed] [Google Scholar]
- 10.Efstathiou, S., S. Kemp, G. Darby, and A. C. Minson. 1989. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J. Gen. Virol. 70:869-879. [DOI] [PubMed] [Google Scholar]
- 11.Englund, J. A., M. E. Zimmerman, E. M. Swierkosz, J. L. Goodman, D. R. Scholl, and H. H. Balfour, Jr. 1990. Herpes simplex virus resistant to acyclovir. A study in a tertiary care center. Ann. Intern. Med. 112:416-422. [DOI] [PubMed] [Google Scholar]
- 12.Farabaugh, P. J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60:103-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudreau, A., E. Hill, H. H. Balfour, Jr., A. Erice, and G. Boivin. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297-303. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths, A., and D. M. Coen. 2003. High frequency phenotypic reversion and pathogenicity of an acyclovir-resistant herpes simplex virus mutant. J. Virol. 77:2282-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths, A., S. Renfrey, and T. Minson. 1998. Glycoprotein C-deficient mutants of two strains of herpes simplex virus type 1 exhibit unaltered adsorption characteristics on polarized or non-polarized cells. J. Gen. Virol. 79:807-812. [DOI] [PubMed] [Google Scholar]
- 16.Horsburgh, B. C., S. H. Chen, A. Hu, G. B. Mulamba, W. H. Burns, and D. M. Coen. 1998. Recurrent acyclovir-resistant herpes simplex in an immunocompromised patient: can strain differences compensate for loss of thymidine kinase in pathogenesis? J. Infect. Dis. 178:618-625. [DOI] [PubMed] [Google Scholar]
- 17.Horsburgh, B. C., H. Kollmus, H. Hauser, and D. M. Coen. 1996. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell 86:949-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang, C. B., B. C. Horsburgh, E. Pelosi, S. Roberts, P. Digard, and D. M. Coen. 1994. A net +1 frameshift permits synthesis of thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc. Natl. Acad. Sci. USA 91:5461-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 20.Kost, R. G., E. L. Hill, M. Tigges, and S. E. Straus. 1993. Recurrent acyclovir-resistant genital herpes in an immunocompetent patient. N. Engl. J. Med. 329:1777-1782. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel, T. A., and K. Bebenek. 2000. DNA replication fidelity. Annu. Rev. Biochem. 69:497-529. [DOI] [PubMed] [Google Scholar]
- 22.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leib, D. A., K. C. Nadeau, S. A. Rundle, and P. A. Schaffer. 1991. The promoter of the latency-associated transcripts of herpes simplex virus type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proc. Natl. Acad. Sci. USA 88:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morfin, F., D. Thouvenot, M. Aymard, and G. Souillet. 2000. Reactivation of acyclovir-resistant thymidine kinase-deficient herpes simplex virus harbouring single base insertion within a 7 Gs homopolymer repeat of the thymidine kinase gene. J. Med. Virol. 62:247-250. [PubMed] [Google Scholar]
- 25.Sacks, S. L., R. J. Wanklin, D. E. Reece, K. A. Hicks, K. L. Tyler, and D. M. Coen. 1989. Progressive esophagitis from acyclovir-resistant herpes simplex. Clinical roles for DNA polymerase mutants and viral heterogeneity? Ann. Intern. Med. 111:893-899. [DOI] [PubMed] [Google Scholar]
- 26.Sarisky, R. T., M. R. Quail, P. E. Clark, T. T. Nguyen, W. S. Halsey, R. J. Wittrock, J. O. Bartus, M. M. Van Horn, G. M. Sathe, S. Van Horn, M. D. Kelly, T. H. Bacon, and J. J. Leary. 2001. Characterization of herpes simplex viruses selected in culture for resistance to penciclovir or acyclovir. J. Virol. 75:1761-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasadeusz, J. J., and S. L. Sacks. 1996. Spontaneous reactivation of thymidine kinase-deficient, acyclovir-resistant type-2 herpes simplex virus: masked heterogeneity or reversion? J. Infect. Dis. 174:476-482. [DOI] [PubMed] [Google Scholar]
- 28.Sasadeusz, J. J., F. Tufaro, S. Safrin, K. Schubert, M. M. Hubinette, P. K. Cheung, and S. L. Sacks. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmit, I., and G. Boivin. 1999. Characterization of the DNA polymerase and thymidine kinase genes of herpes simplex virus isolates from AIDS patients in whom acyclovir and foscarnet therapy sequentially failed. J. Infect. Dis. 180:487-490. [DOI] [PubMed] [Google Scholar]
- 30.Swetter, S. M., E. L. Hill, E. R. Kern, D. M. Koelle, C. M. Posavad, W. Lawrence, and S. Safrin. 1998. Chronic vulvar ulceration in an immunocompetent woman due to acyclovir-resistant, thymidine kinase-deficient herpes simplex virus. J. Infect. Dis. 177:543-550. [DOI] [PubMed] [Google Scholar]
- 31.Tenser, R. B., A. Gaydos, and K. A. Hay. 1996. Reactivation of thymidine kinase-defective herpes simplex virus is enhanced by nucleoside. J. Virol. 70:1271-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wills, N. M., J. A. Ingram, R. F. Gesteland, and J. F. Atkins. 1997. Reported translational bypass in a trpR′-lacZ′ fusion is accounted for by unusual initiation and +1 frameshifting. J. Mol. Biol. 271:491-498. [DOI] [PubMed] [Google Scholar]