Abstract
Adenosine (Ado) kinase (ADK; ATP:Ado 5′ phosphotransferase, EC 2.7.1.20) catalyzes the salvage synthesis of adenine monophosphate from Ado and ATP. In Arabidopsis, ADK is encoded by two cDNAs that share 89% nucleotide identity and are constitutively, yet differentially, expressed in leaves, stems, roots, and flowers. To investigate the role of ADK in plant metabolism, lines deficient in this enzyme activity have been created by sense and antisense expression of the ADK1 cDNA. The levels of ADK activity in these lines range from 7% to 70% of the activity found in wild-type Arabidopsis. Transgenic plants with 50% or more of the wild-type activity have a normal morphology. In contrast, plants with less than 10% ADK activity are small with rounded, wavy leaves and a compact, bushy appearance. Because of the lack of elongation of the primary shoot, the siliques extend in a cluster from the rosette. Fertility is decreased because the stamen filaments do not elongate normally; hypocotyl and root elongation are reduced also. The hydrolysis of S-adenosyl-l-homo-cysteine (SAH) produced from S-adenosyl-l-methionine (SAM)-dependent methylation reactions is a key source of Ado in plants. The lack of Ado salvage in the ADK-deficient lines leads to an increase in the SAH level and results in the inhibition of SAM-dependent transmethylation. There is a direct correlation between ADK activity and the level of methylesterified pectin in seed mucilage, as monitored by staining with ruthenium red, immunofluorescence labeling, or direct assay. These results indicate that Ado must be steadily removed by ADK to prevent feedback inhibition of SAH hydrolase and maintain SAM utilization and recycling.
The adenine (Ade) salvage pathway recycles Ade and adenosine (Ado) to AMP and thereby contributes to the maintenance of cellular energy charge and to the synthesis of a variety of biomolecules including nucleotide cofactors and nucleic acids. In addition, the salvage cycle enzymes fulfill two additional roles in plant metabolism. First, they decrease the intracellular levels of Ade and Ado that may otherwise affect the activity of other enzymes. In vitro, enzyme activities associated with methyl recycling and polyamine biosynthesis are sensitive to inhibition by Ade or Ado, suggesting that salvage of these purines may be important to the maintenance of flux through the transmethylation and polyamine pathways (Poulton and Butt, 1975; Guranowski et al., 1981; Miyazaki and Yang, 1987). Second, plant Ade and Ado salvage enzymes interconvert cytokinin (CK) bases, ribosides, and ribotides in vitro (Burch and Stuchbury, 1987; Moffatt et al., 1991; Mok and Martin, 1994) and may do so in vivo as well. CK interconversion is thought to be a key mechanism for regulating the level of active CK (McGaw and Burch, 1995).
There are two principal routes for the recycling of Ado: direct phosphorylation by Ado kinase (ADK) or hydrolysis to Ade by Ado nucleosidase followed by conversion to AMP, by Ade phosphoribosyltransferase (APT). An initial examination of ADK activity in Arabidopsis revealed that its genome encodes two ADK isoforms that are 92% identical at the amino acid level (Moffatt et al., 2000). These ADK genes appear to be constitutively expressed with the highest transcript levels found in flowers and roots. Both enzymes are similar in their ability to utilize Ado or CK ribosides as substrates although based on in vitro estimates of their Kms, both enzymes prefer Ado (Kms for Ado of 0.3–0.5 μm versus 3–5 μm for N6 (isopentenyl) Ado; Moffatt et al., 2000). The low Kms of the Arabidopsis ADKs for Ado suggest that the intracellular Ado concentration in Arabidopsis cells may be maintained at low concentrations, consistent with those found in other plants (1–50 μm; Wagner and Backer, 1992).
To further assess the physiological significance of Ado salvage in plants, we have generated a set of Arabidopsis plants deficient in ADK activity. Direct selection schemes involving Ado analogs that become toxic only after their conversion to nucleotides by ADK have been used to isolate ADK mutants in other organisms such as Neurospora crassa, Toxoplasma gondii, and yeast (Saccharomyces cerevisiae; Magill et al., 1982; Iltzsch et al., 1995; Iwashima et al., 1995). However, we were unable to isolate Arabidopsis ADK-deficient mutants using these Ado analogs. Instead, transgenic approaches employing sense and antisense expression of an ADK cDNA led to the recovery of several ADK-deficient lines. To our knowledge, these are the first ADK-deficient multicellular organisms to be described. Their phenotype suggests that ADK plays a major role in Ado salvage in plants and that reduced ADK activity leads to inhibition of S-adenosyl-l-homo-Cys (SAH) hydrolase, accumulation of SAH, and limits S-adenosyl-l-Met (SAM) utilization and regeneration.
RESULTS AND DISCUSSION
Isolation of ADK-Deficient Lines
Since the direct selection of ADK mutants using Ado analogs was unsuccessful, we sought to induce ADK deficiency using antisense and sense expression of the ADK1 cDNA from an enhanced 35S promoter. Because of the high nucleotide identity of the two ADK coding regions (89%), it was hoped that the expression of both genes might be down-regulated simultaneously by the introduction of either the sense or antisense ADK1 transgene. Fifteen sense and six antisense transformant lines were generated by vacuum infiltration of wild-type (WT) Arabidopsis with each construct. Of these, eight sense (sADK1–8) and four antisense (aADK1–4) lines, each containing a single transgene insert based on Southern hybridization and kanamycin segregation analysis (data not shown), were retained for further analysis. Both sets of plants had a similar phenotype, although most of the antisense lines were extremely small, limiting our molecular characterization of them.
ADK Levels in Transgenic Lines
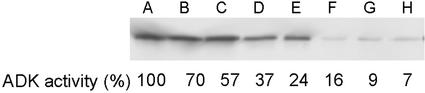
A radiochemical assay of ADK activity in leaf tissue of 4-week-old plants of each transgenic line showed that they contained lower ADK activity levels than WT plants of the same chronological age. However, at each generation about 10% to 15% of the plants appeared morphologically normal and had WT, or higher, ADK activity and protein levels (data not shown). The basis for this consistent loss of gene silencing, which was observed in both sense and antisense lines, is unknown. Excluding these non-silenced plants, the ADK activity in the lines ranged from 7% to about 70% of WT with about 3% to 5% variability between individuals of a specific line (Fig. 1). These activity levels and the loss of silencing in a fraction of the population have been reproducibly stable for at least six generations.
Figure 1.
ADK protein levels and activity in WT and ADK-silenced lines. ADK protein in crude leaf extracts of WT and representative lines were monitored by immunoblotting with an antibody raised against recombinant ADK1 that detects both ADK isoforms. A, WT; B, aADK10; C, aADK3-5; D, sADK8-1; E, sADK7-2; F, sADK5-2; G, sADK9-1; H, sADK4-2. The results of the radiochemical assay of ADK activity in leaf extracts of the same plants expressed as a percentage of WT activity (18.9 nmol mg−1 protein min−1) is shown below each lane.
Immunoblot analysis of leaf extracts using an antibody which detects both Arabidopsis ADKs also showed that the lines contained 13% to 97% less ADK protein than WT (Fig. 1). Thus, the radiochemical assay and immunoblots analysis clearly indicate that the transgenic lines contain lower ADK activity and protein, respectively. The values obtained by the enzyme assay have been used for all further descriptions of these lines.
ADK Gene Expression
Northern analysis was carried out to determine whether the steady-state level of ADK transcripts is reduced by the expression of the ADK1 transgene. ADK transcript abundance in leaves and stems was lower in the ADK sense silenced lines as compared with the WT when the full-length ADK1 cDNA was used as the probe (Fig. 2A). Semiquantitative RT-PCR analysis indicated that transcripts of both genes were decreased to a similar extent in each line in agreement with the RNA blot (Fig. 2B). Thus, overexpression of the ADK1 cDNA reduces, but does not eliminate, ADK transcripts leading to decreases in ADK activity and protein levels.
Figure 2.
Analysis of ADK transcript abundance by RNA blotting and reverse transcription (RT)-PCR. A, Total RNA from leaves and stems of the indicated lines was probed with the full-length ADK1 cDNA. The silenced lines contain reduced ADK transcript levels in leaves and a less moderate decrease in the apparently normal secondary shoots. B, RT-PCR analysis of cDNA prepared from RNA of the indicates a reduction in ADK1 and ADK2 transcript levels in the sADK lines relative to the WT, consistent with the RNA blot results. Transcripts of RNA helicase were amplified as an internal control.
Phenotype of ADK-Deficient Lines
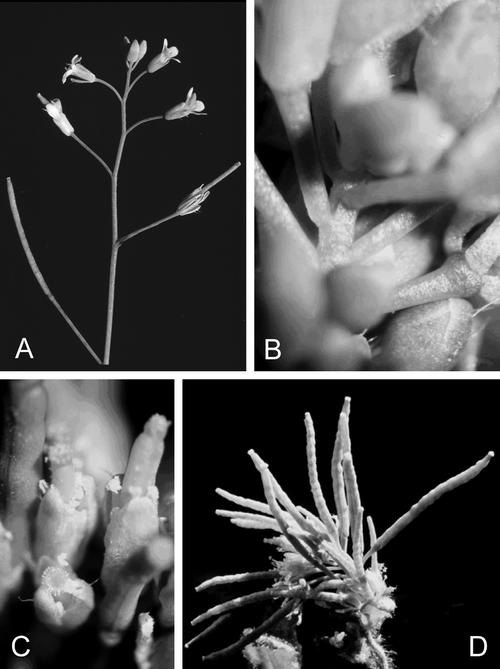
Lines with greater than 50% residual ADK activity appeared to have a normal vegetative morphology. However, those with lower ADK activity had a very distinct phenotype, the severity of which correlated directly with the level of residual ADK in each line (Fig. 3). Lines with less than 10% ADK activity were small with rounded wavy leaves and a very short primary shoot. The internodes on the primary shoot were very close together (Fig. 4, A versus B) and their flowers appeared to extend directly from the rosette. The filaments on the stamens did not extend normally so that self-fertility was reduced even though pollen production appeared normal (Fig. 4C). The siliques that did form on these transgenic plants were bumpy (Fig. 4D). Lines with intermediate ADK activity (15%–25% residual activity) bolted normally but later internodes did not elongate, further causing the siliques to cluster together (Fig. 3). Often, more elongated secondary shoots developed later on these lines. Root growth was inhibited: All the ADK lines tested had a short primary root with an increased number of lateral roots of about the same length (data not shown).
Figure 3.
Representative ADK-deficient lines recovered by gene silencing described on the basis of ADK activity relative to WT. A, Plants grown in soil on a 16-h day length for approximately 1 month are shown. From left to right, WT; sADK2-2 (≈64% ADK activity), which appear morphologically normal; sADK8-1 (≈33% ADK activity), which have bushier rosettes, shortened primary shoots, but normal secondary shoots; sADK5-1 (≈16% ADK activity), which are bushy but often make a few normal secondary shoots later than the WT; and sADK7-4 (≈7% ADK activity), which are smaller with a dense rosette of wavy leaves. B, Three-week-old plant representative of the 30% to 50% plants with some stem elongation ending in a region of shortened internodes. C, sADK7-4 plant showing the dense rosette of wavy leaves and short internodes on both the primary and secondary shoots.
Figure 4.
Altered stem and stamen elongation in ADK-deficient lines. A, WT plant with regular spacing of internodes. B, Region of the primary shoot in sADK7-4 showing the lack of elongation. C, Stamens on the most deficient lines do not elongate normally resulting in dehiscence of the anther below the stigma. D, Close-up view of the cluster of bumpy siliques formed on the primary shoot of sADK7-4 plants.
Ado and Transmethylation
The pleiotropic phenotype of these transgenic lines is perhaps not surprising given the fact that ADK is constitutively expressed (Moffatt et al., 2000) and contributes to maintaining adenylate levels. However, APT has a similar expression pattern and also aids in recycling adenylates, yet mutants with 1% residual APT activity have a normal vegetative morphology (Moffatt and Somerville, 1988). Thus, we hypothesized that the phenotype of the ADK-deficient lines was due more to the lack of Ado salvage in these plants rather than low adenylate levels.
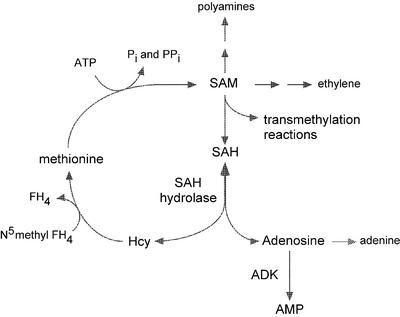
In addition to being released from the breakdown of nucleic acids and various nucleotide cofactors, Ado is a byproduct of SAM-dependent transmethylation reactions. For each methyl group transferred from SAM to the methyl acceptor, one molecule of SAH is produced (Fig. 5). The SAH is hydrolyzed to Ado and Hcy by SAH hydrolase; Met and SAM are regenerated from the Hcy, whereas the Ado is salvaged to the adenylate nucleotide pool.
Figure 5.
Key intermediates of methyl recycling via SAH. SAM is the methyl donor for many transmethylation reactions including those leading to the synthesis of pectin, lignin, phosphotidylcholine, and methylated DNA. The SAH produced from these reactions is an inhibitor of the transmethylases and is thus rapidly metabolized by SAH hydrolase. The methyl moiety is recycled to Met, whereas the adenosyl component is converted to AMP by ADK. The synthesis of polyamines and ethylene also rely on a steady SAM supply. The recycling of methylthioadenosine that is produced as a result of ethylene and polyamine biosynthesis is not shown here but also contributes to the methyl and adenylate pools.
Ado salvage/catabolism are critical for removing SAH because the reaction catalyzed by SAH hydrolase is reversible and its equilibrium lies strongly in the direction of synthesis. This reaction is only drawn in the direction of SAH hydrolysis by the continued metabolism of both products, Hcy and Ado (de la Haba and Cantoni, 1959). Moreover, in vitro SAH hydrolase activity from several organisms, including some plants, is inhibited by Ado (Poulton and Butt, 1975; Duerre and Briske-Anderson, 1981; Poulton, 1981). A similar effect was observed for Arabidopsis SAH hydrolase activity; SAH hydrolysis was reduced 25% by the presence of only 0.5 μm Ado and 70% by 5 μm Ado (Table I). If this inhibition also occurs in vivo, it suggests that hydrolysis of SAH by Arabidopsis SAH hydrolase is sensitive to Ado levels.
Table I.
Inhibition of in vitro SAH hydrolase activity by adenosine
| Ado Concentration at Start of the Assay | SAH Hydrolase Activity | Inhibition |
|---|---|---|
| μm | nmol min−1 mL−1 | % |
| 0 | 5.9 | – |
| 0.5 | 4.4 | 25.4 |
| 1.0 | 2.9 | 50.9 |
| 5.0 | 1.8 | 69.5 |
| 25 | 1.5 | 74.1 |
| 100 | 1.3 | 77.9 |
SAH hydrolase activity (SAH → Ado and Hcy) was measured using crude desalted extracts of Arabidopsis leaves in the presence and absence of added Ado. The starting concentration of SAH in each assay was 150 μm. A representative set of assay results are shown.
Two other enzymes that could metabolize Ado, Ado nucleosidase, and Ado deaminase are generally low or undetectable in plants (Barankiewicz and Paszkowski, 1980; Chen and Kristopeit, 1981; Burch and Stuchbury, 1987; Wagner and Backer, 1992; Edwards, 1996; Dancer et al., 1997). We have not been able to detect Ado deaminase activity in leaf extracts of Arabidopsis (less than 200 pmol min−1 g−1 fresh weight; data not shown) and Auer (1999) found Arabidopsis Ado nucleosidase activity to be low as well. Thus, Arabidopsis may rely primarily on ADK for the recycling of Ado and the phenotype of the ADK-deficient lines may be a reflection of Ado accumulation causing an inhibition of SAH hydrolase activity and an increase in SAH.
Quantification of SAM and SAH levels in leaves of WT and several ADK-deficient lines indicated that although SAM levels were only moderately reduced (less than 2-fold), SAH levels were increased up to 40-fold relative to the WT (Table II) in the lines with less than 20% ADK activity (sADK4-2, 5-1, and 7-4); a line with higher residual ADK activity (sADK9-1; 35% ADK activity) has an SAH level about 14 times higher than WT. The levels of SAM and SAH in WT are in good agreement with those reported previously for other plants (for review, see Ranocha et al., 2001). Thus, ADK deficiency in these plants has led to increases in SAH. This is consistent with the effects of ADK deficiency in mammalian cell cultures and yeast: An increase in Ado results in an increase in the level of SAH (Kredich and Martin, 1977; Duerre and Briske-Anderson, 1981; Iwashima et al., 1995; Lecoq et al., 2001).
Table II.
Degree methylation of polygalacturonic acid of seed mucilage
| Line | Relative ADK Activity | Degree Methyl Esterification |
|---|---|---|
| % of WT | % | |
| WT | 100 | 17.6 ± 1.8 |
| sADK4-5 | 25 | 13.2 ± 0.4 |
| sADK9-3 | 10 | 10.3 ± 0.1 |
| sADK7-4 | 7 | 7.6 ± 1.0 |
The level of methyl esterification of uronic acid residues recovered from ammonium oxalate extracts of WT and ADK-deficient seed. The degree methyl esterification represents the mean of three extractions and the se.
All methyltransferases that use SAM as a methyl donor are inhibited by SAH (de la Haba and Cantoni, 1959; Cantoni et al., 1979). In most cases, the affinity of a methyltransferase for SAH is higher than for its substrate SAM, leading to the proposal that it is the ratio of SAM to SAH that regulates transmethylation activity (Cantoni, 1977; Duerre and Briske-Anderson, 1981). For example, it has been proposed that caffeic acid O-methyltransferase from spinach (Spinacia oleracea) is regulated by the ratio of SAM to SAH in vivo (Poulton and Butt, 1975). The sensitivity of a specific transmethylase to changes in the ratio of SAM to SAH depends upon several factors including its relative affinity for SAM versus SAH, the cellular abundance of the enzyme, and the subcellular compartmentation of the methyltransferase (Cantoni, 1977). Relatively few methyltransferases have been characterized in sufficient detail to make meaningful comparisons regarding their sensitivity to changes in the SAH to SAM ratio, although it is typical for a transmethylase to be sensitive to micromolar levels of SAH in vitro (Poulton and Butt, 1975; Poulton, 1981; Edwards and Dixon, 1991, and refs. therein). Given that the intracellular concentration of SAH is low and of the same order of magnitude as SAM (Poulton, 1981), a relatively small change in the amount of SAH could affect the SAM to SAH ratio sufficiently to reduce transmethylation activity. Reduction of transmethylation activity may affect not only the synthesis of methylated products but also the recycling of Met and subsequent synthesis of SAM for polyamine and ethylene synthesis (Fig. 5).
To test whether SAM-dependent methylation is affected by a reduction in ADK activity, we compared the level of pectin methylation in the mucilage of ADK-deficient and WT seed by staining with ruthenium red (RR). This stain is thought to bind to Ca2+ ions associated with the free carboxyl groups of nonesterified poly-GalUA (Luft, 1971; Hanke and Northcote, 1975). Reduction of the charge density of the carboxyl groups by esterification reduces the intensity of pectin staining by RR (Sterling, 1970). The mucilage of the Arabidopsis seed coat contains pectin (Goto, 1985; Western et al., 2000), the principle constituents of which are methylesterified and nonesterified poly-GalUAs, as well as Rha. When WT Arabidopsis seeds were imbibed in water supplemented with 0.1% (w/v) RR, a bright-pink halo was visible by light microscopy (Fig. 6A). Seed from transgenic lines with 7%, 50%, and 64% residual ADK activity stained more intensely with the RR than did the WT seed, with the seed of the 7% line staining the darkest. Estimation of the staining intensity from a digitized version of Figure 6A indicated an inverse relationship between the amount of bound RR and the residual ADK level (Fig. 6A). The RR staining of these lines suggests that the pectin of the seed mucilage of the ADK-deficient lines is less esterified as compared with that of WT seed.
Figure 6.
Pectin esterification in seed mucilage of WT and ADK-deficient lines. A, Increased RR staining was observed in seed from plants of decreasing ADK activity. Nonesterified pectin moieties within the mucilage layer of Arabidopsis seed bind RR. Seeds were imbibed in water for 30 min and stained in a solution of 0.01% (w/v) RR for 1.5 h and photographed using a 10× objective. Bar = 160 μm. From top to bottom, Representative staining pattern of seed from WT, sADK2-2 (≈64% ADK activity), aADK3-5 (≈50% ADK activity), and sADK7-4 (≈7% ADK activity). Average intensity of staining of each seed stock estimated using NIH Image. From top to bottom: 27, 31, 77, and 123. B through G, Immunofluorescence staining with JIM5 and JIM7 antisera. Seed were incubated with either JIM5 (upper row) or JIM7 (lower row). Decreasing ADK activity is associated with decreased JIM7 staining and increased JIM5 staining. B and E, WT; C and F, sADK 5-1 (≈16% ADK activity); D and G, sADK4-2 (≈7% ADK activity). Bar = 160 μm.
Paired assays of uronic acid and methanol released by saponification from WT and several ADK-deficient lines also showed a direct correlation between ADK activity and methyl-esterification (Table III). The most deficient line tested, sADK7-4, which has about 7% residual ADK activity, had 56.9% less methylesterification in its seed mucilage than WT. Lines with slightly more ADK activity (10% and 25% residual ADK activity; sADK9-3 and sADK4-5, respectively) were less affected (41.7% and 25.4% less uronic acid methylation than WT, respectively.)
Table III.
Quantification of SAM and SAH in leaves of WT and ADK-deficient lines
| Plant Line | SAM | SAH | Average Methyl Index (SAM:SAH) |
|---|---|---|---|
| μm | |||
| WT | 17.39 ± 0.75 | 0.34 ± 0.03 | 50.61 ± 2.19 |
| sADK4-2 | 16.81 ± 0.35 | 12.85 ± 0.77 | 1.31 ± 0.02 |
| sADK5-1 | 14.34 ± 0.25 | 14.75 ± 0.45 | 0.97 ± 0.02 |
| sADK7-4 | 11.19 ± 0.25 | 12.63 ± 0.20 | 0.88 ± 0.03 |
| sADK9-1 | 9.46 ± 0.87 | 4.14 ± 2.08 | 2.28 ± 0.01 |
The SAM and SAH contents of leaf extracts were assayed as their fluorescent isoindoles (Capdevila and Wagner, 1998). A representative set of data from one experiment is presented. Values are averages (n = 3) ± se and have been corrected for the 46% recovery of SAM and SAH.
The JIM5 and JIM7 antisera preferentially react with low- and high-esterified pectin, respectively (Knox et al., 1990). Willats et al. (2001) recently reported the characterization of WT C24 seed mucilage pectin polysaccharides using these antisera. Whereas JIM5 binds to the mucilage pectin closest to the seed coat, JIM7 binds to the outer edge of the mucilage pectin, indicating that the level of esterification of mucilage pectin varies within distinct regions of the mucilage. We analyzed the seed of WT Columbia and two ADK-deficient lines with these antisera. The staining pattern of Columbia WT is similar to that reported for C24: strong JIM5 staining near the seed coat (Fig. 6B) and a halo of JIM7 staining on the outer mucilage (Fig. 6E). Staining of sADK4-2 seed, which has about 7% of the ADK activity of WT, shows almost no reaction with JIM7 (Fig. 6G), but JIM5 binds a wider region of the mucilage than in WT (Fig. 6D). A transgenic line with moderate ADK activity (sADK5-1; 15%) had a staining pattern intermediate between WT and the most deficient line: a moderate reaction with both JIM5 and JIM7 (Fig. 6, C and F).
Thus, the most plausible explanation for the pleiotropic phenotype of these transgenic lines is a defect in methyl recycling and transmethylation activities because of Ado inhibition of SAH hydrolase activity. Decreased transmethylation could affect numerous end products of this pathway, including phosphotidylcholine and cell wall components as well as levels of methylated protein and DNA. In addition to affecting the synthesis of these end products directly, lower methyltransferase activity results in less Hcy production and ultimately lower Met available for the synthesis of SAM. Because SAM is the precursor for both polyamines and ethylene as well as the methyl donor for almost all methyltransferases, ADK deficiency has the potential to affect a large number of cellular activities. Moreover, reduced Ado salvage from SAH would eventually decrease the adenylate pools.
The numerous morphological changes and the developmental variability observed in the ADK-deficient lines may reflect the methyl requirements of different tissues: Those requiring higher rates of transmethylation activity may be the most sensitive to ADK deficiency. Immunofluroscence staining of pectin seed mucilage of these lines as well as direct assay of methyl-esterification of the uronic acid indicates that methylation of pectin is sensitive to reduced Ado salvage. The correlation between the level of ADK activity and increased JIM5 staining of less esterified pectin indicates that ADK activity is limiting pectin methylation in these lines. This suggests that stresses or developmental processes that put an increased demand on the methyl budget may necessitate increased ADK activity to maintain SAH hydrolase activity. It is possible that similar changes in pectin methylation may contribute to the reduction in stem, root, and stamen elongation observed in these lines, perhaps by cross-linking the pectin to a greater extent.
Tobacco (Nicotiana tabacum) lines with reduced methylation capacity because of antisense reduction of SAH hydrolase expression have been described (Tanaka et al., 1997). Although the mechanism causing the change in methylation activity is more direct in these lines, several aspects of their phenotype resemble that of the Arabidopsis ADK-deficient lines described here. Both sets of plants have wrinkled leaves, dwarf statures, and shorter stamen filaments resulting in reduced fertility. Lines with intermediate levels of either ADK or SAH hydrolase expression have normal vegetative morphology in both cases. The level of DNA methylation in the tobacco SAH hydrolase lines is decreased, an effect that is consistent with an inhibition of DNA methyltransferase activity by increased SAH. A similar analysis of DNA methylation using methylation-sensitive restriction endonuclease (HpaII) and its non-methylation-sensitive isochizomer MspI (for review, see Jeddeloh and Richards, 1996) suggests that the genomic DNA of sADK5-1 and sADK 7-1 are also less methylated than WT DNA (data not shown). More quantitative studies using direct HPLC measurement of 5-methylcytosine (Matassi et al., 1992) and analysis of the methylation of specific sequences (Cai et al., 1996) will be necessary to clarify the extent of this decrease and the types of sequences affected.
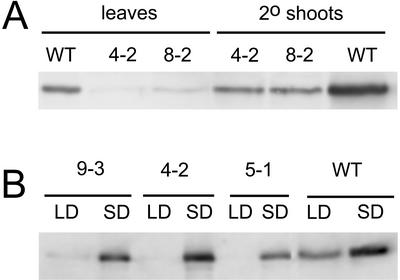
Although their primary shoots are severely stunted, both sense and antisense lines formed secondary shoots that appeared to resemble the WT shoots (Fig. 3A). Northern analysis indicates a moderate increase in transcript levels in the “normal” secondary shoots (Fig. 2A), whereas immunoblots of the same organs showed almost normal levels of ADK (Fig. 7A), suggesting that the silencing of ADK has been eliminated later in development. This raises the question as to whether there is insufficient transmethylation activity to maintain posttranscriptional gene silencing, which relies on methylation of the target and transgene (Finnegan et al., 1998). Although plausible, this hypothesis does not explain why normal secondary shoots are observed also in the antisense lines despite the fact that antisense silencing does not rely on transgene methylation (Finnegan et al., 1998). Perhaps neither sense nor antisense silencing is able to accommodate the increased ADK transcript levels in stems (Fig. 2A). This is consistent with the observed apparent loss in gene silencing in sADK lines grown under a short photoperiod, a condition that requires an increased methyl demand for lignin accumulation (Fig. 7B; Weretilnyk et al., 2000). The apparently normal plants that arise each generation from these homozygous lines may be because of a competition for methyl groups early during development such that silencing does not get established. A developmental and tissue-specific analysis of the levels of key end products of the transmethylation pathway including phosphatidylcholine, pectin, and DNA, as well as other compounds that require SAM such as polyamines and ethylene, should provide greater insight into how the methyl budget is prioritized in plants under conditions of a methyl deficit.
Figure 7.
Developmental and day length-induced changes in ADK protein levels in ADK-silenced lines. A, ADK protein levels in crude extracts of leaves and normal-looking secondary shoots of the indicated sADK lines and WT were monitored by immunoblotting. ADK protein levels are increased in secondary shoots versus leaves in the WT and transgenic lines. B, Extracts of leaves of WT and the indicated sADK lines grown in 16-h (LD) or 8-h (SD) photoperiods were analyzed by immunoblotting. The plants grown in short days have higher ADK protein levels in all cases.
MATERIALS AND METHODS
Seed Material and Germination
Arabidopsis ecotype Columbia WT seeds were either plated directly onto soil or on sterile Murashige and Skoog media (Murashige and Skoog, 1962) as described by Zhang et al. (2002). Plants were grown with 16 h light at 120 μmol m−2 s−1 photosynthetically active radiation.
Nucleic Acid Isolation and Analysis
DNA was extracted from leaves following the procedure of Dellaporta et al. (1983). Restriction digests were separated by electrophoresis through 1.0% (w/v) agarose gels and transferred to nitrocellulose for Southern analysis of transgene copy number. Blots were hybridized with 32P-labeled ADK1 cDNA probes, prepared by random priming, in 5× SSC buffer supplemented with 50% (v/v) formamide at 42°C as previously described (Moffatt et al., 2000). RNA was extracted using Triazol (Roche Biochemicals, Laval, QC) following the manufacturer's instructions. Northern analysis was performed on RNAs that had been separated by electrophoresis through formaldehyde gels and transferred to Hybond N+ membranes (Amersham-Pharmacia, Uppsala). Gene-specific probes were recovered from the 3′-untranslated regions of each cDNA (nucleotides 1,054–1,233 of ADK1 cDNA; AF180894) and nucleotides 1,041 through 1,198 of ADK2 cDNA (AF180895) by PCR using primers described by Moffatt et al. (2000) and cloned into Bluescript KS (Stratagene, La Jolla, CA). The same primers were used for RT-PCR analysis of cDNA obtained from 2 μg of total leaf RNA (MBI Fermentas, Burlington, ON). The reaction products were quantified after 25 cycles, in the linear range of the reaction. The RNA helicase gene (RH4) was used as an internal control (Aubourg et al., 1999).
Creation of Transgenic Plants
The full-length ADK1 cDNA contained in the EST R30128 was linearized at the 5′ end of the predicted coding region with SmaI and ligated to XbaI linkers [d(pTGCTCTAGAGCA); New England Biolabs, Mississauga, ON]. Subsequent digestion with XbaI produced a 1,086-bp fragment flanked by XbaI ends that was cloned into the T-DNA vector pKYLX71 (Schardl et al., 1987) in either the sense (pYS2) or antisense (pYS3) orientation relative to the enhanced 35S promoter. After transformation into the Agrobacterium tumefaciens strain C58 pGV3101, each construct was introduced into Arabidopsis by vacuum infiltration (Bechtold et al., 1993). Transformed plants were identified by spreading sterilized seed from the infiltrated plants on Murashige and Skoog media supplemented with 50 μg mL−1 kanamycin (Sigma, St. Louis). Both the morphological and metabolic phenotype of the individual lines have remained stable through eight generations. Molecular comparisons were done on 4-week-old T4-T6 generation plants.
Assay of ADK Activity and Protein
ADK activity in crude leaf extracts was monitored by a radiochemical assay as described (Moffatt et al., 2000). The assay measures the conversion of radioactive Ado ([2,8-3H]Ado, 20 Ci mmol−1; ICN, Costa Mesa, CA) to AMP in the presence of ATP and MgCl2. In brief, leaf tissue was homogenized in 50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.4, buffer (Weretilnyk et al., 2000) and desalted through Sephadex G25 (medium grade, Amersham-Pharmacia) to remove low-molecular- weight molecules. Normally, 0.1 to 0.3 μg of protein was used in each 5-min incubation, during which time the assay remains linear. AMP was precipitated with lanthanum chloride (Sigma), collected by filtration, and quantified by liquid scintillation counting (LS 1701, Beckman, Fullerton, CA).
ADK protein was detected by immunoblotting using an affinity-purified antibody raised against recombinant ADK1 (Moffatt et al., 2000). Bound primary antibody was detected using a secondary antibody conjugated to alkaline phosphatase (Sigma), reacted with a fluorescent substrate (ECF; Amersham-Pharmacia), and quantified using a Storm 860 phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Assay of SAH Hydrolase and Ado Deaminase
SAH hydrolase activity was measured spectrophotometrically in the hydrolytic direction (Wolfson et al., 1986). Each reaction was conducted at 25°C with 50 mm HEPES-KOH, pH 7.5; 1 mm EDTA, 150 μm SAH, and 100 μm 5,5-dithio-bis(2-nitrobenzoic acid). Ado was included at concentrations of 0, 0.5, 1.0, 5, 25, or 100 μm. The reaction produces a chromophoric thiolate and a corresponding increase in A412. Calculations were made using a molar extinction coefficient of 13,600 m−1 cm−1 and controls containing no SAH.
Two assays were used to investigate the presence of Ado deaminase based on its production of inosine from Ado. In the first assay, desalted leaf extracts were incubated with [2,8-3H]Ado at 0.28 μm (a tracer dose; 0.5 μCi) or 1 mm (a physiological dose; 0.5 μCi); 50 mm HEPES-KOH, pH 7.5; and 2 mm dithiothreitol at 30°C for 0 (control) or 60 min and the reaction was terminated by boiling. Five microliters of each 50-μL stopped reaction was applied to a fluorescein-containing Silica Gel G 0.25-mm plate (Machery-Nagel, Düren, Germany) and developed in n-butanol:acetic acid:water (12:3:5 [v/v]). Inosine, located as a nonfluorescent spot on the plate, was scraped into a scintillation vial and its radioactive content determined (Guranowski and Jakubowski, 1987). A check for Ado deaminase was done by carrying out the ADK assay in the absence and presence of the Ado deaminase inhibitor deoxycoformycin (Nipent, SuperGen, Dublin, CA). No significant difference was found in the ADK activity under these two conditions following multiple trials, as determined by a Student's t test.
RR Staining of Seed Coat
Seeds were imbibed in warm water at room temperature with continuous shaking for 30 min. The seeds were then stained with 0.01% (w/v) RR (Sigma), prepared in sterile water at room temperature, for 1.5 h. After two washes with water, seeds were photographed with the 10× or 5× objective of an Axiophot microscope (1/30-s exposure time; Carl Zeiss, North York, ON).
Assay of Pectin Methylation in Seed Mucilage
Seed (100 mg) was stirred gently in 1.5 mL of 0.5% (w/v) ammonium oxalate (Sigma) at 80°C for 1 h. After centrifugation (13,000g, 20 min) the supernatant was poured into 5 volumes of ethanol and centrifuged again (2,300g, 20 min). The pellet was dissolved in a minimal amount of water (about 5 mL), placed in dialysis tubing (10,000 molecular weight cutoff), and dialyzed against running tap water for 20 h and running distilled water for another 20 h, after which it was lyophilized. Two colorimetric assays for saponifiable methanol and GalUA were used to determine the degree of methyl esterification (Filisetti-Cozzi and Carpita, 1991; Kim and Carpita, 1992) with the minor modifications that sodium tetraborate was not added to the H2SO4 and 10 μL of carbazole (Sigma) in ethanol (1 mg mL−1) was used for color development in place of m-hydroxydiphenyl. Both assays were scaled down by a factor of 10 to preserve plant material. Results were quantified based on standard curves generated using methanol and poly-GalUA (Sigma).
Immunostaining of Low- and High-Methylesterfied Pectin with JIM5 and JIM7
Approximately 10 mg of seed from WT and ADK-deficient lines was imbibed in 20 mm HEPES, pH 7.0, for 24 h at room temperature with gentle shaking, then incubated with either JIM5, which reacts preferentially with low methylesterified pectin, or JIM7, which recognizes high methylesterified pectin (Knox et al., 1990). This incubation was done in 300 μL of buffer at room temperature for 12 h; bound antibodies were detected using FITC (Sigma) as described by Willats et al. (2001).
Quantitation of SAM and SAH
Leaf tissue was frozen in liquid nitrogen, ground to a powder, and resuspended in 20% (w/v) trichloroacetic acid (1 μL trichloroacetic acid mg fresh weight−1). Aftera low-speed centrifugation to remove debris, SAM and SAH were measured as their fluorescent isoindoles as described by Capdevila and Wagner (1998). Recovery of SAM and SAH was estimated to be between 47% and 56% from samples spiked with 25 to 100 pmol SAM and SAH (Sigma).
ACKNOWLEDGMENTS
The authors appreciate Nick Carpita's (Purdue University, West Lafayette, IN) help with pectin methylation assays and Heather Root's (Utrecht, The Netherlands) and Kirsten Alexander's (Toronto) characterization of the ADK-deficient lines; Dung Tiet (Toronto) did the immunoblots of secondary shoots. Paul Knox's (University of Leeds, UK) generous gift of JIM5 and JIM7 sera and his helpful technical assistance are gratefully acknowledged. SuperGen (Dublin, CA) generously provided the Nipent used in this research.
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (research grants to B.A.M. and E.A.W.), by the Public Health Service (grant nos. DK15289 and DK54859 to C.W.), and by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (Merit Revue award to C.W.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010880.
LITERATURE CITED
- Aubourg S, Kreis M, Lecharny A. The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acid Res. 1999;27:628–636. doi: 10.1093/nar/27.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer C. The Arabidopsis mutation cym changes cytokinin metabolism, adenosine nucleosidase activity and plant phenotype. Biol Plant. 1999;42:S-3. [Google Scholar]
- Barankiewicz J, Paszkowski J. Purine metabolism in mesophyll protoplasts of tobacco (Nicotiana tabacum) leaves. Biochem J. 1980;186:343–350. doi: 10.1042/bj1860343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold NB, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Burch LR, Stuchbury T. Activity and distribution of enzymes that interconvert purine bases, ribosides and ribotides in the tomato plant and possible implications for cytokinin metabolism. Physiol Plant. 1987;69:283–288. [Google Scholar]
- Cai Q, Guy CL, Moore GA. Detection of cytosine methylation and mapping of a gene influencing cytosine methylation in the genome of Citrus. Genome. 1996;39:235–242. doi: 10.1139/g96-032. [DOI] [PubMed] [Google Scholar]
- Cantoni GL. S-adenosylmethionine: present status and future perspectives. In: Salvatore F, Borek E, Zappia V, Williams-Ashman HG, Schlenk F, editors. The Biochemistry of Adenosylmethionine. New York: Columbia University Press; 1977. pp. 557–577. [Google Scholar]
- Cantoni GL, Richards HH, Chiang PK. Inhibitors of S-adenosylhomocysteine hydrolase and their role in the regulation of biological methylation. In: Usdin E, Borchardt RT, Creveling ER, editors. Transmethylation. New York: Elsevier; 1979. pp. 155–164. [Google Scholar]
- Capdevila A, Wagner C. Measurement of plasma S-adenosylmethionine and S-adenosylhomocysteine as their fluorescent isoindoles. Anal Biochem. 1998;264:180–184. doi: 10.1006/abio.1998.2839. [DOI] [PubMed] [Google Scholar]
- Chen C-M, Kristopeit SM. Metabolism of cytokinin: deribosylation of cytokinin ribonucleoside by adenosine nucleosidase from wheat germ cells. Plant Physiol. 1981;68:1020–1023. doi: 10.1104/pp.68.5.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancer JE, Hughes RG, Lindell SD. Adenosine-5′-phosphate deaminase: a novel herbicide target. Plant Physiol. 1997;114:119–129. doi: 10.1104/pp.114.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Haba G, Cantoni GL. The enzymatic synthesis of S-adenosyl-l-homocysteine from adenosine and homocysteine. J Biol Chem. 1959;234:603–608. [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Duerre JA, Briske-Anderson M. Effect of adenosine metabolites on methyltransferase reactions in isolated rat livers. Biochem Biophys Acta. 1981;678:275–282. doi: 10.1016/0304-4165(81)90217-8. [DOI] [PubMed] [Google Scholar]
- Edwards R. S-adenosyl-l-methionine metabolism in alfalfa cell cultures following treatment with fungal elicitors. Phytochemistry. 1996;43:1163–1169. [Google Scholar]
- Edwards R, Dixon RA. Purification and characterization of S-adenosyl- l-methionine:caffeic acid 3-O-methyltransferase from suspension cultures of alfalfa (Medicago sativa L.) Arch Biochem Biophys. 1991;287:372–379. doi: 10.1016/0003-9861(91)90492-2. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Genger RK, Peacock WT, Dennis ES. DNA methylation in plants. Annu Rev Plant Mol Biol Plant Physiol. 1998;49:223–241. doi: 10.1146/annurev.arplant.49.1.223. [DOI] [PubMed] [Google Scholar]
- Filisetti-Cozzi TMCC, Carpita NC. Measurement of uronic acids without interference from neutral sugars. Anal Biochem. 1991;197:157–162. doi: 10.1016/0003-2697(91)90372-z. [DOI] [PubMed] [Google Scholar]
- Goto N. A mucilage polysaccharride secreted from testa of Arabidopsis thaliana. Arabidopsis Inf Serv. 1985;22:143–145. [Google Scholar]
- Guranowski AB, Chiang PK, Cantoni GL. 5′-Methylthioadenosine nucleosidase: purification and characterization of the enzyme from Lupinus luteus seeds. Eur J Biochem. 1981;114:293–299. doi: 10.1111/j.1432-1033.1981.tb05148.x. [DOI] [PubMed] [Google Scholar]
- Guranowski A, Jakubowski H. Adenosylhomocysteine from yellow lupine. Methods Enzymol. 1987;143:430–434. doi: 10.1016/0076-6879(87)43075-9. [DOI] [PubMed] [Google Scholar]
- Hanke DE, Northcote DH. Molecular visualization of pectin and DNA by ruthenium red. Biopolymers. 1975;14:1–17. doi: 10.1002/bip.1975.360140102. [DOI] [PubMed] [Google Scholar]
- Iltzsch MH, Uber S, Tankersley KO, Kouni MH. Structure-activity relationship for the binding of nucleoside ligand to adenosine kinase from Toxoplasma gondii. Biochem Pharmacol. 1995;49:1501–1512. doi: 10.1016/0006-2952(95)00029-y. [DOI] [PubMed] [Google Scholar]
- Iwashima A, Ogata M, Nosaka K, Nishimura H, Hasegawa T. Adenosine kinase-deficient mutant of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1995;127:23–28. doi: 10.1111/j.1574-6968.1995.tb07444.x. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Richards EJ. mCCG methylation in angiosperms. Plant J. 1996;9:579–586. doi: 10.1046/j.1365-313x.1996.9050579.x. [DOI] [PubMed] [Google Scholar]
- Kim J-B, Carpita NC. Changes in esterification of the uronic acid groups of cell wall polysaccharides during elongation of maize coleoptiles. Plant Physiol. 1992;98:646–653. doi: 10.1104/pp.98.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta. 1990;181:512–521. doi: 10.1007/BF00193004. [DOI] [PubMed] [Google Scholar]
- Kredich NM, Martin DW., Jr Role of S-adenosylhomocysteine in adenosine mediated toxicity in cultured mouse T lymphoma cells. Cell. 1977;12:931–938. doi: 10.1016/0092-8674(77)90157-x. [DOI] [PubMed] [Google Scholar]
- Lecoq K, Belloc C, Desgranges C, Daignan-Fornier B. Role of adenosine kinase in Saccharomyces cerevisiae: identification of the ADO1 gene and study of the mutant phenotypes. Yeast. 2001;18:335–342. doi: 10.1002/1097-0061(20010315)18:4<335::AID-YEA674>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Luft JH. Ruthenium red and violet I: chemistry, purification, methods of use for electron microscopy and mechanism of action. Anat Rec. 1971;171:347–368. doi: 10.1002/ar.1091710302. [DOI] [PubMed] [Google Scholar]
- Magill JM, Dalke P, Lyda TS, Magill CW. Adenosine kinase-deficient mutant of Neurospora crassa. J Biol Chem. 1982;152:1292–1294. doi: 10.1128/jb.152.3.1292-1294.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassi G, Melis R, Kuo K, Macaya G, Gehrke CW, Bernardi G. Large-scale methylation patterns in the nuclear genomes of plants. Gene. 1992;12:239–245. doi: 10.1016/0378-1119(92)90211-7. [DOI] [PubMed] [Google Scholar]
- McGaw BA, Burch LR. Cytokinin biosynthesis and metabolism. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry and Molecular Biology. Boston: Kluwer Academic Press; 1995. pp. 101–117. [Google Scholar]
- Miyazaki JH, Yang SF. Inhibition of the methionine cycle enzymes. Phytochemistry. 1987;26:2655–2660. [Google Scholar]
- Moffatt BA, Pethe C, Laloue M. Metabolism of benzyladenine is impaired in a mutant of Arabidopsis thaliana lacking adenine phosphoribosyltransferase activity. Plant Physiol. 1991;95:900–908. doi: 10.1104/pp.95.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Somerville CR. Positive selection for male-sterile mutants of Arabidopsis lacking adenine phosphoribosyl transferase activity. Plant Physiol. 1988;86:1150–1154. doi: 10.1104/pp.86.4.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Wang L, Allen M, Stevens Y, Qin W, von Schwartzenberg K. Adenosine kinase of Arabidopsis thaliana: kinetic properties and gene expression. Plant Physiol. 2000;124:1775–1785. doi: 10.1104/pp.124.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DWS, Martin RC. Cytokinin metabolic enzymes. In: Mok DWS, Mok MC, editors. Cytokinins. Chemistry, Activity and Function. Boca Raton, FL: CRC Press; 1994. pp. 129–137. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Poulton JE. Transmethylation and demethylation reactions in the metabolism of secondary plant products. In: Conn EE, editor. The Biochemistry of Plants. Vol. 7. New York: Academic Press Inc.; 1981. pp. 667–723. [Google Scholar]
- Poulton JE, Butt VS. Purification and properties of S-adenosyl-l-methionine: caffeic acid O-methyltransferase from leaves of spinach beet (Beta vulgaris L.) Biochim Biophys Acta. 1975;403:301–314. doi: 10.1016/0005-2744(75)90060-1. [DOI] [PubMed] [Google Scholar]
- Ranocha P, McNeil SC, Ziemak MJ, Li C, Tarczynski MC, Hanson AD. The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J. 2001;25:575. doi: 10.1046/j.1365-313x.2001.00988.x. : 584. [DOI] [PubMed] [Google Scholar]
- Schardl SJ, Byrd A, Benzion G, Altshuler M, Hildebrand D, Hunt A. Design and construction of a versatile system for the expression of foreign genes in plants. Gene. 1987;61:1–11. doi: 10.1016/0378-1119(87)90359-3. [DOI] [PubMed] [Google Scholar]
- Sterling C. Crystal-structure of ruthenium red and stereochemistry of its pectin stain. Am J Bot. 1970;57:172–175. [Google Scholar]
- Tanaka H, Masuta C, Uehara K, Kataoka J, Koiwai A, Noma M. Morphological changes and hypomethylation of DNA in transgenic tobacco expressing antisense RNA of the S-adenosyl-l-homocysteine hydrolase gene. Plant Mol Biol. 1997;35:981–986. doi: 10.1023/a:1005896711321. [DOI] [PubMed] [Google Scholar]
- Wagner KG, Backer AI. Dynamics of nucleotides in plants studied on a cellular basis. Int Rev Cytol. 1992;134:1–84. [Google Scholar]
- Weretilnyk EA, Alexander KJ, Drebenstedt M, Snider J, Summer PS, Moffatt BA. Maintaining methylation activities during salt stress: the involvement of adenosine kinase. Plant Physiol. 2000;125:856–865. doi: 10.1104/pp.125.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL, Skinner DJ, Haughn GW. Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 2000;122:345–355. doi: 10.1104/pp.122.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Knox JP. In situ analysis of pectic polysaccharides in seed mucilage and at the root surface of Arabidopsis thaliana. Planta. 2001;213:37–44. doi: 10.1007/s004250000481. [DOI] [PubMed] [Google Scholar]
- Wolfson G, Chishold J, Tashjian AH, Jr, Fish S, Abeles RH. Actions of neplanocin A on pituitary cells. J Biol Chem. 1986;261:4492–4498. [PubMed] [Google Scholar]
- Zhang C, Guinel F, Moffatt BA (2002) A comparative ultrastructural study of pollen development in Arabidopsis thaliana ecotype Columbia and the male sterile mutant apt 1-3. Protoplasma (in press) [DOI] [PubMed]