Abstract
Many reports now describe the manipulation of plant metabolism by suppressing the expression of single genes. The potential of such work could be greatly expanded if multiple genes could be coordinately suppressed. In the work presented here, we test a novel method for achieving this by using single chimeric constructs incorporating partial sense sequences for multiple genes to target suppression of two or three lignin biosynthetic enzymes. We compare this method with a more conventional approach to achieving the same end by crossing plants harboring different antisense transgenes. Our results indicate that crossing antisense plants is less straightforward and predictable in outcome than anticipated. Most progeny had higher levels of target enzyme activity than predicted and had lost the expected modifications to lignin structure. In comparison, plants transformed with the chimeric partial sense constructs had more consistent high level suppression of target enzymes and had significant changes to lignin content, structure, and composition. It was possible to suppress three target genes coordinately using a single chimeric construct. Our results indicate that chimeric silencing constructs offer great potential for the rapid and coordinate suppression of multiple genes on diverse biochemical pathways and that the technique therefore deserves to be adopted by other researchers.
The directed engineering of plant metabolism has been a major focus for both academic and applied plant research in recent years. An enormous amount of work describes the use of transgenic technologies to manipulate genes on important biochemical pathways. In the vast majority of cases, single genes have been manipulated, often by down-regulating their activity using antisense RNA or cosuppression. Although this work has been extremely successful and illuminating, full exploitation of the potential for plant metabolic engineering will likely necessitate the manipulation of multiple genes.
We have been researching the lignin biosynthetic pathway for many years and have encountered a number of situations where it would be desirable to manipulate the expression of multiple genes in a coordinate fashion. A major aim of our research is to understand how the lignin biosynthetic pathway operates in vivo so as to elucidate fundamental questions such as the exact sequence and identity of enzymes involved. Although the basic pathway was outlined many years ago, recent data have prompted some revisions and multiple alternative routes have been suggested for the synthesis of certain intermediates. Lignin has significant commercial importance and modified-lignin transgenics can also provide improved raw materials for industrial and agricultural uses. Manipulating combinations of genes involved in monolignol biosynthesis offers the best potential for gaining additional evidence on the organization of the pathway in planta and may give rise to novel, commercially valuable lignins.
A small number of reports recently have described the combined suppression of two unrelated lignin genes. Crossing tobacco (Nicotiana tabacum cv Samsun NN) parents harboring different antisense transgenes provided an apparently simple method for producing progeny suppressed in the activity of two target genes (Chabannes et al., 2001; Pinçon et al., 2001). Retransformation has also proved to be a useful strategy in tobacco (Zhong et al., 1998) and particularly in species where combining transgenes by sexual crossing is impossible or impractical, such as trees (Lapierre et al., 1999). However both sexual crossing and retransformation are time-consuming strategies that take a number of plant generations to complete. Although plants down-regulated in two genes are ultimately produced, the two transgenes involved are not linked and will segregate in subsequent progeny generations. Maintaining plant lines suppressed in both target genes becomes labor intensive and this effectively limits the number of transgenes that can be combined in this way (for discussion, see Halpin et al., 2001). An alternative strategy using a single chimeric transgene to simultaneously effect suppression of two non-homologous genes was described over 8 years ago for tomato (Lycopersicon esculentum). This strategy was rather inadvertently discovered in an attempt to test the role of the presequence of an enzyme involved in fruit ripening, polygalacturonase (PG). A chimeric DNA construct was assembled where the presequence for PG was attached to the mature protein sequence of pectin esterase. When expressed in tomato fruit from the 35S cauliflower mosaic virus (CaMV) promoter, this construct unexpectedly suppressed expression of both PG and pectin esterase (Seymour et al., 1993). Despite further exploration of this phenomenon by the same research group (Jones et al., 1998), the strategy has not been adopted by others.
In this report, we test this alternative strategy to determine whether it can be effective for the suppression of combinations of genes on another pathway (lignin biosynthesis) and in another species (tobacco). We also extend the strategy by determining whether a single chimeric transgene can be used to suppress the expression of three genes simultaneously. Single chimeric transgenes were produced by fusing partial sense sequences for all possible combinations of two, or of all three, of the lignin genes caffeate/5-hydroxyferulate O-methyltransferase (COMT), cinnamoyl-coenzyme A reductase (CCR), and cinnamyl alcohol dehydrogenase (CAD). The transgenes were placed under the control of the 35S CaMV promoter. Results from this experiment are compared with those obtained when sexual crossing was used as a means to combine distinct antisense transgenes for the same target genes. Our results demonstrate that single chimeric transgenes work in tobacco, as they do in tomato, to effect simultaneous suppression of multiple target genes. This strategy can likely be applied universally to achieve more effective, and far more rapid, down-regulation of combinations of genes than can be achieved by conventional methods such as sexual crossing.
RESULTS
Conventional Crossing of Antisense Plants
We expected that crossing plants carrying different antisense transgenes would provide a quick and easy method of obtaining plants modified in the activity of more than one gene. Tobacco plants down-regulated in CAD (Halpin et al., 1994) or COMT (Atanassova et al., 1995) were crossed to obtain plants simultaneously suppressed in both genes. The parent lines had shown stable high level suppression of target gene expression through a number of generations and were homozygous for a single transgenic locus (see Table I). Two CAD reduced lines (J40 and J48) were selected and crossed with a COMT down-regulated line (B10) using each plant as both male and female in reciprocal crosses.
Table I.
Previously characterized CAD and COMT antisense plant lines
| Line | Activity of Target Enzyme
|
Transgenic Loci | Transgene Copy No. | |
|---|---|---|---|---|
| Hemizygotea | Homozygoteb | |||
| % wt | ||||
| COMT B10 | 8 | 5 | 1 | 3 |
| CAD J40 | 20–30 | 15–20 | 1 | 1 |
| CAD J48 | 50 | 20–30 | 1 | 1 |
wt, Wild type.
Hemizygous lines were obtained by out-crossing homozygous lines to wild type.
Homozygous lines were used as parents in the crosses described here.
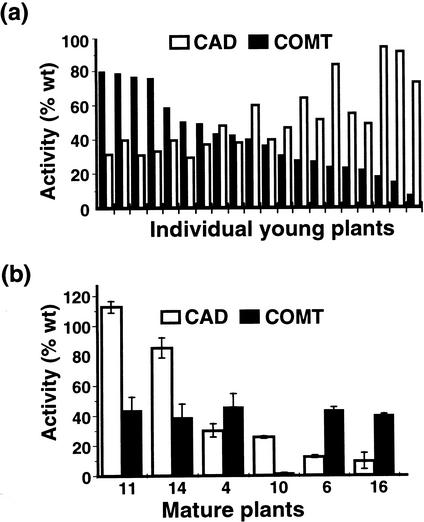
Twenty, tissue culture-grown, 6-week-old progeny from each cross were assayed for CAD and COMT activity along with 20 wild-type plants. It was surprising that activity for each enzyme varied from 8% to 100% of average wild-type values in individual progeny, a result very different from the uniform 8% COMT activity and 20% to 30% CAD activity that was expected in these hemizygous plants (see Table I). Combining the two transgenes had apparently destabilized the previously stable suppression of both CAD and COMT target genes. The data for one progeny population is shown in Figure 1a, but exactly the same variable pattern of activity was observed in every cross. In total, 80 progeny were assayed and none displayed coordinate suppression of both CAD and COMT to levels below 40% of wild-type values. Few plants displayed suppression of either target gene to the level expected in plants hemizygous for that transgene.
Figure 1.
Target enzyme activity in progeny of the cross between COMT B10 and CAD J40. The activities of CAD and COMT were measured in wild-type plants to determine the normal mean activity (100%) for each enzyme (n = 20, young plants; n = 3, mature plants). CAD and COMT activities assayed in individual young (a) and mature (b) progeny of the cross were expressed as a percentage of this mean wild-type value. Results in a are shown with individual plants ranked from highest (left) to lowest (right) CAD activity. Assays on mature plants were performed on three clonal plants per line and error bars indicate the se between assays.
To determine whether the observed lack of coordinate suppression of CAD and COMT would also be evident in adult plants, progeny from one cross (♂B10 × ♀J40) were grown in the greenhouse for 10 weeks. Assays for CAD and COMT showed that CAD activity was highly variable between plants (10%–100% of wild-type values), whereas COMT activity was consistently reduced to around 40%, except for plant 10, which had only 2% of residual COMT activity (Fig. 1b). Thus suppression of two lignin genes by crossing parents harboring different antisense transgenes gave unpredictable results and was only moderately successful.
Chimeric Partial Sense Transgenes
To test the alternative strategy of using chimeric partial sense constructs to suppress the expression of multiple lignin genes, four different constructs were made. Each construct incorporated partial sense sequences of two, or all three, of the lignin genes CAD, COMT, and CCR, expressed from the 35S CaMV promoter (Fig. 2). After transformation into tobacco, 14 to 35 independent transgenic plants were produced for each construct and grown for 6 weeks before determining CAD and COMT levels by enzyme assay and western blotting. CCR expression was monitored routinely by northern blotting, with enzyme assays being performed on just a few samples (neither the radiolabeled assay substrate nor CCR-specific antibodies are easily available).
Figure 2.
Chimeric constructs for suppressing multiple lignin biosynthetic genes. Constructs were prepared by fusing together partial sense sequences for CAD, COMT and CCR, expressing these from the 35S CaMV promoter. Nos 3′ indicates the nos terminator.
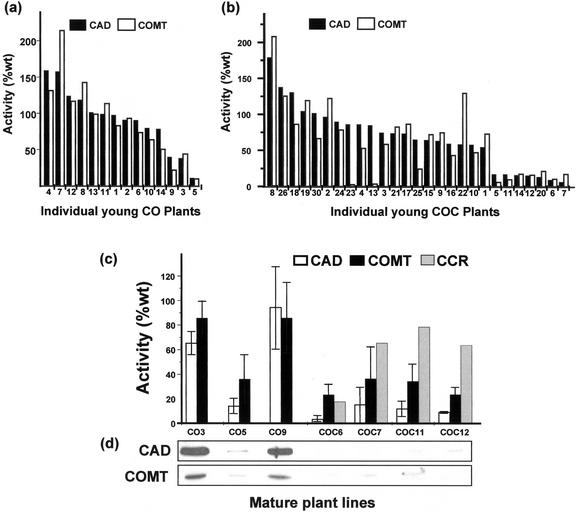
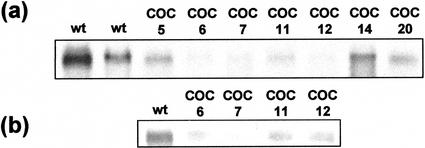
Results on target gene suppression were similar in every population and, in each case, plants coordinately suppressed in all target genes were identified. Detailed results for only two populations (CAD-COMT and CAD-COMT-CCR) are presented here. The activities of CAD and COMT measured in wild-type plants were averaged to provide a mean “control” activity (100%) for each enzyme, with ses of 14.2% for CAD, and 9.5% for COMT (n = 10 plants). Plants transformed with the CAD-COMT construct (hereafter referred to as CO plants) displayed a range of activities for the target enzymes but three plants (plants CO 3, 5, and 9) showed levels of activity well below that of wild-type plants (Fig. 3a). In any given plant, both target enzymes were suppressed to similar levels, indicating coordinate down-regulation. The larger population transformed with the CAD-COMT-CCR construct (hereafter referred to as COC plants) contained a similar proportion of down-regulated plants. Seven plants (plants COC 5, 6, 7, 11, 12, 14, and 20) displayed CAD and COMT activity considerably below wild-type values (Fig. 3b). Again, down-regulation appeared to be coordinate, with the exception of two plants (plants COC 23 and 13). A northern blot of total RNA from the same plants was hybridized with a riboprobe that recognized the 1.3-kb CCR mRNA in wild-type plants (Fig. 4a). COC plants that had shown significant down-regulation of CAD and COMT all showed considerably reduced signals for CCR mRNA, with plants COC6, COC7, and COC12 showing particularly low expression.
Figure 3.
Target enzyme activity in plants transformed with CO or COC partial sense chimeric constructs. The activities of CAD and COMT were measured in wild-type plants to determine the normal mean activity (100%) for each enzyme (n = 10, young plants; n = 3, mature plants). CAD and COMT activities were assayed in individual young CO (a) or COC (b) plants. Selected plants were clonally propagated to yield small populations (“lines”) of genetically identical individuals. c, Clonal plant lines were grown to maturity and assayed for CAD, COMT, and CCR (as appropriate). Error bars in c indicate the se between assays on different clonal individuals of the same plant line. d, Enzyme assay data for CAD and COMT activity in mature plants were confirmed by western blotting.
Figure 4.
Northern blots of COC lines to monitor CCR expression. Total RNA from young (a) and mature (b) plants transformed with the CAD-COMT-CCR construct was subjected to northern blotting and hybridized to a CCR riboprobe. wt, Wild-type plant.
Target Enzyme Activity in Mature Plants Harboring Chimeric Transgenes
Three plants (CO3, CO5, and CO9) from the CAD-COMT population, and four plants (COC6, COC7, COC11, and COC12) from the CAD-COMT-CCR population were selected for further study. These were clonally propagated to yield small populations or “lines” of genetically identical individuals that were grown to maturity in the greenhouse (10 weeks). Assays for CAD and COMT were performed on all plants, whereas one plant from each line was assayed for CCR activity (because of limited substrate availability). Results showed that some lines had maintained very significant levels of target gene suppression throughout development. Line CO5 had only 15% of wild-type CAD activity and 36% of wild-type COMT activity at maturity (Fig. 3c). Suppression of CAD and COMT in mature CO3 plants was less efficient than it had been in young plants, whereas mature CO9 plants showed no significant enzyme suppression. Suppression of target gene activity was better maintained in the CAD-COMT-CCR populations. Line COC6 stood out as having substantial reductions in all three enzymes, with CAD reduced to 4%, COMT to 24%, and CCR to 18% of wild-type levels (Fig. 3c). CAD was reduced to below 15% in all the lines, whereas residual COMT activity varied between 24% and 35%. Western blots confirmed this assay data (Fig. 3d). On the basis of the single CCR assay performed for each line, CCR down-regulation appeared to be less consistent, with typical activities of around 70% of wild-type levels (except for line COC6). However, other data indicated that CCR was significantly suppressed in both COC6 and COC7. Northern blots showed little detectable CCR transcript in mature COC6 and COC7 plants (Fig. 4b). These lines also had serious growth defects similar to, but more severe than, those previously described for CCR-suppressed plants (Piquemal et al., 1998). All COC6 and COC7 plants were stunted and could not support themselves as they grew (Fig. 5d), whereas other transgenic lines grew normally. CO primary transformants were shorter than wild type (Fig. 5, a and c), but supported themselves normally and grew to wild-type heights in subsequent generations, similar to plants of the B10 × J40 cross (Fig. 5b).
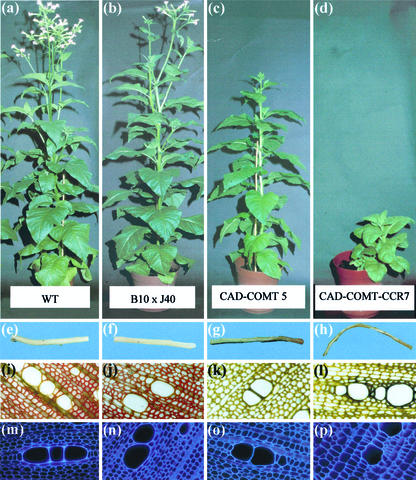
Figure 5.
Phenotypes and histochemistry of transgenics. Relative heights of plants at maturity are shown in a through d. The color of woody stem sections (with bark and cortex removed) are shown in e through h. Results of Maüle staining (i–l) and UV autofluorescence (m–p) of thin-stem sections is shown. Wild-type plants are shown in column a through m, B10 × CAD J40 are shown in column b through n, CO5 plants are shown in column c through o, and COC7 plants are shown in column d through p.
Wood Characteristics and Histochemistry
Wood from the most promising B10 × J40 plants (plants 6, 10, and 16) and from the CO5, COC6, and COC7 lines, was examined for evidence of the effects of CAD, COMT, or CCR suppression on lignin composition and structure.
Significant reduction of CAD activity has been shown previously to result in the production of red colored wood (Halpin et al., 1994, 1998; Baucher et al., 1996, 1999; MacKay et al., 1997), thought to be because of the accumulation of cinnamyl aldehydes in lignin (Higuchi et al., 1994). Wood from progeny of the B10 × J40 cross looked like wild type (Fig. 5f), supporting the assay data indicating only moderate CAD reduction and suggesting that there is little incorporation of cinnamyl aldehydes into lignin in these plants. In contrast CO5, COC6 and COC7 plants had bronze-colored wood, often with bright-red patches at the base (Fig. 5, g and h). This is consistent with good levels of CAD suppression and associated changes to lignin composition that include the incorporation of cinnamyl aldehydes. The fact that much of the wood was bronze rather than red reflects the influence of concomitant suppression of COMT and, in the case of COC plants, CCR. Severe suppression of CCR alone is also known to influence color of wood in tobacco (Piquemal et al., 1998).
Plants with severely suppressed COMT activity produce greatly reduced amounts of syringyl (S) lignin monomers, and this can be detected by specific staining of S units with Maüle reagent (Atanassova et al., 1995). To determine whether our plants had changes to S lignin, basal stem cross sections were stained with Maüle reagent. Lignin in the woody xylem of wild-type plants stained a bright red, as did the xylem of progeny of the B10 × J40 cross (Fig. 5, i and j), suggesting similar levels of S lignin units. In contrast, lignin in CO5 and COC7 showed greatly reduced staining, indicating a severe reduction in S lignin units (Fig. 5, k and l), confirming modifications to lignin to be consistent with significant COMT suppression.
The UV autofluorescence of lignin in xylem sections has been used previously to detect a “deformed” xylem vessel phenotype in plants with severe CCR down-regulation (Piquemal et al., 1998). Figure 5 shows that vessels in sections from wild-type, B10 × J40, and CO5 plants look normal (Fig. 5, m–o) but those from COC7 show irregular cell walls and appear similar to the “deformed” vessels described by Piquemal et al., (1998). This result further indicates that CCR is severely suppressed in these plants and suggests that lignin content in vessel cell walls may be reduced.
These results on wood characteristics and histochemistry suggest that plants of the CO5, COC6, and COC7 lines have major changes to lignin composition and structure, consistent with significant suppression of CAD, COMT, and, in COC lines, CCR activities. Conversely, progeny of the B10 × J40 cross showed no evidence of gross changes to lignin composition.
Lignin Structure Evaluated by Thioacidolysis Experiments
Histochemical methods are fairly crude and will only detect major alterations to lignin. To complete our characterization of the cross progeny, we wanted to determine whether combining transgenes by this method had been partially successful, causing subtle changes to lignin. Progeny of the cross and their parents therefore were subjected to thioacidolysis, an analytical degradation method providing information on lignin structure. Thioacidolysis specifically generates guaiacyl (G) and S monomers from G and S lignin units that are only involved in β-O-4 bonds. The yield of these monomers therefore reflects the frequency of β-O-4 linked lignin units. In agreement with previous work (Halpin et al., 1994; Atanassova et al., 1995), thioacidolysis revealed changes in lignin structure in both of the parent lines, with monomer yields markedly reduced because of an increase in the proportion of resistant interunit condensed bonds in lignin (Table II). The COMT B10 parent also showed the expected massive decrease in β-O-4-linked S units and the accumulation in lignin of unusual 5-OH G units that accounted for more than 5% of the total lignin-derived monomers (Table II). All of these structural changes were lost in the progeny of the B10 × J40 cross, which showed thioacidolysis yields and profiles similar to wild-type plants.
Table II.
Analysis of lignin-derived monomers by thioacidolysis
| Line | Total Yield | G/S/5-OH G |
|---|---|---|
| Wild type | 1,996 | 53/46.5/0.5 |
| 1,830 | 55/44.5/0.5 | |
| 1,810 | 52/47.5/0.5 | |
| CAD J40 | 1,555* | 67/32.6/0.4* |
| 1,441* | 65/34.6/0.4* | |
| 1,428* | 67/32.7/0.3* | |
| COMT B10 | 1,289* | 85/10/5* |
| 1,269* | 85/10/5* | |
| 1,222* | 83/10/7* | |
| ♂B10 × ♀J40 | 1,787 | 52/47.5/0.5 |
| 1,732 | 54/45.4/0.6 | |
| 1,778 | 59/40.2/0.8 |
Thioacidolysis was performed on extract-free xylem cell wall material from mature plants of the parental and ♂B10 × ♀J40 cross. The total yield and relative frequency in G, S, and 5-OH G monomers were determined for three clonal plants per line. The se between duplicate thioacidolysis experiments run on the same sample is lower than 5%. An asterisk indicates data that are significantly different (P < 0.05) from data of wild-type plants as determined by the Student's t test.
The fact that the cross progeny did not maintain the structural changes to lignin present in the parental lines was further confirmed by the following observation. Plants with less than 30% of normal CAD activity have marked increases in lignin solubility in alkali because of a specific structural change—the enrichment in free phenolic groups (Lapierre et al., 1999). Mild alkaline hydrolysis confirmed the presence of this modification in the parental CADJ40 line (49% of lignin solubilized), whereas lignin from progeny of the B10 × J40 cross had similar alkali solubility (29% of lignin solubilized) to wild-type plants (27% of lignin solubilized). This further illustrates the absence of significant structural changes to lignin in these plants.
Lignin Determination
To complete the analysis of potential lignin modifications in our transgenics, total lignin contents were estimated by two techniques using acetyl bromide and Klason lignin procedures. Because the plants produced by conventional crossing and those transformed with the chimeric partial sense constructs were grown in the greenhouse at slightly different times, each must be compared with its own set of concomitantly grown wild-type plants (Table III). The results showed that, consistent with previous data (Halpin et al., 1994), suppression of CAD or COMT alone (in parental genotypes CAD J40 and COMT B10) did not change lignin content to any great extent. Similarly, selected progeny of the COMTB10 × CADJ40 cross (plants 6, 10, and 16) had normal levels of lignin. Plant lines CO3 and CO9, which had only moderate reduction of CAD and COMT at maturity, also had normal levels of lignin. In contrast, line CO5, the line with most severe and consistent suppression of CAD and COMT, had dramatically reduced lignin content. The amount of lignin was reduced by 20% to 30%, depending on the method used for lignin determination. This confirms the superior efficiency of the chimeric construct strategy for achieving stable suppression of multiple genes and indicates a synergistic action of coordinate CAD and COMT suppression in controlling flux of metabolites through the lignin pathway. Lignin analysis of plant lines of the COC population also showed changes in lignin content in lines COC6 and COC7, the plants with stunted phenotypes. Lines COC11 and COC12, the lines that grew normally and showed higher levels of CCR transcript at maturity (see Fig. 4), showed no significant change in lignin.
Table III.
Lignin contents of extract-free mature plants
| Line | Acetyl Bromide Lignin | Klason Lignin |
|---|---|---|
| % wt | ||
| Wild type | 19.2 | 19.5 ± 0.28 |
| CAD J40 | 18.3 | 18.8 ± 0.29 |
| COMT B10 | 18.3 | 17.3 ± 0.20 |
| ♂B10 × ♀J40 | 19.4 | 19.5 ± 0.13 |
| Wild type | 21.18 ± 0.78 | 19.26 ± 0.07 |
| CO 9 | 20.8 ± 1.31 | nd |
| CO 3 | 23.25 ± 1.48 | nd |
| CO 5 | 17.0 ± 1.71 | 13.8 ± 0.81 |
| COC 6 | 18.63 ± 0.83 | 16.28 ± 0.18 |
| COC 7 | 19.54 ± 0.32 | 17.83 ± 0.36 |
| COC 11 | 20.74 ± 0.95 | nd |
| COC 12 | 19.01 ± 1.23 | nd |
Data are shown for the ♂B10 × ♀J40 cross and parental genotypes (upper part of Table) and for the CAD-COMT (CO)- and CAD-COMT-CCR (COC)-transformed plants (lower part of Table). Lignin contents were determined by two independent methods according to acetyl bromide and Klason procedures carried out on extract-free samples. Data that are significantly different from data of wild-type plants as determined by the Student's t test are indicated by asterisks (*, P < 0.05; **, P < 0.1; n = 3). nd, Not determined. ses reflect the variability between individual plants of each line.
DISCUSSION
The work presented here clearly demonstrates the superiority of chimeric partial sense constructs over conventional methods such as sexual crossing for rapid production of plants coordinately suppressed in multiple genes. Plants with reduced activity of two lignin biosynthetic genes, CAD and COMT, were produced by two methods: (a) sexual crossing of parents harboring different antisense genes, and (b) transformation of wild-type plants with a single construct that expresses partial sense sequences for both genes from a single promoter. Crossing of CAD and COMT antisense plants was only partially successful. Although some plants were identified that had reasonable levels of suppression of both target genes, most plants had much higher activity than expected. Analysis of lignin content and structure in these plants showed that there were no significant changes to lignin. In contrast, plants transformed with chimeric partial sense constructs had more consistent suppression of target enzymes and had significant changes to lignin content and structure. The orientation of the target sequences within chimeric transgenes is apparently not important and chimeric antisense constructs work equally well (M. Legrand, personal communication). Moreover, our work shows that it is possible to suppress three, or possibly more, genes coordinately using chimeric transgenes. A chimeric partial sense construct targeted to down-regulate the expression of three lignin genes, CAD, COMT, and CCR, was successful in suppressing all three targets and gave rise to plants with modified lignin and severe phenotypes.
The problems we encountered in obtaining double antisense plants by crossing were unexpected. We assumed that, using well-characterized homozygous parents with previously consistent transgene expression, we could expect progeny to have similar levels of activity to hemizygous plants of the parental genotypes (i.e. 8% COMT and 20%–30% CAD compared with wild-type activity). However, none of the progeny had such low enzyme levels and activities were highly variable across the population, particularly in young plants. In mature plants, suppression of COMT was more stable with most plants having 40% of wild-type activity (still considerably higher than the 8% expected and too high to modify lignin), but CAD activity remained variable. These data suggest that the efficacy of suppressing transgenes can be modified by genetic background, particularly when a transgene is introduced into background already containing another, partly homologous, transgene. This phenomenon, particularly the variability of target enzyme activity between genetically identical progeny of the same cross, has many of the hallmarks of transgene silencing. One plausible explanation for our data, therefore, is that silencing of both antisense transgenes has been triggered by combining them. The degree of silencing could vary in different individual progeny, giving rise to widely different levels of target enzyme activity in genetically identical plants. Silencing might be exacerbated by the repeated use of the 35S CaMV promoter on both transgenes in our experiments.
The problem of silencing is not unique to the B10 × J40 cross. We have substituted a different CAD antisense line (CADJ48) and different COMT-suppressed lines (COMTA17 and COMTD6) as parents in crosses and in every case found a variable pattern of target gene suppression in young progeny. No cross yielded progeny with predictable levels of activity for both target enzymes. In most cases, enzyme activity was higher than expected, but in one case (CADJ48 × COMTA17) activity for CAD was lower than expected, whereas COMT activity varied. Many of the crossing experiments described here were carried out in tandem in two different laboratories in Dundee and Strasbourg with similar results, suggesting that these phenomena are triggered by genetic rather than environmental factors.
In general, successful experiments where transgenes have been combined by crossing, have involved protein-coding transgenes, and usually the levels of transgene expression in progeny have not been investigated (e.g. Nawrath et al., 1994; Ma et al., 1995). It is likely that combining transgenes aimed at suppressing target plant genes is always going to be more difficult than combining protein-coding genes. Effective suppression is only ever seen in a small proportion of transformants because target gene activity often has to be severely reduced (by 80%–90% or more) before physiological changes become evident. As a consequence, even partial loss of transgene expression, where only a small proportion of target gene activity is restored, can result in loss of the physiological effect of the transgene. This point is further illustrated by independent experiments where other lignin-modified antisense plants have recently been crossed to generate plants suppressed in two activities (Chabannes et al., 2001; Pinçon et al., 2001). Although suppression of target enzymes was more marked and more stable than in the crossing experiment reported here, in both cases levels of one target enzyme were significantly higher than anticipated for hemizygous plants. Consequently, progeny had a qualitative lignin profile similar to wild-type plants (Chabannes et al., 2001) or to plants suppressed in one gene alone (Pinçon et al., 2001). In the crossing experiments described here, the higher-than-predicted levels of both target enzymes in progeny resulted in complete loss of the expected modifications to lignin. The problem of reduced efficacy of suppressing transgenes after crossing may potentially be alleviated by using double-stranded RNAs or hairpin-forming RNAs as recently described (Chuang and Meyerowitz, 2000; Wesley et al., 2001). These types of transgene are apparently more efficient (in the numbers of gene-suppressed plants recovered) and more effective (in the degree of suppression obtained) than conventional antisense or sense gene-silencing constructs. To date, they have only been used to suppress single genes in plants, but it seems likely that they will also prove invaluable in multiple gene suppression, perhaps even incorporating chimeric transgenes like those described here.
In terms of enzyme activity, plants harboring chimeric partial sense transgenes showed greater degrees of down-regulation of CAD and COMT than the corresponding plants produced by crossing. At maturity, line CO5 had similar levels of enzyme activity to some of the cross progeny, but it had much lower levels of both enzymes (less than 10%) at earlier developmental stages (see Fig. 3a). The very significant changes to lignin content in this line suggest that these low levels of activity were maintained for much of plant development. In contrast, young progeny of the cross had variable but relatively high levels of activity for target genes that only stabilized at a lower level in mature plants, perhaps too late to have an impact on lignin synthesis. Lignin in CO5 plants also had significantly altered structure. Individual plants had a strong red-brown color at the base of stems, indicative of the incorporation of cinnamyl aldehydes into the polymer and characteristic of severe CAD suppression (Halpin et al., 1994; Higuchi et al., 1994; Stewart et al., 1997; Ralph et al., 1998). The effects of COMT suppression were evident in the reduced staining with Maüle reagent indicative of a marked decrease in S lignin. Both of these structural changes were also clearly revealed by thioacidolysis analysis of CO5 lignin (data not shown). The COC lines 6 and 7 showed similar lignin changes as a consequence of CAD and COMT down-regulation and additionally showed growth defects and deformed xylem vessels, both phenotypes associated with severe CCR suppression (Piquemal et al., 1998). These data suggest that lignin in the plants suppressed in two or three lignin genes contains a combination of the modifications characteristic of suppression of activity of each target enzyme.
The significant (20%–30%) reduction in lignin content in CO5 plants was unexpected because suppression of CAD or COMT alone does not cause substantial alterations to lignin content. This result suggests some synergistic effect, when CAD and COMT are suppressed together, to inhibit flux through the lignin pathway. Thus, suppressing combinations of genes can induce novel modifications to lignin that cannot be predicted by reference to existing data on the suppression of single genes. These new combinations of modifications to lignin composition, structure, and content in phenotypically normal plants may offer potential for improving plant raw materials for specific industrial and agricultural purposes. In contrast with the data on CO5 plants, reductions in lignin content in the lines COC6 (18% reduction) and COC7 (8% reduction) were expected because severe CCR suppression reduces lignin content. It is perhaps surprising that lignin content is not more substantially altered in these lines, given their severe phenotypes. The very big reductions in the amount of xylem tissue produced by these plants (data not shown) suggests a greater inhibition of lignin production than is obvious from the lignin content data alone. Further work is currently under way to investigate this point further and to more comprehensively analyze all the changes to lignin structure in CO5, COC6, and COC7 plants.
Overall, our results support the findings of previous work by the group that initially described and used the chimeric transgene system for the manipulation of fruit ripening genes (Seymour et al., 1993; Jones et al., 1998). By extending this work and successfully using the system to manipulate lignin biosynthesis, a pathway quite distinct from fruit ripening, we illustrate that the technique should be applicable to the manipulation of any combinations of genes on any pathway. The strategy has many advantages over conventional methods of combining transgenes by crossing, cotransformation, or retransformation. It is rapid because plants suppressed in multiple genes can be produced in one generation, and subsequent breeding to produce homozygotes is simpler than it is with methods where combined transgenes can resegregate. Most importantly, linking the suppression sequences in a single transcriptional unit ensures coordinate expression while avoiding duplication of regulatory sequences such as promoters, thereby reducing the likelihood of transgene silencing and favoring stable transgene expression. Chimeric constructs therefore offer great potential for the rapid and coordinate suppression of multiple genes and the technique deserves to be more widely adopted by other researchers.
MATERIALS AND METHODS
Crossing of CAD and COMT Antisense Plants
CAD and COMT antisense transgenes were combined in tobacco (Nicotiana tabacum cv Samsun NN) plants by crossing homozygous parents carrying CAD (Halpin et al., 1994) or COMT (Atanassova et al., 1995) antisense transgenes. Each parent was used as male and female partner in reciprocal crosses.
Chimeric Constructs
Oligonucleotide primers were used in polymerase chain reactions to amplify appropriate sections of the tobacco CAD, COMT, and CCR cDNAs while at the same time engineering convenient unique restriction sites (XmaI or XbaI) onto the ends of these fragments. Using these methods, a 987-bp CAD fragment, a 902-bp COMT fragment, and a 622-bp CCR were generated and assembled together in appropriate combinations in a pUC19-based cloning vector (pJR19) using the PCR-engineered restriction sites. pJR19 already had the 35S CaMV promoter inserted between the EcoRI and KpnI sites of the pUC19 polylinker, whereas the nos 3′ terminator had been inserted between the PstI and HindIII sites. As a consequence, once the cDNA fragments had been assembled between the 35S promoter and nos3′ terminator in sense orientation, the entire EcoRI-HindIII plant expression cassette could be moved over into a Bin19-based plant transformation vector in one step. Using these techniques, four chimeric constructs were prepared (see Fig. 2) and used for Agrobacterium tumefaciens-mediated transformation of tobacco seedlings using the method of Tinland et al. (1995).
Extracts and Enzyme Assays
Basal stem sections (1–2 cm) were either homogenized in a microfuge tube using a micropestle (tissue culture plants) or frozen in liquid N2, crushed, and ground using a mortar and pestle (greenhouse plants) then extracted in 300 μL to 2.5 mL of extraction buffer (100 mm Tris-HCl, pH 7.5; 20 mm β-mercaptoethanol; and 5% [w/v] polyvinylpolypyrrolidone). Extracts were clarified by centrifugation twice (21,000g; 4°C) before use. Protein concentrations were determined using the Bio-Rad reagent (Bio-Rad Laboratories, Hemel Hempstead, Herts, UK).
CAD assays were carried out in 1 mL of 100 mm Tris-HCl pH 8.8, containing 50 to 100 μL of plant extract, 0.1 mm coniferyl alcohol, and 0.02 mm NADP and monitored at 400 nm over 15 min at 30°C. COMT was assayed by the protocol of Fukuda and Komamine (1982) with minor modifications. CCR assays (350 μL) contained 100 mm KH2PO4-Na2HPO4, pH 6.25; 5 mm dithiothreitol; 100 μm NADPH; 30 μm 14C feruloyl coenzyme A (13,000 dpm); and 30 μL of plant extract. Control reactions were carried out by omitting NADPH. Reactions were incubated at 30°C for 10 min, stopped with 1 mL of ethyl acetate, vortexed, and partitioned by centrifugation. Five hundred microliters of the organic phase was counted in 4.5 mL of Ecoscint scintillator (National Diagnostics, Hull, Herts, UK).
Histochemistry
Hand sections of woody xylem were made using a razor blade. For UV autofluorescence, sections were observed at an excitation wavelength of 340 to 380 nm using a 430-nm barrier filter. Phloroglucinol staining was performed by mixing two parts 2% (w/v) phloroglucinol in 95% (v/v) ethanol with one part concentrated HCl, and adding drop wise to the tissue. Sections were photographed after 2 min. Maüle staining was performed by incubating sections in 1% (w/v) potassium permanganate for 5 min, then washing twice with water before adding 3% (v/v) HCl and waiting for the color to change. Sections were washed again then immersed for 2 min in ammonium hydroxide.
Lignin Analysis
All lignin analyses were performed according to previously described protocols. Thioacidolysis and Klason analyses were carried out as described by Lapierre et al. (1995). Acetyl bromide lignin determinations were carried out according to Iiyama and Wallace (1990).
Western and Northern Blotting
Western blots were performed using standard procedures. Ten micrograms of plant extracts were separated on 12% (w/v) SDS-PAGE, then transferred to nitrocellulose membrane. Blots were probed with primary antibody (rabbit anti-CAD or anti-COMT sera at 1:10,000 or 1:5,000 [v/v] dilution) followed by HRP-conjugated anti-rabbit IgG (1:10,000) and developed using the Phototope-HRP western Blot Detection Kit (New England Biolabs, Hitchin, Herts, UK). Northern blots were performed as previously described (Halpin et al., 1998).
ACKNOWLEDGMENTS
We are grateful to Frédéric Legée for Klason lignin determinations, to Prof. Alain Boudet for the CCR assay substrate, and to Jess Searle for care of the plants.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (funding to C.H.) and by the European Communities (FAIR–CT95–0424, funding to M.L. and C.L.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010698.
LITERATURE CITED
- Atanassova R, Favet N, Martz F, Chabbert B, Tollier MT, Monties B, Fritig B, Legrand M. Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 1995;8:465–477. [Google Scholar]
- Baucher M, Vailhe MAB, Chabbert B, Besle J-M, Opsomer C, Van Montagu M, Botterman J. Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol Biol. 1999;39:437–447. doi: 10.1023/a:1006182925584. [DOI] [PubMed] [Google Scholar]
- Baucher M, Chabbert B, Pilate G, VanDoorsselaere J, Tollier MT, PetitConil M, Cornu D, Monties B, VanMontagu M, Inzé D et al. Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiol. 1996;112:1479–1490. doi: 10.1104/pp.112.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabannes M, Barakate A, Lapierre C, Marita JM, Ralph J, Pean M, Danoun S, Halpin C, Grima-Pettenati J, Boudet AM. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 2001;28:257–270. doi: 10.1046/j.1365-313x.2001.01140.x. [DOI] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:4985–4990. doi: 10.1073/pnas.060034297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H, Komamine A. Lignin synthesis and its related enzymes as markers of tracheary element differentiation in single cells isolated from the mesophyll of Zinnea elegans. Planta. 1982;155:423–430. doi: 10.1007/BF00394471. [DOI] [PubMed] [Google Scholar]
- Halpin C, Barakate A, Askari BM, Abbott JC, Ryan MD (2001) Enabling technologies for manipulating multiple genes on complex pathways. Plant Mol Biol (in press) [PubMed]
- Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakate A, Foxon GA. Brown-midrib maize (bm1): a mutation affecting the cinnamyl alcohol-dehydrogenase. Plant J. 1998;14:545–553. doi: 10.1046/j.1365-313x.1998.00153.x. [DOI] [PubMed] [Google Scholar]
- Halpin C, Knight ME, Foxon GA, Campbell MM, Boudet AM, Boon JJ, Chabbert B, Tollier M-T, Schuch W. Manipulation of lignin quality by downregulation of cinnamyl alcohol dehydrogenase. Plant J. 1994;6:339–350. [Google Scholar]
- Higuchi T, Ito T, Umezawa T, Hibino T, Shibata D. Red-brown color of lignified tissues of transgenic plants with antisense CAD gene: wine-red lignin from coniferyl aldehyde. J Biotechnol. 1994;37:151–158. [Google Scholar]
- Iiyama K, Wallis AFA. Determination of lignin in herbaceous plants by an improved acetyl bromide procedure. J Sci Food Agric. 1990;51:145–161. [Google Scholar]
- Jones CG, Scothern GP, Lycett GW, Tucker GA. The effect of chimeric transgene architecture on co-ordinated gene silencing. Planta. 1998;204:499–505. [Google Scholar]
- Lapierre C, Pollet B, Petit-Conil M, Toval G, Romero J, Pilate G, Leple J-C, Boerjan W, Ferret V, De Nadai V et al. Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the effeciency of industrial craft pulping. Plant Physiol. 1999;119:153–163. doi: 10.1104/pp.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Pollet B, Rolando C. New insight into the molecular architecture of hardwood lignins by chemical degradative methods. Res Chem Intermediates. 1995;21:397–412. [Google Scholar]
- Ma JK-C, Hiatt A, Hein MB, Vine ND, Wang F, Stabila P, van Dolleweerd C, Mostov K, Lehner T. Generation and assembly of secretory antibodies in plants. Science. 1995;268:716–719. doi: 10.1126/science.7732380. [DOI] [PubMed] [Google Scholar]
- MacKay JJ, OMalley DM, Presnell T, Booker FL, Campbell MM, Whetten RW, Sederoff RR. Inheritance, gene expression, and lignin characterization in a mutant pine deficient in cinnamyl alcohol dehydrogenase. Proc Natl Acad Sci USA. 1997;94:8255–8260. doi: 10.1073/pnas.94.15.8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Poirier Y, Somerville C. Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation. Proc Natl Acad Sci USA. 1994;91:12760–12764. doi: 10.1073/pnas.91.26.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinçon G, Chabannes M, Lapierre C, Pollet B, Ruel K, Joseleau J-P, Boudet A-M, Legrand M. Simultaneous down-regulation of caffeic/5-hydroxy ferulic acid-O-methyltransferase I and cinnamoyl-coenzyme A reductase in the progeny from a cross between tobacco lines homozygous for each transgene: consequences for plant development and lignin synthesis. Plant Physiol. 2001;126:145–155. doi: 10.1104/pp.126.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal J, Lapierre C, Myton K, O'Connell A, Schuch W, Grima-Pettenati J, Boudet AM. Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 1998;13:71–83. [Google Scholar]
- Ralph J, Hatfield RD, Piquemal J, Yahiaoui N, Pean M, Lapierre C, Boudet AM. NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamyl-alcohol dehydrogenase and cinnamoyl-CoA reductase. Proc Natl Acad Sci USA. 1998;95:12803–12808. doi: 10.1073/pnas.95.22.12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Fray RG, Hill P, Tucker GA. Down-regulation of 2 nonhomologous endogenous tomato genes with a single chimeric sense gene construct. Plant Mol Biol. 1993;23:1–9. doi: 10.1007/BF00021414. [DOI] [PubMed] [Google Scholar]
- Stewart D, Yahiaoui N, McDougall GJ, Myton K, Marque C, Boudet AM, Haigh J. Fourier-transform infrared and raman spectroscopic evidence for the incorporation of cinnamaldehydes into the lignin of transgenic tobacco (Nicotiana tabacum L.) plants with reduced expression of cinnamyl alcohol dehydrogenase. Planta. 1997;201:311–318. doi: 10.1007/s004250050072. [DOI] [PubMed] [Google Scholar]
- Tinland B, Schoumacher F, Gloeckler V, Bravoangel AM, Hohn B. The Agrobacterium tumefaciens virulence D2 protein Is responsible for precise integration of T-DNA into the plant genome. EMBO J. 1995;14:3585–3595. doi: 10.1002/j.1460-2075.1995.tb07364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Zhong R, Morrison WH, Negrel J, Ye Z-H. Dual methylation pathways in lignin biosynthesis. Plant Cell. 1998;10:2033–2045. doi: 10.1105/tpc.10.12.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]