Abstract
Expansins are a group of extracellular proteins that directly modify the mechanical properties of plant cell walls, leading to turgor-driven cell extension. Within the completely sequenced Arabidopsis genome, we identified 38 expansin sequences that fall into three discrete subfamilies. Based on phylogenetic analysis and shared intron patterns, we propose a new, systematic nomenclature of Arabidopsis expansins. Further phylogenetic analysis, including expansin sequences found here in monocots, pine (Pinus radiata, Pinus taeda), fern (Regnellidium diphyllum, Marsilea quadrifolia), and moss (Physcomitrella patens) indicate that the three plant expansin subfamilies arose and began diversifying very early in, if not before, colonization of land by plants. Closely related “expansin-like” sequences were also identified in the social amoeba, Dictyostelium discoidium, suggesting that these wall-modifying proteins have a very deep evolutionary origin.
The availability of information from genome sequencing programs now offer a new route to understanding multigene families within and across different species. Several recent studies have demonstrated the usefulness of phylogenetic analysis to complement parallel investigations of gene function in vivo (Sanderfoot et al., 2000; Kellogg, 2001; Li et al., 2001; Ross et al., 2001). The present analysis makes use of the completely sequenced Arabidopsis genome (The Arabidopsis Genome Initiative, 2000), together with comprehensive searches of GenBank and expressed sequence tag (EST) databases (maintained at the National Center for Biotechnology Information, NCBI), to determine the phylogeny of the plant cell wall protein, expansin.
Expansins play a variety of roles in vivo by modifying the cell wall matrix during growth and development (for review, see Cosgrove, 2000a; Darley et al., 2001). Initially identified by their unique ability to induce the pH-dependent extension of plant cell walls in vitro (McQueen-Mason et al., 1992), expansins appear to increase polymer mobility in the cell wall, allowing the structure to slide apart during extension (McQueen-Mason et al., 1993; McQueen-Mason and Cosgrove, 1994, 1995; Whitney et al., 2000). To date, expansin remains the only protein to demonstrate cell wall extension in vitro and in vivo. In addition to roles in plant cell growth, expansins are also now believed to play key roles in the early development of leaf primordia (Fleming et al., 1997), fruit softening (Civello et al., 1999; Rose et al., 2000), plant reproduction (Cosgrove et al., 1997), and wall disassembly (Cho and Cosgrove, 2000).
Growing tissues from a wide range of plants, including dicotyledons (Rayle and Cleland, 1977), grasses (Kutschera, 1994), gymnosperms (Kim et al., 2000), and green algae (Metraux and Taiz, 1977), have been shown to undergo acid-induced extension. As it is now generally accepted that expansins are the chief agents responsible for acid-induced extension, these data suggest that expansins may be found in all land plants and probably algae. In support of this, there are now abundant reports of expansins from a variety of angiosperms (for reviewed, see Cosgrove, 2000b).
RESULTS AND DISCUSSION
The Arabidopsis Expansin Multigene Family
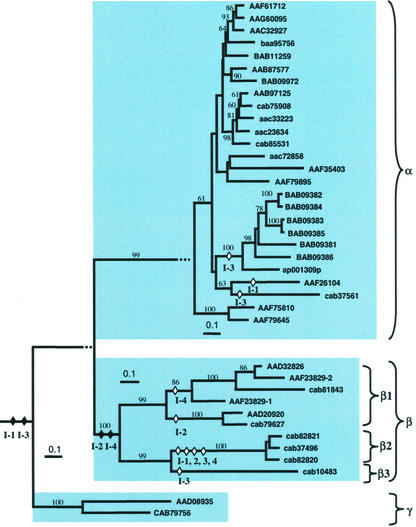
Previous classifications of the expansin gene family divided proteins into two subfamilies, α and β, based on substrate specificity and sequence similarity (Cosgrove, 1997). However, to date, classification has been based on only limited sequence information. The present study is the first to analyze all expansin-like sequences in the entire genome of a single plant species, and reveals that expansins exist as a large multigene family. Database searches revealed a total of 38 expansin or expansin-like sequences in Arabidopsis. These expansins fall into the two previously identified major subfamilies and a novel third subfamily based on overall sequence conservation and shared structural features (Fig. 1). These subfamilies are strongly supported by phylogenetic analysis (≥99% bootstrap; Fig. 2). Following convention (Cosgrove, 1999), we used Greek letters to signify these three subfamilies as α-, β-, and, γ-expansins. Two sequences were identified as pseudogenes on the basis of duplication and interruption of the open reading frame (ORF). Corrections to the database annotations of Arabidopsis expansin sequences held at NCBI are given in Table I. A majority (54%) of the 38 Arabidopsis expansins were also present as ESTs or full-length cDNAs, indicating that most of these genes are expressed at some stage of the Arabidopsis life cycle (Table II).
Figure 1.
Alignment of representative α-, β-, and γ-expansins from Arabidopsis. The shaded boxes highlight the conserved areas used in our analyses. α and β insertions within the central core of the expansins and signal sequences are indicated. Intron positions are arrowed, and residues are highlighted at the point of insertion into the predicted exon.
Figure 2.
Phylogenetic analysis of the Arabidopsis expansin gene family shows three major groups: α, β, and γ. The composite tree was derived by neighbor-joining distance analysis using ClustalX version 1.8. The main backbone of the tree was calculated with the 90-residue conserved core of amino acid positions alignable among all 38 sequences. The three subtrees were then calculated separately based on additional amino acid positions shared within each group, and were appended to the main backbone tree as indicated by broken horizontal lines. Only bootstrap values over 60% are indicated above the nodes. The major expansin subfamilies α, β, and γ are highlighted by shaded boxes. Hypothetical intron gains and losses are indicated by diamonds followed by intron number. Postulated intron gains are indicated by filled diamonds, and intron losses are indicated by unfilled diamonds. All branches are drawn to scale as indicated by the scale bar (=0.1 substitutions/site).
Table I.
Corrections to the database annotations of Arabidopsis expansin sequences
| Arabidopsis Expansin-Like Genes | Corrections |
|---|---|
| AAF26104-m, | Different intron 3 positions predicted (confirmed by EST) |
| BAB09381-m, | Different intron 1 positions predicted |
| BAB09386-m | Different first exon/intron predicted |
| AAF23829 | Two ORFs were predicted from this single ORF annotation as AAF23829-1 and AAF23829-2P (pseudogene) |
| cab81843 | Different intron 3 positions predicted |
| cab79627 | Last intron/exon were not predicted (confirmed by EST) |
| cab10483 | Different ORF predictions (confirmed by ESTs) |
| AAD08935 | Missed intron 3 and last exon (confirmed by EST) |
| CAB79756 | Different intron 1 positions predicted, missed intron 3 and last exon |
Table II.
The Arabidopsis expansin gene family
| New Annotation | Previous Name | Chromosome | Accession No. (protein) | Expressed Gene? |
|---|---|---|---|---|
| Ath-Expα-1.1 | At-EXP10 | I | AAF61712 | Yes (cDNA) |
| Ath-Expα-1.2 | At-EXP1 | I | AAG60095 | Yes (cDNA) |
| Ath-Expα-1.3 | At-EXP15 | II | AAC32927 | Yes (EST) |
| Ath-Expα-1.4 | At-EXP5 | III | BAA95756 | Yes (cDNA) |
| Ath-Expα-1.5 | At-EXP14 | V | BAB11259 | |
| Ath-Expα-1.6 | At-EXP4 | II | AAB97125 | Yes (EST) |
| Ath-Expα-1.7 | III | CAB75908 | ||
| Ath-Expα-1.8 | At-EXP6 | II | AAC33223 | Yes (cDNA) |
| Ath-Expα-1.9 | At-EXP3 | II | AAC23634 | Yes (EST) |
| Ath-Expα-1.10 | At-EXP9 | V | CAB85531 | Yes (EST) |
| Ath-Expα-1.11 | At-EXP8 | II | AAB87577 | Yes (EST) |
| Ath-Expα-1.12 | At-EXP2 | V | BAB09972 | Yes (cDNA) |
| Ath-Expα-1.13 | IV | AAC72858 | ||
| Ath-Expα-1.14 | At-EXP11 | I | AAF79895 | Yes (EST) |
| Ath-Expα-1.15 | V | BAB09382 | ||
| Ath-Expα-1.16 | V | BAB09384 | ||
| Ath-Expα-1.17 | V | BAB09383 | ||
| Ath-Expα-1.18 | V | BAB09385 | Yes? (EST) | |
| Ath-Expα-1.19 | V | BAB09386 | ||
| Ath-Expα-1.20 | V | BAB09381 | ||
| Ath-Expα-1.21p | III | AP001309 | Pseudogene | |
| Ath-Expα-1.22 | III | AAF26104 | Yes (EST) | |
| Ath-Expα-1.23 | IV | CAB37561 | ||
| Ath-Expα-1.24 | At-EXP12 | III | AAF35403 | Yes (EST) |
| Ath-Expα-1.25 | I | AAF75810 | Yes (EST) | |
| Ath-Expα-1.26 | At-EXP7 | I | AAF79645 | |
| Ath-Expβ-1.1 | II | AAD32826 | ||
| Ath-Expβ-1.2p | I | AAF23829 | Pseudogene | |
| Ath-Expβ-1.3 | III | CAB81843 | ||
| Ath-Expβ-1.4 | I | AAF23829 | ||
| Ath-Expβ-1.5 | β-Expansin | II | AAD20920 | Yes (cDNA) |
| Ath-Expβ-1.6 | IV | CAB79627 | Yes (EST) | |
| Ath-Expβ-2.1 | III | CAB82821 | Yes (EST) | |
| Ath-Expβ-2.2 | IV | CAB37496 | Yes (EST) | |
| Ath-Expβ-2.3 | III | CAB82820 | ||
| Ath-Expβ-3.1 | IV | CAB10483 | Yes (EST) | |
| Ath-Expγ-1.1 | IV | CAB79756 | ||
| Ath-Expγ-1.2 | II | AAD08935 |
Alignment of representative α-, β-, and γ-expansins (Fig. 1) clearly shows that α- and β- can be defined by characteristic “motifs” in the central domain of the predicted proteins that, based on phylogeny, are probably insertions (Fig. 2). The “α-insertion” is about 14 residues long and contains four highly conserved residues at its 3′ end, “GWCN.” The “β-insertion” is present in all β-expansins and is usually seven amino acid residues without obvious conservation, but often containing two or more charged residues. Both of these insertions are absent in γ-expansins. The major defining characteristic of the γ-group is that they terminate shortly after the conserved intron-3 exon boundary, resulting in predicted proteins lacking the C-terminal domain (alignment of all Arabidopsis expansins is available in Fig. S1 in the supplementary material).
Conserved intron patterns within genomic sequences give further general support for three distinct subfamilies. Our analyses indicate that the common ancestor of all α-, β-, and γ-expansins possessed introns at present day positions 1 and 3 (Fig. 2). α-Expansins are generally defined by the presence of intron 1 and 3; however, in some cases, they possess only a single intron (1 or 3). It is intriguing to note that within the α subfamily, apparently unrelated gene lineages have lost the same intron.
β-Expansins are generally defined by intron gains at positions 2 and 4. Although, as with α-expansins, unrelated β-expansins also appear to have lost the same intron independently (i.e. β1 and β3 have lost intron 3). The presence of an entirely intronless group (β2) is interesting. It is possible that this group arose from a single recombination event with a reverse-transcribed DNA copy of a fully spliced mRNA (Frugoli et al., 1998).
α-Expansins represent the largest subfamily within Arabidopsis, with 26 members. These form a tight distinct cluster that is separated from β and γ by a long primary branch (Fig. 2). Sequence conservation within this subfamily ranges from 52% to 99% deduced protein sequence identity. α-Expansins appear to be further divided into a number of subgroups; however, their generally high amino acid sequence identity results in short internodes and, hence, poor bootstrap support (Fig. 2).
The β-expansin subfamily of Arabidopsis is smaller than the α's, with only 10 members. These show greater sequence divergence, as indicated by branch length on the phylogenic tree (Fig. 2). Phylogenetic analysis suggests that β-expansins can be further divided into three subgroups (bootstrap values ≥ 99; Fig. 2), which we designated as β1 through β3. Intron gain and loss also support the subdivision of β-expansins. For example, all β2 sequences are devoid of introns at positions 1 to 4 (indicating two intron losses in the early evolution of this subgroup). Sequence divergence within the β-expansins may reflect a wide range of biological functions for the encoded proteins. However, as none of these proteins have yet been characterized in Arabidopsis, functional divergence can only be speculated upon.
Our analysis also revealed a small (two members) but distinct subfamily of expansin-like sequences in Arabidopsis that we have named γ-expansins. The predicted amino acid sequences of these putative proteins contain up to 49% similarity and 31% identity to α-expansins, and similar levels of similarity/identity with some β-expansins. Their identification as expansin-like is further supported by conservation of intron-exon patterns. Genomic sequence analysis of the two Arabidopsis γ-expansins reveals that the predicted position of intron 1 and intron 3 sites are similar to those for α- and β-expansins (Fig. 1). One intriguing characteristic of γ-expansins is that the reading frame of their final exon terminates one residue after the intron 3 site. The deduced amino acid sequences thus predicts that γ-expansins encode a truncated expansin-like protein.
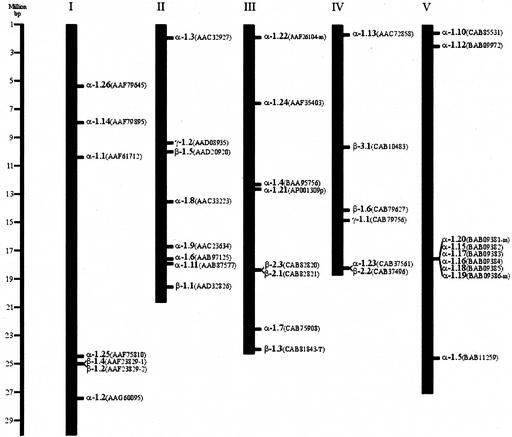
Mapping of the Arabidopsis expansins revealed that these genes are scattered throughout all five chromosomes (Fig. 3). A cluster of five α-expansin genes in tandem on chromosome V shows a high degree of sequence similarity at the nucleotide and protein level (approximately 77% identity; BAB093-82 plus −84 and BAB093-83 plus −5). This cluster most likely has evolved from a series of relatively recent unequal recombination events. The Arabidopsis Genome Initiative (2000) has mapped transpositional gene duplications during the evolution of the Arabidopsis genome. Using this data, it would appear that at least one other expansin pair (AAD20920 and CAB79627) may have arisen from recent transpositional gene duplication. This is further supported by their closeness in a terminal subgroup of the phylogenetic tree (Fig. 2).
Figure 3.
Deduced chromosomal positions of Arabidopsis expansin (Ath-Exp) genes. Genes are annotated by accession number (protein) and specific name based on the proposed nomenclature system. The positions are given according to the nearest recombinant inbred marker. Genetic distance was calculated using the proportions suggested by the Lister and Dean recombinant map (Lister and Dean, 1993).
At present, Arabidopsis expansins are named purely by their chronological clone identity. Thus, At-EXP1 and At-EXP10 represent the first and the 10th cDNAs identified from Arabidopsis, respectively. On the basis of our phylogenic analysis, we propose a systematic nomenclature for the Arabidopsis expansin gene family. We suggest that Arabidopsis expansins be identified by a three-letter species identification (Ath), followed by the group annotation (α, β, or γ) and then further identified by any subgroup into which they fall (Table II). This is consistent with recent nomenclature trends using phylogenetic classification of Arabidopsis multigene families (Lacombe et al., 2001; Li et al., 2001).
Why are there so many expansins in Arabidopsis? It is possible that each expansin (or group of expansins) is involved in wall-loosening processes, in different cell types, and in response to different stimuli. Limited biochemical data from other plant species has shown that α- and β-expansins exhibit similar rheological effects on the cell wall, but have different substrate specificities and wall-binding affinities (Cosgrove et al., 1997). Indirect evidence also hints at some cell-specific function within the Arabidopsis α-expansin family (Cho and Cosgrove, 2000), but it remains to be shown if the Arabidopsis expansins subfamilies mediate tissue-specific, or even polymer-specific, wall-loosening events. In theory, because the present expansin multigene family has arisen from ancient gene duplication events, many existing family members must have experienced strong selection pressure. This would support the hypothesis that individual expansin isoforms have specific physiological roles in vivo to avoid gene silencing (Lynch and Conery, 2000).
The functional role of the genes that we are denoting as γ-expansins is also unknown. ESTs for these putative proteins were found in Arabidopsis and a number of other plant species, indicating that these genes are widely expressed and thus presumably have some physiological function in planta. Our analysis also identified a blight-associated protein from Citrus jambhiri as a γ-expansin. This small (12 kD), soluble plant protein accumulates in the leaves and stems of plants infected with citrus blight (Ceccardi et al., 1998). Anecdotal evidence has indicated that this blight-associated protein does not exhibit wall-loosening activity in reconstitution assays (Cosgrove, 2000b). However, as a thorough analysis on a range of wall substrates is still lacking, the biological role of γ-expansins is uncertain.
Phylogenetic Analysis of Plant Expansin Gene Families
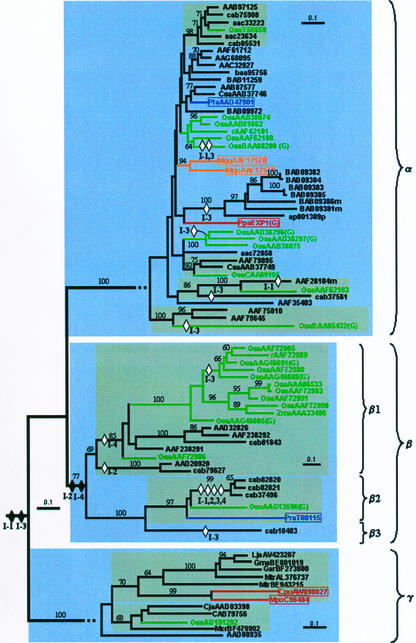
During our searches of the public databases, we also identified expansins in pine (Pinus radiata, Pinus taeda), fern (Regnellidium diphyllum, Marsilea quadrifolia), and moss (Physcomitrella patens). These sequences, along with all available rice (Oryza sativa) and several additional dicot sequences cover most of the taxonomic depth of land plants. To gain further insight into the evolution and origins of plant expansins, we included these sequences into our phylogenetic analysis. Inclusion of these sequences does not alter the basic shape of the phylogenetic tree, and continues to strongly support the division of expansins into three subfamilies (99% bootstrap; Fig. 4). Within these subfamilies, the strong amino acid sequence conservation of the α-expansins remains evident from the short branch lengths. This striking conservation of sequence over hundreds of millions of years of evolution suggests an equally remarkable conservation of function. Likewise, the inclusion of β-expansins from other plant species highlights the greater sequence divergence in this subfamily and also further supports the subdivision of β-expansins into three distinct subgroups (β1–β3; Fig. 4). To obtain a reasonable number of sequences for analysis within the γ-expansin group, it was necessary to incorporate EST data. Analysis including these data confirmed that γ-expansins are widespread throughout land plants (Fig. 4).
Figure 4.
Phylogenetic tree for the plant expansin gene family shows an ancient origin of α-, β-, and γ-expansins. Bootstrap values over 60% are indicated above the nodes. The tree was constructed as described in “Materials and Methods.” The major groups are indicated by light-shaded boxes and were strongly supported (>80% bootstrap). Subgroups containing sequences from mixed plant groups in the major branches are further highlighted with dark-shaded boxes. Major plant taxonomic divisions are indicated by accession number color thus: dicot, black; monocot, green; pine, blue; fern, orange; and bryophyte, red. Hypothetical intron gains and losses are indicated as described in Figure 1b. Branches are drawn to scale as indicated by the scale bar (= 0.1 substitutions/1,000 residues). Accession numbers for non-Arabidopsis sequences have a species-specific indentifier as follows: Osa, rice (Oryza sativa); Csa, cucumber (Cucumis sativa); Pta, pine (Pinus taeda); Mqu, Marsilea quadrifolia; Ppa, moss (P. patens); Zma, maize (Zea mays); Pra, Pinus radiata; Lja, Lotus japonicus; Gma, soybean (Glycine max); Gar, Gossypium arborium; Mtr, Medicago truncatula; Cpu, Ceratodon purpureus; Mpo, Marchantia polymorpha; Cja, C. jambhiri; Mcr, common ice plant (Mesembryanthemum crystallinum).
The distribution of moss, fern, and pine sequences throughout the expansin superfamily tree indicates that all three expansin subfamilies arose very early in land plant evolution, if not before. This is certainly the case for the α- and γ-expansins, where completely typical sequences of each are found in the moss, P. patens (Fig. 4). Furthermore, that these sequences do not branch below the base of their respective subtrees indicates that at least some of the diversity within these groups was also already present in the common ancestor of moss and “higher” land plants (Fig. 4). Therefore, we predict that other α- and γ-expansins will be found in P. patens once genome sequencing is complete. Likewise, the placement of the single identified pine β-expansin embedded well within the β-expansin subtree indicates that this group, and at least some of its diversity, also dates back at least to the origin of seed plants (Fig. 4). Furthermore, the presence of strongly supported mixed subgroups, including rice and Arabidopsis in the α-expansin subtree (Fig. 4), indicates that the gene duplications giving rise to these subgroups must have predated the monocot/dicot split.
Intron positions within Arabidopsis, rice, and moss expansins sequences are also generally conserved, and they further support the integrity of α, β, and γ subgroups (Fig. 4). Some of the intron positions appear to be very old, in evolutionary terms, especially 1 and 3, which were probably present in the common ancestor all plant expansins. Introns 2 and 4 are unique to β-expansins, and are probably the result of insertion events dating shortly after the α-β split. Despite the conservation of the intron positions, our analyses also identified numerous isolated instances of intron loss scattered through the tree with no apparent pattern (Fig. 4).
Nonplant Expansin-Like Sequences
Iterative profile searches of GenBank and EST databases also revealed a limited number of expansin-like sequences from a diversity of taxa showing substantial similarity to plant expansins based on E values (≤0.002) and global pairwise alignment. These included one expansin-like sequence from blue mussel (Mytillus edulis), four bacterial sequences (Clavibacter michiganensis [two sequences], Bacillus subtillus, and Xylella fastiosa), a single fungal sequence (Trichoderma reesii), and a small multigene expansin-like family in Mycetozoa (Dictyostelium discoideum).
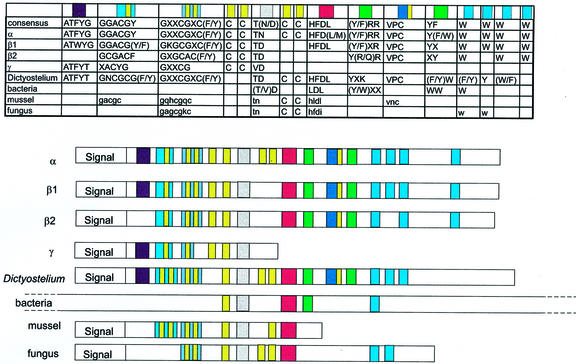
Examination of the aligned protein sequences revealed a number of conserved motifs within plant expansins and expansin-like sequences from nonplant sources (Fig. 5). The size of the predicted mature protein varied from 131 (citrus blight γ-expansin) to 712 (C. michiganensis sequence) residues. The plant α- and β-expansins and the D. discoideum expansin-like sequences are of similar size (approximately 300 amino acids), and we suggest that this is the predominant form for an expansin. In contrast, the plant γ-expansins and the mussel and fungal sequences are significantly shorter and, in the bacterial sequences, the expansin-like domain is generally present within a much larger polypeptide (Laine et al., 2000).
Figure 5.
A schematic illustration of the presence of conserved motifs in expansin-like proteins. Positions of conserved motifs and residues are not presented to scale, and are based on information in the comprehensive alignment of protein sequences used in this paper (Fig. S1 in the supplementary material). The table at the top lists the motifs for each class of expansins. Schematics below show the physical arrangements of the motifs within the deduced sequences.
All of the predicted proteins contain an N-terminal signal peptide that shows no significant sequence conservation, but implies that all of the predicted proteins are targeted for secretion (Fig. 5). Following the signal peptide, a major characteristic in the N-terminal one-half of the proteins is a series of conserved Cys, suggesting that expansins may have a similar tertiary structure involving the formation of disulphide bonds. The first three of these Cys occur in two motifs and are present in all the proteins apart from the bacterial group (Fig. 5). The next pair of Cys are separated by only one residue in the α and β1 groups, four residues in the β2 group, and are contiguous in γ-expansins. This pair of Cys is absent from all nonplant sequences. A third pair occurs in α-expansins and in the D. discoideum sequences, but not in any other groups included in this analysis. A final Cys residue occurs in a conserved RVPC motif (KVPC in D. discoideum) found after the HFD box (see below).
One particularly notable motif is the HFD box, which has been considered a characteristic of expansins (Cosgrove, 1999). This motif lies near the center of the mature protein and has been described as a putative catalytic motif on the basis of similarity to the catalytic core of microbial endoglucanases (Davies et al., 1995; Cosgrove, 1999). We found that this motif is present in the majority of expansin groups. However, there are a number of expansins in which the HFD box is absent or incompletely conserved. For example, a cluster of three α-expansins, i.e. AAF26104, CAB37561, and rAAF62183, have HFV, HFL, or HLE, respectively (Fig. S1 in the supplementary material). All β2-expansins we examined lacked the HFD box, although there is a conserved D residue in the same general area (Fig. S1). The other group lacking an HFD box in general is the γ-expansin group (Fig. 5). In this group, some sequences possess similar triplets such as GFD or AFD, but there is clearly no strict conservation of HFD. This suggests that the functional significance of this motif is far from clear and will remain so until a greater diversity of expansin-like proteins have been characterized biochemically.
With the notable exception of the γ-expansins (which have a truncated terminal exon), and the mussel sequence that is similarly short, the C-terminal halves of the proteins are characterized by a series of noncontiguous conserved Trp residues. The spacing between these residues resembles that found in the cellulose-binding domain of some cellulases (Gilkes et al., 1991). This is consistent with speculation that this region may be responsible for expansin binding to cellulose and related wall glycans (Shcherban et al., 1995).
The existence of expansin-like sequences in nonplant species is intriguing. As there are now complete genomic sequences for 38 bacteria, covering most of the known bacterial diversity, it is notable that our comprehensive searches only revealed four bacterial expansin-like sequences. All of these sequences, with the exception of the B. subtilis sequence, were found as part of larger complex proteins, which most likely possess cellulase and expansin-like activity in vivo (Laine et al., 2000). Of these bacteria, both of the C. michiganensis species and X. fastiosa are plant pathogens, whereas B. subtilis is known to contain many genes encoding plant wall-degrading enzymes (Kunst et al., 1997). It is tempting to speculate that an expansin-like domain within these complex proteins may serve to loosen plant wall material, facilitating cellulytic digestion. It is interesting that these three groups of bacteria represent quite disparate parts of the bacterial kingdom; X. fastiosa belongs to the proteobacteria group and C. michiganensis and B. subtilis belong to high and low GC gram-positive bacteria, respectively. An evolutionary explanation for the diverse bacterial species distribution and the context in which the expansin-like epitope occurs can only be speculated upon. The possibility of the preservation of ancestral form in only a small number of plant-pathogenic or plant-degrading bacteria seems rather less likely than horizontal transfer (Doolittle, 2000).
A similar picture emerges for expansin-like sequences from animals and fungi. Again, despite iterative searches with a variety of probes and search algorithms, we found very few expansin-like sequences in either group. A single animal sequence was identified from the mussel, M. edulis, and its corresponding protein has been reported to possess limited expansin activity (Xu et al., 2000). Mussels are active degraders of plant material, which suggests a digestive role for this expansin-like protein in vivo. Likewise, we found a single fungal expansin-like sequence in the Ascomycete, Trichoderma reesii. T. reesii is a highly active degrader of plant material, and this protein is most probably active in cell wall digestion (Saloheimo et al., 1994). Thus, as with the bacterial expansin-like sequences, those in animals and fungi appear to be restricted to organisms involved in plant pathogenesis or plant cell wall digestion.
The D. discoideum Expansin Gene Family
An exception to the association of nonplant expansin-like sequences with plant pathogenesis or degradation is the occurrence of these sequences in the slime mold, D. discoideum. Detailed analysis of D. discoideum genomic data (D. discoideum Genome Project, http://www.uni-koeln.de/dictyostelium/project.shtml) revealed that there are at least five expansin-like genes in this organism, three of which are also represented in EST collections. The deduced amino acid sequences revealed that these putative proteins are conserved among themselves (up to 60% identity and 73% similarity). From the nonplant sequences, it is apparent that the D. discoideum sequences show by far the closest similarity to plant expansins. Not only is overall similarity greater than 30% to α- and β-expansins, but they are also very similar in length and share many of the major conserved expansin motifs.
D. discoideum is an extensively studied model organism because of its fascinating life cycle. These organisms initially live as free-ranging amoebae, but associate to form a multicellular slug of more than 100,000 individual cells when nutrients become limiting. Cells in the slug subsequently differentiate to form a rapidly elongating stalk supporting a spore-containing fruiting body. The role of an expansin-like protein in D. discoideum physiology is intriguing. Because D. discoideum feeds primarily on bacteria, these expansin-like sequences probably do not serve a digestive function in this organism. However, the slug stage and the stalk of the fruiting body produce an extracellular cellulosic matrix. This matrix has some resemblance to plant cell walls and is particularly prominent in elongating stalk cells. The matrix presumably imparts mechanical strength to the rapidly growing stalk. Thus, D. discoideum expansins may serve to lubricate the movement of the cellulose microfibrils during cell growth and wall extension and/or they may serve to maintain the fluid state of the slug cell wall. We are currently characterizing the D. discoideum expansin-like sequences and their role in D. discoideum physiology and development.
The Evolution of Plant Expansins
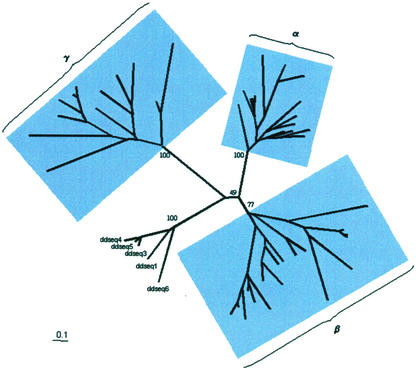
Phylogenetic analysis using the five D. discoideum sequences as an outgroup tentatively places the root for the plant expansins between α/β and γ (50% bootstrap; Fig. 6). Alternative rooting between β/γ and α or γ/α and β received considerably less support (30% and 8.5% bootstrap, respectively). These results combined with those in Figure 4 allow us to make some speculation about the order of evolution within the plant expansin super family. The presence of derived α- and γ-expansins in moss suggests that the three subfamilies were already well established in the common ancestor of moss and all the “higher” plants. Thus, we propose that the α/β-γ split and the subsequent α-β split predated the origin of land plants, as did at least some of the diversification within them. Within the β subfamily, the position of the pine sequence in the β2 subgroup suggests that at least three β-expansins (β1–β3) were present in the common ancestor of angiosperms and gymnosperms. Within the α subfamily, the presence of at least four strongly supported mixed rice and Arabidopsis α-expansin subgroups indicates the presence of at least five α-expansins in the common ancestor of monocots and dicots. We propose that the earliest land plants had a minimum of two αs, one β, and one γ, by the time of the evolution of gymnosperms, this would have increased to three βs, and by the origin of angiosperms, the family would have had at least minimum of five α-expansins, four β-expansins, and two γ-expansins. Thus, representatives of all three subfamilies should be found in all land plants, including moss.
Figure 6.
Phylogenetic analysis suggests the “best-guess” root of the expansin family is between the α/β and γ groups. The analysis used five D. discoidium sequences as an outgroup. Only representative sequences from α, β, and all γ were included in this analysis (see Table S1 in the supplementary material). Bootstrap values are indicated against the appropriate branches. The major groups, α, β, and γ, are grouped as highlighted boxes. The lower support for the β subgroup (56% versus 77%) is due to the smaller number of alignable positions used in theses analyses.
It is interesting to note that significant diversification of the expansin gene family is so far found only in D. discoideum and plants. This makes sense as both are characterized by the presence of cellulose-containing cell walls that need to be elongated during growth. Why expansins are maintained as multigene families remains an unanswered question, but hints at the universal complexity of the problem of cell wall modification.
MATERIALS AND METHODS
Database Search
Expansin and expansin-like sequences were retrieved by tBLASTn and psiBLAST searches of the nonredundant EST database (dbEST) and the finished and unfinished genome databases held at the NCBI (http://www.ncbi.nlm.nih.gov/). These searches were conducted using protein sequences for α-expansins, CsExp1 (AAB37746) and CsExp2 (AAB37749), and a β-expansin from maize (AAA33496). To ensure accuracy in annotations, Arabidopsis ORFs previously annotated as expansins were re-examined by multiple sequence alignment using ClustalX version 1.8, by comparing their genomic coding sequences with corresponding EST sequences, and by analyzing genomic coding sequences for correct identification of ORFs and introns using the program NetGene2 (http://www.cbs.dtu.dk/services/ NetGene2).
As a result of our analyses, 38 unique Arabidopsis expansins were identified. Two of these sequences were identified as pseudogenes, one is a truncated ORF (AAF23829-2), and the other contains a non-sense mutation (AP001309).
The positions of introns were decided by visual scanning of the exon boundaries for Arabidopsis intron splice-site consensus sequences. These predicted sites were further supported by analysis via NetGene2. As a result of our analyses, several corrections were made to the database annotations of Arabidopsis expansin-like genes. These are summarized in Table I.
All non-Arabidopsis sequences were retrieved from incomplete genome projects (i.e. rice [Oryza sativa]) or from individual cDNA or genomic clones (i.e. fern [Regnellidium diphyllum, Marsilea quadrifolia] and moss [Physcomitrella patens]) and, as such, is not completely comprehensive. The alignment of all plant expansins used in our analysis can be found in the supplementary material to this paper (Fig. S1). Where available, we made use of genomic sequences, as this provides added information with regard to the conservation of intron positions in expansin genes. In rice, eight genomic sequences were re-examined, and a single moss sequence was included in the analysis as genomic and full-length cDNA sequences.
As only one γ-expansin sequence (in addition to those from Arabidopsis) was detected in psiBLAST searches of non-redundant databases, this sequence was then used in tBLASTn searches of NCBI EST collections. As a result, an additional eight full-length γ-expansin cDNAs were assembled from ESTs and were included in the analysis.
Three Dictyostelium discoideum EST sequences showing significant similarity to the plant expansins were also identified in GenBank searches. These EST sequences were used in BLAST searches of the D. discoideum Genome Database [http://www.uni-koeln.de/dictyostelium/project]. Five D. discoideum genomic sequences were assembled from overlapping fragments. The intron positions of these genomic sequences were determined by comparing the EST sequences or by prediction using the NetGene2 program.
Sequence Alignment and Phylogenetic Analysis
All deduced amino acid sequences were aligned using ClustalX version 1.8 (with default gap penalties; Thompson et al., 1994). The alignments were then reconciled and further adjusted by eye to minimize insertion/deletion events. The alignment gave a total of 90 conserved residue positions that were used as the dataset for the construction of the initial comprehensive tree (shown as shaded bocks in the alignment Fig. S1 in the supplementary material to this paper). Distance analyses used the program ProtDist of the Phylip 3.5c package with a PAM250 substitution matrix. Phylogenetic trees were then calculated from the matrices by the neighbor-joining algorithm. Parsimony was also used to calculate the phylogeny and the resulting strict consensus trees are consistent with the topology of the distance trees (data not shown). Bootstrap analyses consisted of 1,000 to 5,000 replicates using the same protocol. Strongly supported subgroups (α, β, and γ) were analyzed further with a larger dataset, including additional alignable positions to refine the subtrees. In the analysis investigating the relationship of α, β, and γ subgroups using D. discoideum sequences as an outgroup, only a representative subset of α and β sequences were used. These sequences can be found in the supplementary data (Table S1).
ACKNOWLEDGMENT
We thank Yifang Wang for initial data collection.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (grant nos. 87/P12844, 87/P11582, and 87/G13911 to Y.L., C.P.D., and S.L.B., respectively). V.O. is funded by Fundacion YPF (Argentina). S.J.M.-M. was supported by a Royal Society University Research Fellowship.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010658.
LITERATURE CITED
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–813. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Ceccardi TL, Barthe GA, Derrick KS. A novel protein associated with citrus blight has sequence similarity to expansin. Plant Mol Biol. 1998;38:775–783. doi: 10.1023/a:1006039016393. [DOI] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:9783–9788. doi: 10.1073/pnas.160276997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civello PM, Powell ALT, Sabehat A, Bennett AB. An expansin gene expressed in ripening strawberry fruit. Plant Physiol. 1999;121:1273–1279. doi: 10.1104/pp.121.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol. 1997;13:171–201. doi: 10.1146/annurev.cellbio.13.1.171. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:391–417. doi: 10.1146/annurev.arplant.50.1.391. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Expansive growth of plant cell walls. Plant Physiol Biochem. 2000a;38:109–124. doi: 10.1016/s0981-9428(00)00164-9. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. New genes and new biological roles for expansins. Curr Opin Plant Biol. 2000b;3:73–78. doi: 10.1016/s1369-5266(99)00039-4. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA. 1997;94:6559–6564. doi: 10.1073/pnas.94.12.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley CP, Forrester AM, McQueen-Mason SJ. The molecular basis of plant cell wall expansion. Plant Mol Biol. 2001;47:179–195. [PubMed] [Google Scholar]
- Davies GJ, Tolley SP, Henrissat B, Hjort C, Schulein M. Structures of oligosaccharide-bound forms of the endoglucanase V from Humicola insolensat 1.9 angstrom resolution. Biochemistry. 1995;34:16210–16220. doi: 10.1021/bi00049a037. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. The nature of the universal ancestor and the evolution of the proteome. Curr Opin Struct Biol. 2000;10:355–358. doi: 10.1016/s0959-440x(00)00096-8. [DOI] [PubMed] [Google Scholar]
- Fleming AJ, McQueen-Mason SJ, Mandel T, Kuhlemeier C. Induction of leaf primordia by the cell wall protein expansion. Science. 1997;276:1415–1418. [Google Scholar]
- Frugoli JA, McPeek MA, Thomas TL, Robertson McClung C. Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics. 1998;149:355–365. doi: 10.1093/genetics/149.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes NR, Henrissat B, Kilburn DG, Miller RC, Warren RAJ. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. Evolutionary history of the grasses. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Cho HT, Kende H. α-Expansins in the semiaquatic ferns Marsilea quadrifolia and Regnellidium diphyllum: evolutionary aspects and physiological role in rachis elongation. Planta. 2000;212:85–92. doi: 10.1007/s004250000367. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Kutschera U. The current status of the acid growth hypothesis. New Phytol. 1994;126:549–569. [Google Scholar]
- Lacombe B, Becker D, Hedrich R, DeSalle R, Hollmann M, Kwak JM, Schroeder JI, Le Novere N, Nam HG, Spalding EP, Tester M et al. The identity of plant glutamate receptors. Science. 2001;292:1486–1487. doi: 10.1126/science.292.5521.1486b. [DOI] [PubMed] [Google Scholar]
- Laine MJ, Haapalainen M, Wahlroos T, Kankare K, Nissinen R, Kassuwi S, Metzler MC. The cellulase encoded by the native plasmid of Clavibacter michiganensisspp sepedonicus plays a role in virulence and contains an expansin-like domain. Physiol Mol Plant. 2000;57:221–233. [Google Scholar]
- Li Y, Baldauf S, Lim E-K, Bowles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem. 2001;276:4338–4343. doi: 10.1074/jbc.M007447200. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotype markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1154. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. Disruption of hydrogen-bonding between plant-cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. Expansin mode of action on cell walls: analysis of wall hydrolysis, stress-relaxation, and binding. Plant Physiol. 1995;107:87–100. doi: 10.1104/pp.107.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell-wall extension in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Fry SC, Durachko DM, Cosgrove DJ. The relationship between xyloglucan endotransglycosylase and in vitro cell wall extension in cucumber hypocotyls. Planta. 1993;190:327–331. doi: 10.1007/BF00196961. [DOI] [PubMed] [Google Scholar]
- Metraux P, Taiz L. Cell wall extension in Nitellaas influenced by acids and ions. Proc Natl Acad Sci USA. 1977;74:1565–1569. doi: 10.1073/pnas.74.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. Control of plant cell enlargement by hydrogen ions. Curr Topic Dev Biol. 1977;11:187–214. doi: 10.1016/s0070-2153(08)60746-2. [DOI] [PubMed] [Google Scholar]
- Rose JKC, Cosgrove DJ, Albersheim P, Darvill AG, Bennett AB. Detection of expansin proteins and activity during tomato fruit ontogeny. Plant Physiol. 2000;123:1583–1592. doi: 10.1104/pp.123.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E-K, Bowles DJ. Higher plant glycosyltransferases. Genome Biol. 2001;2:3004.1–3004.6. doi: 10.1186/gb-2001-2-2-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saloheimo A, Henrissat B, Hoffren AM, Teleman O, Penttila M. A novel small endoglucanase gene, EGL5, from Trichoderma reeseiisolated by expression in yeast. Mol Microbiol. 1994;13:219–228. doi: 10.1111/j.1365-2958.1994.tb00417.x. [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Assaad FF, Raikhel NV. The Arabidopsis genome: an abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 2000;124:1558–1569. doi: 10.1104/pp.124.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular-cloning and sequence-analysis of expansins: a highly conserved, multigene family of proteins that mediate cell-wall extension in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SEC, Gidley MJ, McQueen-Mason SJ. Probing expansin action using cellulose/hemicellulose composites. Plant J. 2000;22:327–334. doi: 10.1046/j.1365-313x.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- Xu B, Hellman U, Ersson B, Janson JC. Purification, characterization and amino-acid sequence analysis of a thermostable, low molecular mass endo-β-1,4-glucanase from blue mussel, Mytilus edulis. Eur J Biochem. 2000;267:4970–4977. doi: 10.1046/j.1432-1327.2000.01533.x. [DOI] [PubMed] [Google Scholar]