Extensins are abundant proteins presumed to determine physical characteristics of the plant cell wall. We have cloned a cDNA encoding LeExt1 from a tomato (Lycopersicon esculentum Mill.) root hair cDNA library. The deduced sequence of the LeExt1 polypeptide defined a novel type of extensin-like proteins in tomato. Patterns of mRNA distribution indicated that expression of the LeExt1 gene was initiated in the root hair differentiation zone of the tomato rhizodermis. Cloning of the corresponding promoter and fusion to the β-glucuronidase (GUS) reporter gene allowed detailed examination of LeExt1 expression in transgenic tomato plants. Evidence is presented for a direct correlation between LeExt1 expression and cellular tip growth. LeExt1/GUS expression was detectable in trichoblasts (=root hair-bearing cells), but not in atrichoblasts of the tomato rhizodermis. Both hair formation and LeExt1 expression was inducible by the plant hormone ethylene. Comparative analysis of the LeExt1/GUS expression was performed in transgenic tomato, potato (Solanum tuberosum), tobacco (Nicotiana tabacum), and Arabidopsis plants. In the apical/basal dimension, GUS staining was absent from the root cap and undifferentiated cells at the root tip in all species investigated. It was induced at the distal end of the differentiation zone and remained high proximally to the root/hypocotyl boundary. In the radial dimension, GUS expression was root hair specific in the solanaceous species. Whereas LeExt1 mRNA was exclusively detectable in the rhizodermis, root hair-specific expression correlated with GUS expression in germinating pollen tubes. This is correlative evidence for a role of LeExt1 in root hair tip growth.
Rhizodermal cells differentiate in the root meristem after asymmetric division from initial cells (Dolan et al., 1993). During rhizodermis development in Arabidopsis, these cells specialize further into two distinct cell types, rhizodermal cells without root hairs (atrichoblasts) and root hair-bearing cells (trichoblasts). Once initiated, root hair growth is characterized by oriented tip growth, comparable with that in a growing pollen tube. The zone of growth is restricted to the tip of the growing cell (Sievers and Schnepf, 1981). As a consequence of oriented cell elongation, the hair extends into the yet-unexplored rhizosphere, where it acquires water and nutrients from the soil solution to sustain plant growth.
Root hair development is amenable to genetic dissection and has proved in the past to be a useful model system to study the molecular mechanisms regulating cell differentiation in Arabidopsis (Schiefelbein and Somerville, 1990). Several loci have been reported to be involved in rhizodermal cell patterning (Galway et al., 1994; Wada et al., 1997) and root hair initiation (Schiefelbein and Somerville, 1990; Masucci and Schiefelbein, 1994; Schneider et al., 1997, 1998). A minimum of five genes, rhd2, rhd3, rhd4, tip1, and cow1 are involved in hair elongation (Grierson et al., 1997; Ryan et al., 1998). Mutations within these genes lead to abnormalities in root hair shape and elongation. It is interesting that Arabidopsis tip1 mutants exhibit disruption of both root hair and pollen tube growth, suggesting that the TIP1 protein is important for tip growth (Schiefelbein et al., 1993).
During rapid root hair expansion, the synthesis of the plasma membrane and cell wall material must represent a major metabolic load. The site of deposition of this material is confined to the expanding tip, thus leading to the tubular morphology. Several factors are involved in the regulation of root hair growth. Oriented tip growth correlates well with a steep [Ca2+]cyt gradient in the growing hair (Wymer et al., 1997). Upon cessation of growth, the gradient dissipates. Vice versa, disruption of the tip-focused gradient of [Ca2+]cyt inhibits root hair growth. Another component involved in the regulation of tip growth is the microtubule cytoskeleton. Microtubule-depolymerizing agents cause a waving growth habit of the growing hairs and the formation of multiple growing points (Bibikova et al., 1999). Other components contributing to the elongation or the morphology of tip-growing cells are likely to be located in the cell wall (Gilroy and Jones, 2000).
The cell wall affects cell form and function. The protein component of the cell wall contains both enzymes and structural proteins. So far, the best characterized structural cell wall protein is extensin (Showalter, 1993; Cassab, 1998), a member of the family of Hyp-rich glycoproteins (HRGPs) that are among the most abundant proteins present in the cell wall of higher plants. Gene expression of HRGPs is developmentally regulated in a tissue-specific manner (Ye and Varner, 1991). For example, the tobacco HRGPnt3 gene is specifically expressed in a subset of the pericycle and endodermal cells from which a lateral root initiates (Keller and Lamb, 1989). A soybean (Glycine max) extensin gene, SbHRGP3, is expressed in hypocotyl and roots of seedlings (Ahn et al., 1996). This was shown by using an SbHRGP3 promoter-GUS chimeric gene that was expressed in the rhizodermal cells of the zone from which the lateral root was to be initiated. mRNAs of extensin-like proteins recently have been shown to be highly abundant in root hairs of tomato and cowpea (Vigna unguiculata; Arsenijevic-Maksimovic et al., 1997; Bucher et al., 1997). HRGPs are subject to extensive posttranslational modification. Pro residues are hydroxylated by prolyl hydroxylase. Carbohydrates subsequently are attached to the Hyp residues and probably serve to stabilize the protein into a rigid rod-like structure (Showalter, 1993). The mature extensin protein is generally rich in Hyp and Ser and some combination of the amino acids Tyr, Lys, Val, and His. Extensins of dicot plants usually contain the repeating pentapeptide Ser-Hyp4, often within the context of other larger repeating motifs. Isodityrosine linkages that are presumably formed by peroxidases (Schnabelrauch et al., 1996) have been suggested to cross-link extensins in the cell wall, thus leading to insolubilization of the proteins and cell wall strengthening, e.g. as a response to pathogen attack (Epstein and Lamport, 1984; Brisson et al., 1994) or to confer mechanical resistance to load-bearing cells (Keller and Lamb, 1989; Tiré et al., 1994).
In this study, we describe LeExt1 (accession no. AJ417830), a novel gene encoding an extensin-like protein in tomato. Its expression correlates with tip growth, which suggests a role of the LeExt1 protein in root hair expansion. Moreover, comparative studies of four different transgenic plant species expressing an LeExt1/GUS chimeric gene indicate the existence of common mechanisms involved in the regulation of apical/basal polarity in root gene expression.
RESULTS
LeExt1 Encodes a Putative Cell Wall Protein from Tomato
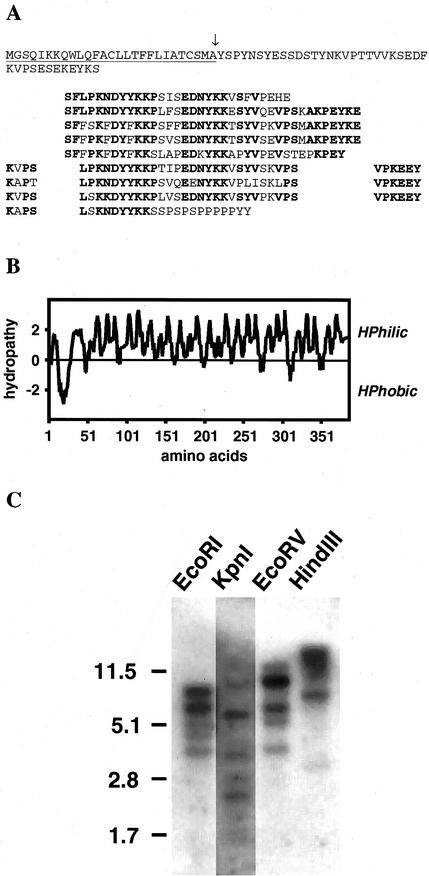
The LeExt1 cDNA was identified in the course of a differential screening of a tomato root hair-specific cDNA library, which was set up to identify root hair-specific genes (Bucher et al., 1997). The cDNA is 1,419 bp long, including a putative initiator ATG at its 5′ end and a poly(A+) tail. The deduced amino acid sequence revealed the repetitive nature of the polypeptide (Fig. 1A). Hydropathy analysis (Kyte and Doolittle, 1982) indicated that the encoded protein is hydrophilic, carrying a hydrophobic N-terminal leader sequence (Fig. 1B). The hydrophobic segment at the N terminus has the characteristics typical of a signal peptide for translocation into the endoplasmic reticulum, and based on the rules of von Heijne (1986), the cleavage site is located carboxyterminal of an Ala (marked with an arrow in Fig. 1A). The predicted mature protein consists of 385 amino acids and has a predicted molecular mass of 42.2 kD and a calculated pI of 9.2. Assuming that cleavage does occur after the designated Ala, mature LeExt1 extending from amino acids 28 through 385 is rich in Lys (19%), Ser (14%), Pro (14%), Tyr (12%), Glu (10%), and Val (8%). Thus, these seven amino acids together comprise 77 mol % of the protein. Similar to other HRGPs the polypeptide is composed of several repeating motifs rich in Lys, Tyr, Pro, and Ser (Fig. 1A).
Figure 1.
Peptide structure of LeExt1 and genomic DNA gel-blot analysis. A, Deduced LeExt1 amino acid sequence. Repetitive amino acid units (indicated in bold) are arranged to emphasize various amino acid repeat units and their periodicity. The signal peptide is underlined and the putative cleavage site is marked with an arrow. B, Hydropathy plot of LeExt1 polypeptide. Hydrophilicity and hydrophobicity values are indicated at left. C, Genomic DNA was digested with the designated restriction enzymes. The LeExt1 cDNA was used as a radioactive probe. The positions of DNA marker fragments and their lengths in kb are indicated at left.
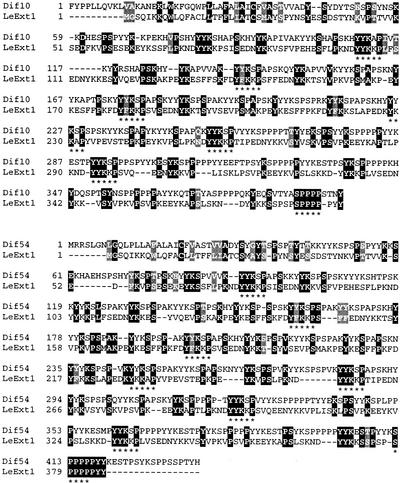
A sequence similarity search through the GenEMBL database revealed highest similarities of LeExt1 to the published Dif10 and Dif54 extensin-like proteins from tomato root hairs (Bucher et al., 1997), to an Arabidopsis periaxin-like protein (accession no. CAB89377) exhibiting similarity to periaxin from rat (Rattus norvegicus) (Gillespie et al., 1994), and to the marine mussel (Mytilus edulis) polyphenolic adhesive protein (Filpula et al., 1990). Expect (E) values according to the BLAST search results were 8e−20, 20e−15, 3e−14, and 9e−13, respectively (an E value of 1 meaning that in a database of the current size, one might expect to see one match with a similar score simply by chance). All these proteins are rich in at least some of the amino acids Tyr, Pro, Lys, Ser, Val, and Glu. Comparison with both Dif10 and Dif54 extensin-like proteins revealed that specifically spaced repetitive elements contribute to the sequence similarity rather than identity across a large region of LeExt1 that is similar with the compared proteins (Fig. 2). These elements are characterized by the following di-, tri-, and pentapeptides: YK, PS, YYK, and YY/F/KKS/K/AP, where bold faced letters in the latter designate conserved amino acids in the one-letter code.
Figure 2.
Alignment of the deduced amino acid sequence of LeExt1 with that of Dif10 and Dif54. Identical amino acids are shaded in black, similar amino acids are shaded in gray. The conserved pentapeptides YxKxP and SPPPP are underlined.
To get an indication of the number of related genes in the tomato genome, genomic DNA was digested with four different restriction enzymes of which EcoRI, EcoRV, and HindIII do not cut within the cDNA sequence, and the restricted DNA was subjected to genomic DNA gel-blot analysis (Fig. 1C). The radioactively labeled LeExt1 cDNA hybridized to up to five DNA fragments under stringent conditions, thus suggesting the existence of a small multigene family of the corresponding gene in tomato. This was substantiated by the cloning of two λ-clones with high similarity to the LeExt1 cDNA sequence (see below). These results qualify LeExt1 as a novel extensin-like protein that is most likely localized in the cell wall.
LeExt1 Transcripts Accumulate in Rhizodermal Cells
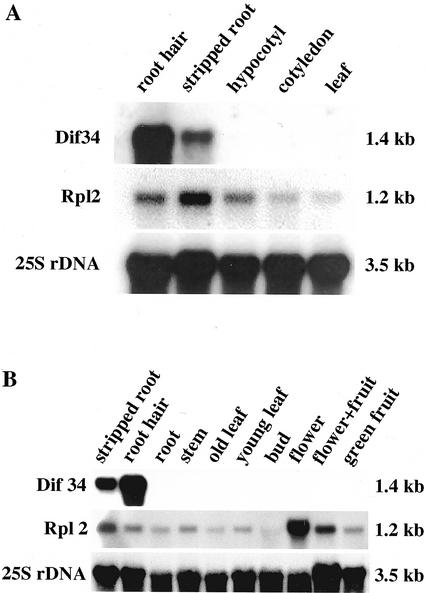
The radioactively labeled LeExt1 cDNA was used for the analysis of corresponding transcript levels. It hybridized to a single band of 1.4 kb on the RNA gel blot (Fig. 3, A and B). Transcript levels were severalfold higher in root hairs as compared with primary roots with their hairs stripped off (designated stripped roots; Fig. 3A). No signals were detected in the hypocotyl, the cotyledons, and leaves. As a control, Rpl2 transcripts (encoding ribosomal protein L2) were detectable in all organs. Rpl2 is a relatively well-characterized housekeeping gene that should reflect constitutive expression (Fleming et al., 1993). Its expression fluctuated somewhat between samples, probably representing different translational activities in the respective tissues. Thus, the results indicated high abundance of LeExt1 mRNA in root hairs of the tomato seedling.
Figure 3.
LeExt1 transcript abundance in seedlings and flowering plants. A, RNA gel-blot analysis was performed with 5 μg of total RNA from seedling root hairs, stripped roots, hypocotyls, cotyledons, and mature leaves and hybridized with randomly labeled cDNA probes, as indicated at left. A cDNA encoding 25S rDNA from tomato served as a control for equal loading of total RNA on the gel and equal transfer to the membrane. Transcript sizes are indicated at right. B, RNA gel-blot analysis was performed with 5 μg of total RNA from stripped seedling roots and root hairs, and with 10 μg of total RNA from the organs indicated and hybridized with randomly labeled cDNA probes, as indicated at left. Transcript sizes are indicated at right.
To investigate whether LeExt1 is expressed in shoot organs of tomato at later stages in development, total RNA from various organs of soil-grown plants from the greenhouse was run on a gel (Fig. 3B). No transcripts of the gene were detected in these organs, whereas Rpl2 mRNA was detectable in all lanes on the blot. No signal was detectable with RNA from roots of soil-grown plants (lane 3). This can be explained by the microscopic observation that removal by careful washing of soil particles bound to the roots before RNA extraction also removed the root hairs. Thus, RNA gel-blot analysis clearly indicated root-specific expression of LeExt1 in tomato roots with preferential expression in root hairs.
In situ hybridization studies further allowed cell-specific localization of LeExt1 mRNA in tomato primary roots (Fig. 4). LeExt1 transcripts were exclusively detected in rhizodermal cells in the differentiation zone (Fig. 4, A and B). No signals were observed in root cap, meristematic, and elongating cells at the root tip. Sense RNA as a hybridizing probe did not give rise to a signal (Fig. 4C), and Rpl2 transcripts were abundant in all cells (Fig. 4D). In general, staining occurred in vacuolated rhizodermal cells adjacent to the elongation zone of young seedling roots. Thus, high LeExt1 transcript abundance correlates with differentiation of rhizodermal cells.
Figure 4.
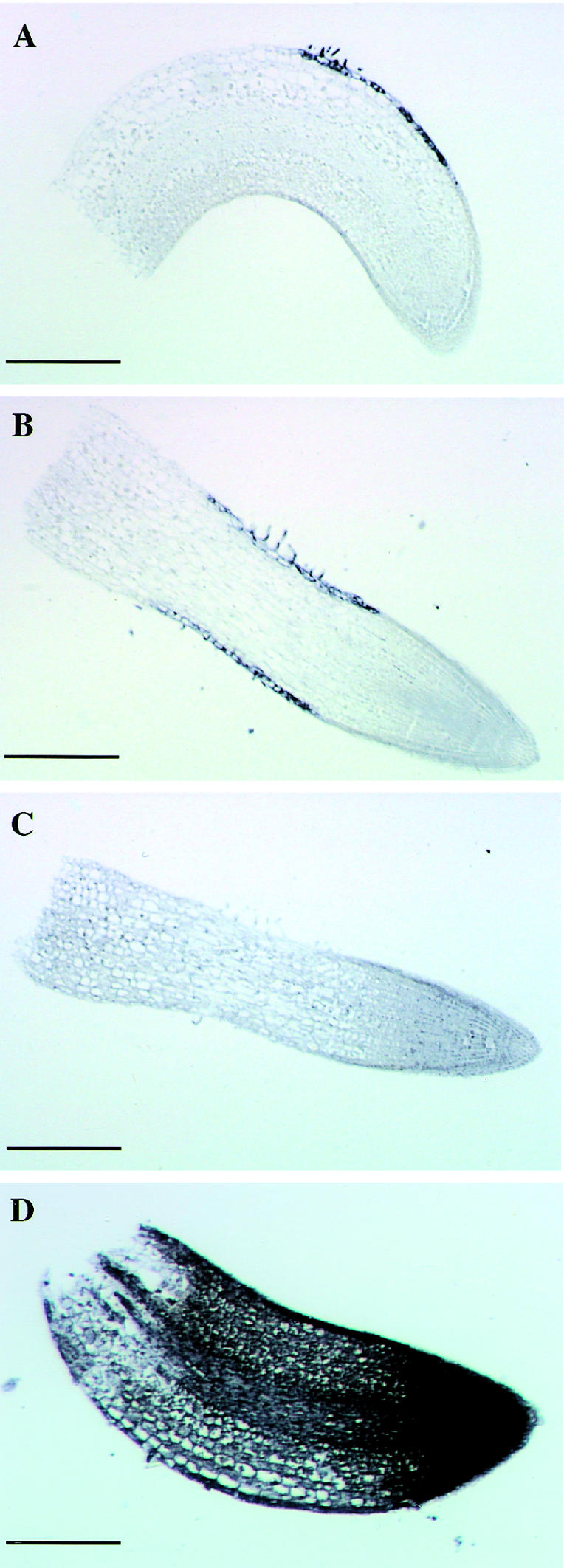
Localization of LeExt1 transcripts in tomato seedling roots. A through D, Bright-field microscopy of root sections. Shown in A and B are sections hybridized with the LeExt1 antisense probe. The purple dye reflects LeExt1 mRNA. C, Section hybridized with the LeExt1 sense probe as a negative control. D, Section hybridized with an antisense Rpl2 probe as a positive control. Bar = 0.25 mm in A through C; bar = 0.125 mm in D.
Cloning of the LeExt1 Promoter
To allow a more thorough study of LeExt1 gene expression, we isolated a λ-clone of 3,417 bp from a tomato genomic library. The sequence determined had an identical overlap at its 3′ end with 204 bp of the 5′ end of the LeExt1 cDNA (Fig. 5) and thus was assumed to contain the LeExt1 promoter. Two additional clones were partially sequenced and exhibited >80% sequence similarity to the LeExt1 cDNA (data not shown). Sequence comparison of the putative LeExt1 promoter-containing fragment with the respective cDNA revealed a stop codon upstream of the putative initiator ATG. Approximately 80 bp upstream of the putative start ATG, the sequence TATATAAA resembling a TATA box is present. A search for transcription factor-binding sites using the TFSEARCH algorhithm with a threshold score of greater than 85 revealed that the 3,181 bp upstream of the LeExt1 ORF contained the conserved motifs of Athb-1-, MYB.Ph3-, P-, and SBF-1-binding boxes (Fig. 5).
Figure 5.
Upstream sequence of the LeExt1 gene. Bold letters underlined with dashed arrows indicate putative binding sites of transcription factors P, MYB.Ph3, SBF-1, and Athb-1 as determined using TFSEARCH. Bold arrows on top of a base indicate the start of the different promoter fragments as indicated to the right. The TATA box is underlined and indicated in bold. An asterisk indicates a stop codon upstream of the open reading frame (ORF) in the genomic sequence. Amino acids encoded by the ORF are given in the one letter code. Bold letters designate the N terminus of the LeExt1 protein.
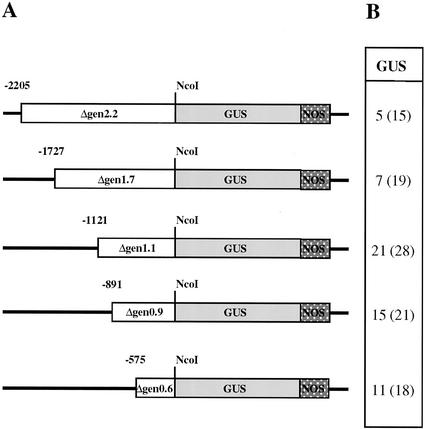
Tobacco and tomato plants transformed with the full promoter sequence fused to the GUS gene did not produce visible GUS staining (data not shown). This led us to speculate that silencing protein factors binding to any of the promoter regions might lead to GUS supression. Next, exact translational fusions at the initiator ATG were constructed between the GUS reporter gene and serial deletions of the LeExt1 promoter sequence (Fig. 6A). Transgenic tobacco plants were raised and the level of GUS activity in roots of at least 15 independent lines that had been transformed with each of the constructs shown in Figure 6 was microscopically estimated. This analysis revealed strong GUS staining in 75% of the lines transformed with the Δgen1.1/GUS gene (Fig. 6B). Roots carrying the longer Δgen1.7 and Δgen2.2 fragments, respectively, hardly stained blue and roots carrying the two shorter fragments Δgen0.9 and Δgen0.6 exhibited weaker staining. In F1 tobacco seedlings, strong GUS expression was detectable in young regions of the root including the root hairs and to lower degrees in the hypocotyl, petioles, and the margin of leaf blades. GUS was normally absent from the root tip. Plants showing GUS staining in root hairs also gave rise to GUS activity in dry pollen and germinating pollen tubes (data not shown). No changes in developmental regulation of the GUS expression driven by the different promoters were observed, i.e. the spatial pattern of expression was the same.
Figure 6.
Qualitative GUS assay in Δgen/GUS tobacco plants. A, Schematic drawing of the constructs used. The drawing is to scale. Negative numbers indicate the length of the promoter fragments. NcoI designates the fusion site at the ATG of the GUS gene. NOS is the nopaline synthase transcriptional terminator. Bold lines indicate vector sequence. B, Visual evaluation of GUS staining in roots of T0 lines, containing the promoter constructs as indicated in A. The number of independently transformed lines exhibiting strong GUS staining is listed. The total number of lines analyzed is given in parentheses.
Tissue Specificity of LeExt1 Expression
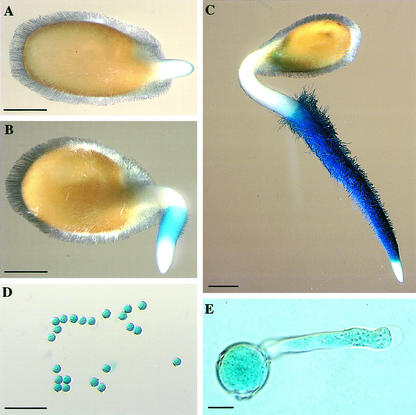
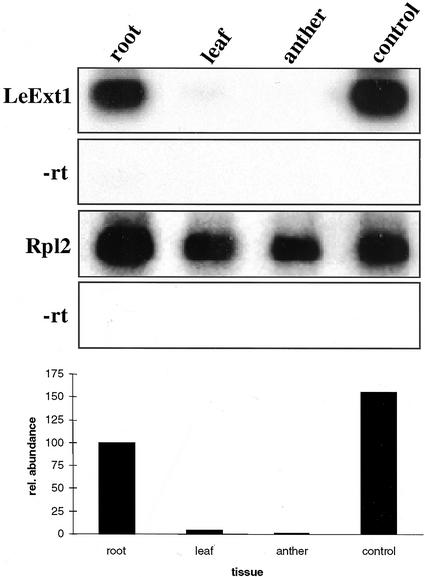
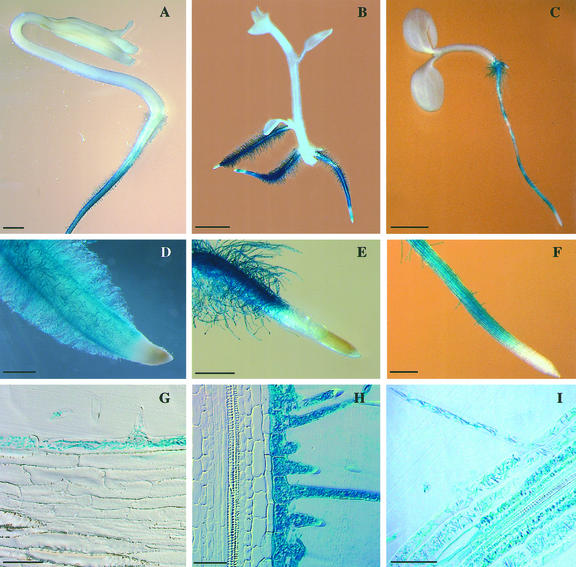
The generation of deletions of the λ-clone proved to be essential to assay histochemical GUS expression in more details and the Δgen1.1 fragment was selected as the strongest promoter for further studies. The Δgen1.1/GUS chimeric gene was then introduced into tomato, potato, and Arabidopsis. Similar to the situation in tobacco, GUS staining was absent from the emerging radicle in germinating tomato seedlings (Fig. 7A). Whereas expression of the GUS gene was readily detectable in the region where root hairs were being formed (Fig. 7B), there was no expression in the root tip and the hypocotyl (Fig. 7C). The pattern of gene expression of the chimeric gene thus corresponded to the pattern of mRNA distribution (Fig. 4). Similar to tobacco, GUS expression was observed in dry pollen and the germinating pollen tube of tomato (Fig. 7, D and E) and potato, but not in Arabidopsis pollen (data not shown). In contrast to Δgen1.1 promoter activity in pollen, RNA gel-blot analysis using 30 μg of total RNA from tomato anthers and the LeExt1 cDNA as a probe did not result in detectable signals. Reverse transcription (RT)-PCR failed to detect significant LeExt1 transcript concentrations in anthers and leaves, whereas a strong signal was obtained with RNA extracted from either wild-type roots or leaves from transgenic tomato plants constitutively expressing LeExt1 (Fig. 8). Used as a positive control, Rpl2 transcripts were abundant in all organs investigated. Non-reverse transcribed template RNA did not give rise to detectable signals on the blot. This demonstrated negligible relative abundance of LeExt1 mRNA in pollen and leaf as compared with the root, indicating that the endogenous LeExt1 gene is not or only very weakly expressed in pollen. Root specificity of the promoter in vegetative organs was conserved between tomato, potato, and Arabidopsis (Fig. 9, A–C). In all three species, GUS staining was absent from developing cells at the primary root tip and was initiated at the distal end of the differentiation zone where hairs were being formed (Fig. 9, D–F). The same staining pattern was observed in lateral roots. The indigo dye never accumulated in cells above the root/hypocotyl junction. Cross sections of GUS-stained roots revealed rhizodermis-specific expression in the two solanaceous species, whereas in Arabidopsis, GUS activity was detectable in rhizodermal cells, in the cortex including the endodermis, and in the stele (Fig. 9, G–I). Rhizodermis specificity was also observed in hairy roots of Medicago truncatula after inoculation with Agrobacterium rhizogenes that were transformed with a binary vector carrying the Δgen1.1/GUS chimeric gene (R. Geurts, personal communication; data not shown). Therefore, we conclude that the regulatory mechanisms that control the apical/basal polarity of Δgen1.1 promoter activity are similar in different plant species. The regulation of rhizodermis-specific expression is conserved within the solanaceous species (and M. truncatula), whereas absence of rhizodermis-specific expression correlates with absence of expression in tip-growing pollen in Arabidopsis.
Figure 7.
Histochemical localization of GUS activity in germinating tomato seeds and pollen. Tomato plants were transformed with the Δgen1.1/GUS construct. A through C, F1 seeds at various stages of germination assayed for GUS activity. D and E, Pollen harvested from dehisced anthers and germinated in vitro. Bar in A through C = 1 mm; bar in D = 0.1 mm; bar in E = 0.01 mm.
Figure 8.
Comparison of LeExt1 and Rpl2 transcript levels in tomato tissues by RT-PCR. RT-PCR was performed with total RNA from roots, leaves, and anthers and hybridized with randomly labeled cDNA probes, as indicated at left. RNA from leaves of transgenic plants constitutively expressing LeExt1 served as a control for primer specificity (control). RNA samples that were not subjected to reverse transcription (−rt) served as a control to determine residual amounts of genomic DNA in the samples (absent in −rt treatments). Black columns indicate relative abundance of transcripts as determined by phosphorimager analysis. The signal for root RNA was arbitrarily set to 100%.
Figure 9.
Histochemical localization of GUS activity in transgenic tomato, potato, and Arabidopsis. A, D, and G, Expression of the Δgen1.1/GUS chimeric gene in seedlings of tomato; B, E, and H, potato; C, F, and I, Arabidopsis. D through F, Stereomicroscopy images of GUS-stained root tips. G through I, Bright-field microscopy images of resin-imbedded cross sections of stained roots using Nomarski optics. Bar in A and B = 2 mm; bar in C = 1 mm; bar in D and E = 0.5 mm; bar in F = 0.2 mm; bar in G through I = 0.05 mm.
Trichoblast-Specific Expression of LeExt1 in the Rhizodermis
The plant hormones ethylene and auxin have been reported to affect the production of root hair and hairless cells in the Arabidopsis root (Masucci and Schiefelbein, 1996). We examined the influence of auxin and ethylene on tomato root development and Δgen1.1/GUS expression. Root exposure to auxin and auxin transport inhibitors gave rise to shorter roots, but no clear difference in root hair length or intensity of GUS staining was observed when compared with controls (data not shown). Exposure to l-α-(2-aminoethoxyvinyl)Gly (AVG), an inhibitor of ethylene synthesis, reduced root elongation (data not shown) and inhibited root hair development in tomato primary roots, thus giving rise to the formation of atrichoblasts as was determined by microscopical analysis (Fig. 10B). GUS staining in these roots was absent from atrichoblasts and was only observed in trichoblasts with hairs being formed before or shortly after the transfer to AVG-containing medium, whereas untreated roots expressed the GUS gene in all rhizodermal cells (Fig. 10A). Upon exposure to the ethylene-releasing agent ethephon in the medium, root hairs were ectopically induced near the root tip of controls and in the hair-free zone of AVG-treated roots. These ectopic hairs uniformly expressed GUS (Fig. 10C). LeExt1 promoter-driven GUS expression remained unchanged after infection of transgenic potato roots with either Phytophtora spp. or arbuscular-mycorrhizal fungi (V. Karandashov, personal communication; data not shown). Tomato var. Moneymaker lines expressing the LeExt1 cDNA under the control of the 35S CaMV constitutive promoter were raised. Plants constitutively expressing the transgene as shown by RNA gel-blot analysis (data not shown, see also Fig. 8) were further investigated for visible phenotypes. All phenotypic parameters analyzed, including root and root cell expansion, shoot growth, leaf epidermal cell expansion, and pollen tube growth, remained unchanged in comparison with wild type (data not shown). Moreover, constitutive expression of LeExt1 in the roots did not mediate an increased resistance to root nematodes (D. Trudgill, personal communication; data not shown). Thus, overall we can conclude that LeExt1 expression correlated with root hair formation and hair expansion and is most likely not involved in pathogen defense.
Figure 10.
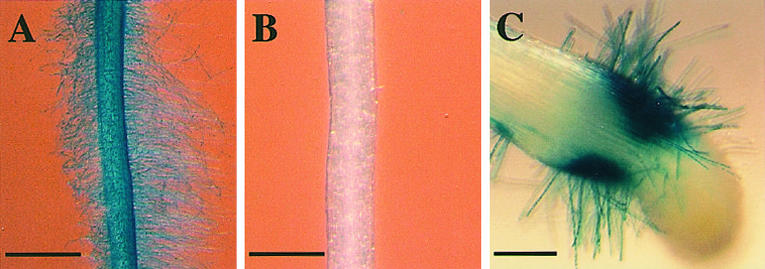
Modulation of ethylene biosynthesis and root hair development. A, Δgen1.1/GUS tomato seeds were germinated on control medium. At a primary root length of 3 to 5 mm, control seedlings were transferred to fresh medium for 2 d before GUS activity was assayed. B, Seedling root grown in presence of 20 μm AVG and stained for GUS activity after pretreatment as described in A. C, Ectopic root hairs 2 d after transfer to medium containing AVG and 1 mm ethephon. Bar in A = 1 mm; bar in B = 0.5 mm; bar in C = 0.2 mm.
DISCUSSION
In this study, the tomato LeExt1 cDNA was cloned via differential screening of a root hair-specific cDNA library. The corresponding upstream genomic sequence was subsequently obtained from a genomic tomato library screen. RNA localization and promoter-GUS fusion studies in tomato primary roots revealed that LeExt1 expression is associated with root hair formation and elongation and is controlled by ethylene. The tissue specificity of the promoter in roots was conserved in three solanaceous species, whereas in Arabidopsis, GUS expression was detectable in all differentiated root cells. Ectopic expression of LeExt1 in cells other than root hairs is not sufficient to phenotypically change cell shape or elongation.
In our efforts to clone the LeExt1 promoter, we have encountered the problem that the longer genomic fragments did not give rise to strong GUS expression in the roots, which might be because of silencer elements within these sequences (Figs. 5 and 6). Transformation of tobacco with truncated versions of the λ-clone fused to GUS has allowed us to perform GUS assays with roots from primary transformants (Fig. 6) and subsequent selection of the Δgen1.1 fragment as the qualitatively strongest promoter for transformation of tomato, potato, and Arabidopsis.
All rhizodermal cells in tomato may form a hair. These cells are long and because many are hairless, the hairs appear to be sparsely distributed over the whole surface. Hairs are more densely developed for a short distance close to the tip. In this region, the cells are much shorter than those developed later and almost every cell forms a long hair (Cormack, 1945). Both in situ hybridization (Fig. 4) and promoter-GUS analysis with tomato seedling roots (Fig. 7) indicated that LeExt1 expression is absent from the root tip, strongly induced in the root hair zone, and remained high proximally to the root-hypocotyl junction. This tip-to-base polarity of Δgen1.1/GUS expression in the root is maintained between the different plant species (Fig. 9), which indicates the existence of common regulatory mechanisms determining apical/basal polarity of gene expression in vascular plants. The tight correlation between LeExt1 expression and root hair development (Figs. 7, 9, and 10) indicates that expression of LeExt1 is regulated by developmental pathways involved in root hair formation. A key player in this regulatory network is the plant hormone ethylene, which strongly promotes root hair development in tomato (Fig. 10) and other plants. Therefore, it is likely that regulatory proteins that are involved in epidermal cell patterning direct root hair-specific gene expression of structural cell wall proteins such as LeExt1 at later stages of tomato epidermis development during hair expansion. Consistent with our results from Figure 10, the hormone ethylene would then act downstream of such regulators to promote LeExt1 expression and root hair outgrowth in tomato. A similar regulatory pathway recently has been suggested for the expression of the Arabidopsis Pro-rich protein AtPRP3 (Bernhardt and Tierney, 2000).
In a search for transcription factor-binding sites in the LeExt1 promoter, putative binding sites for four regulatory proteins were displayed (Fig. 5). Three of them, P, MYB.Ph3, and SBF-1, are involved in flavonoid biosynthesis (Lawton et al., 1991; Grotewold et al., 1994; Solano et al., 1995). Some regulatory genes are known to affect several independent phenotypes in Arabidopsis. For example, the TTG protein not only regulates the accumulation of purple anthocyanins in leaves and stems but also trichome and root hair development (Walker et al., 1999). The fourth protein putatively binding to the LeExt1 promoter is Athb1, which, like the transcription factor GL2, is a member of the large homeodomain-Leu zipper protein family of Arabidopsis (Sessa et al., 1993). GL2 and several MYB transcription factors are required for regulation of root hair development (DiCristina et al., 1996; Lee and Schiefelbein, 1999). Thus, it is tempting to speculate that regulatory proteins may coordinately regulate root hair development and LeExt1 expression in tomato.
All plant species displaying root hair-specific GUS activity also exhibited GUS staining in pollen and germinating pollen tubes (Fig. 7). A similar staining pattern was observed with the shorter fragments Δgen0.9 and Δgen0.6, although at lower intensities (data not shown). These results show that the Δgen1.1 promoter activity is correlated with cellular tip growth rather than specifically with root hair expansion. In contrast to the GUS expression data, we were unable to detect LeExt1 mRNA in pollen (Fig. 8), probably because of rapid degradation of LeExt1 mRNA or suppression of the endogenous gene in pollen. The LeExt1 promoter analysis suggests that transcriptional regulators exist in plants that direct the expression of specific genes in tip-growing cells. The absence of a similar regulation of GUS expression in Arabidopsis (Fig. 9) may be because of the fact that the corresponding regulatory proteins in Arabidopsis do not interact with the tomato Δgen1.1 promoter fragment. In the future, the LeExt1 promoter will serve as an essential tool in attempts to modify root hair gene expression in solanaceous species.
LeExt1 encodes a novel extensin-like protein that belongs to a small multigene family (Fig. 1). The protein shares high similarity with the two recently identified two-domain extensin-like proteins Dif10 and Dif54 from tomato because of the presence of repetitive elements contributing to the sequence similarity (Fig. 2). The Ser-Pro4 motif is usually abundant in extensins and seems to be significant for the structure (Cassab, 1998). In contrast to Dif10 and Dif54, which contain eight and five Ser-Pro3-6 domains in their C-terminal part, respectively (Bucher et al., 1997), LeExt1 contains a single Ser-Pro5 domain at the C terminus (Figs. 1A and 2). A putative N-terminal signal peptide for translocation into the endoplasmic reticulum indicates that LeExt1 is secreted into the apoplast and thus may play a role in determining physicochemical characteristics of the root hair cell wall. Cell wall strength, rigidity, and extensibility are critical factors determining tip growth of a root hair. Memelink et al. (1993) reported on the overexpression and antisense repression of a tobacco extensin that did not result in an altered phenotype in transgenic plants, although the encoded protein constituted the majority of HRGPs in roots, stems, and leaves. Thus, despite the large number of cloned extensins, the function(s) of single extensin genes are still elusive. Constitutive overexpression of LeExt1 in transgenic tomato failed to give conclusive results on the function of the protein. Thus, the biological role of LeExt1 remains to be determined. Down-regulation strategies for LeExt1 gene expression may shed more light on LeExt1 function in the future.
Materials And Methods
Plant Growth Conditions
Various organs were collected from flowering tomato (Lycopersicon esculentum var. Moneymaker) plants grown in soil in the greenhouse and were used for RNA extraction. Tomato seedlings used for root hair isolation were grown as described (Bucher et al., 1997). Seeds were rinsed in 70% (v/v) ethanol, washed in 1.4% (v/v) bleach with Triton X-100, and finally thoroughly washed with sterile water. Tomato seedling roots were used for in situ hybridization or GUS staining, and were grown on filter paper that had been soaked in one-half-strength Hoagland solution under sterile conditions in petri dishes. Potato plantlets and tomato seedlings were grown in vertically oriented petri dishes on Murashige and Skoog medium (Murashige and Skoog, 1962) with 2% (w/v) Suc and 1% (w/v) agarose. Arabidopsis seedlings were grown on Murashige and Skoog medium, pH 5.8, 1% (w/v) Suc, and 1.2% (w/v) agarose. After incubation of the seeds in the dark at 4°C for 2 to 4 d, the seedlings were grown in a vertical orientation in a plant tissue culture incubator (Forma Scientific, Inc., Brouwer AG, Luzern, Switzerland). For the in vitro pollen tube assay, pollen was collected from LeExt1 promoter/GUS transgenic lines and allowed to germinate in germination medium {292 mm Suc, 1 g L−1 [w/v] casein hydrolysate [Difco, Chemie Brunschwig AG, Basel], 213.25 mm MES [2-(N-morpholino)-ethanesulfonic acid]-KOH, pH 5.9, 1 mm CaCl2, 1 mm KCl, 0.8 mm MgSO4, 30 μm CuSO4, and 1.6 mm H3BO4}.
To investigate the influence of AVG and ethylene on LeExt1 expression, tomato seedlings containing the Δgen1.1/GUS construct were first grown on 2× Murashige and Skoog medium (2% [w/v] Suc). At a primary root length of 3 to 5 mm, seedlings were transferred to medium containing 20 μm AVG (Sigma, Buchs, Switzerland) for an additional 2 d before GUS activity was assayed. Alternatively, the seeds were germinated on plates containing 2× Murashige and Skoog medium supplemented with 50 μm AVG for 2 d before they were transferred to medium containing AVG and 1 mm 2-chloroethyl-phosphonic acid (ethephon, Sigma). GUS activity was assayed between 1 and 2 d later after an additional growth of about 1 cm.
For the analysis of LeExt1 overexpression in tomato, the expansion of root, root hair, and leaf epidermal cell was microscopically investigated using seedling roots and epidermal strips from the abaxial side of tomato cotyledons and leaves from greenhouse-grown transgenic plants that were shown to constitutively express LeExt1 via RNA gel-blot analysis (data not shown). Pollen tube growth was evaluated microscopically with pollen from homozygous transgenic plants germinated for 4 h in germination medium (see above).
Plasmids, Gene Constructs, and Plant Transformation
A differential screen of a root hair-specific cDNA library led to the isolation of LeExt1 cDNA in the plasmid pBluescript SK− (Stratagene, Amsterdam; Bucher et al., 1997). This cDNA then was used to screen a tomato genomic DNA library (CLONTECH Laboratories, Heidelberg) according to standard protocols (Sambrook et al., 1989). λ-DNA was prepared according to Locket (1990). A genomic DNA fragment of about 3.4 kb in length was isolated and cloned into pBluescript SK−. Sequencing was performed using T7 DNA polymerase and revealed a 204-bp overlap of the genomic fragment with the 5′ end of the LeExt1 cDNA sequence and extension into the 5′-upstream non-coding region of the LeExt1 gene. PCR-directed amplification using Pyrococcus furiosus DNA polymerase (Stratagene) yielded a 3.3-kb genomic fragment that was finally cloned into pBluescript SK−. Serial deletions with exonuclease III and S1 nuclease according to the manufacturer's protocol (Fermentas, Vilnius. Lithuania) finally yielded five fragments of approximately 2.2, 1.7, 1.1, 0.9, and 0.6 kb, respectively. These fragments were subsequently named Δgenx, where x represents the approximate length of the fragments as listed above. Each of these five fragments was then cloned with an exact fusion at the putative initiator ATG of the LeExt1 gene to the GUS marker gene (Jefferson et al., 1987), flanked by the nos 3′ terminator (Depicker et al., 1982) into the binary vector Bin19 (Bevan, 1984). To construct the chimeric LeExt1 gene for overexpression in tomato under the contol of the 35S cauliflower mosaic virus constitutive promoter, the SmaI-BamHI LeExt1 cDNA insert was inserted in its forward orientation into the EcoRV-BamHI sites of a plant expression cassette containing the 35S cauliflower mosaic virus promoter and the T-DNA octopine synthase gene terminator in the binary vector Bin19 (Franck et al., 1980; Bevan, 1984; Gielen et al., 1984). These constructs were then introduced in Agrobacterium tumefaciens strain C58C1 containing the pGV2260 plasmid (Deblaere et al., 1985) via electroporation. Transformation of tobacco var. Samsun, the miniature tomato cv Micro-Tom (Meissner et al., 1997) and tomato cv Moneymaker, and potato var. Désirée was performed using A. tumefaciens-directed gene transfer essentially as described by Rosahl et al. (1987) for tobacco, by Fillatti et al. (1987) for tomato, and by Rocha-Sosa et al. (1989) for potato. Arabidopsis ecotype C24 was transformed using the in planta transformation method (Bechtold and Pelletier, 1993; Katavic et al., 1994).
Computational Analysis
Sequence analysis was performed by using the Genetics Computer Group software package (Madison, WI; Devreux et al., 1984). The hydropathy analysis was performed according to Kyte and Doolittle (1982) with a window of nine amino acids. TFSEARCH (vers. 1.3) was used to search for putative transcription factor-binding sites (Heinemeyer et al., 1998). The program is available at http://pdap1.trc.rwcp.or.jp/research/db/TFSEARCH.html. Multiple sequence alignment was done using ClustalW, accessible at http://circinus.ebi.ac.uk:6543/cgi-bin/clustalw.cgi. Pretty printing and shading of multiple alignment files was done using Boxshade 3.21 at http://www.ch.embnet.org/software/BOX_form.html.
Gel-Blot Analysis
Genomic DNA and RNA gel-blot analysis were performed as described (Bucher et al., 1997).
RT-PCR
RT-PCR was done according to Borner et al. (2000). Reverse transcription using Superscript II RT (Gibco, Bascel) was performed with 1 μg of total RNA, which was extracted as described previously (Bucher et al., 1997). A 335-bp LeExt1 fragment was then amplified by PCR from the cDNA library using the oligonucleotides AATCCAATTCTTCTCAATGGGAAGC and GCTTCTTGTAGTAGTCATTCTTTGG. As a control, a 432-bp Rpl2 fragment was amplified using the oligonucleotides GAGAGGTGCACCACTTGCGC and GGCCAGCAGTTCCTCTTCAC. All PCR reactions were performed with 40 cycles. As a negative control, PCR reactions were performed with non-reverse transcribed total RNA to exclude fragment amplification because of the presence of genomic DNA in the samples. Fragments were separated on an agarose gel, blotted to a membrane, and hybridized with either radiolabeled LeExt1 or Rpl2 cDNA, respectively.
In Situ Hybridization
In situ hybridization of RNA was performed with paraffin-embedded roots of tomato var. Moneymaker as described earlier (Daram et al., 1998).
GUS Assay and Histochemical Analysis
Plant material was incubated in 0.1% (w/v) X-Gluc (Biosynth, Staad, Switzerland) and 0.1% (v/v) Triton X-100 (Fluka Chemie AG, Buchs, Switzerland) in 0.05 m sodium phosphate buffer, pH 7.2, at 37°C. Methanol was included in the pollen assay at 20% (v/v), because it was shown to almost completely suppress an endogenous GUS activity and thus to abolish background staining in pollen (Kosugi et al., 1990). The stained material was directly assayed for GUS activity or was fixed in ethanol:acetic acid (3:1, v/v) overnight at 4°C. Complete removal of chlorophyll from the tissue was performed in 100% ethanol. Samples were then embedded in Technovit 7100 (Kulzer, Wehrheim, Germany) and 5-μm-thick sections were mounted on glass slides. The indigo precipitate was visualized using a stereomicroscope (Olympus SZX12, Olympus Optical Schweiz AG, Volketswil, Switzerland) or by light microscopy (Olympus AZ70) in combination with Nomarski optics (GUS activity appears indigo blue).
ACKNOWLEDGMENTS
We thank Dr. Claus Frohberg (PlantTec Biotechnology, Potsdam, Germany) for primary analysis of Δgen1.1/GUS potato plants and valuable discussions; Romy Ackermann and Dr. Babette Regierer (Max-Planck-Institute of Molecular Plant Physiology, Potsdam) for Arabidopsis transformation and initial analysis; Drs. Vladimir Karandashov (Institute of Plant Physiology, Russian Academy of Sciences, Moscow) and David Trudgill (Scottish Crop Research Institute, Dundee, Scotland) for Phytophtora spp., mycorrhiza, and nematode infection studies; Drs. Rene Geurts and Ton Bisseling (Wageningen University, The Netherlands) for promoter/GUS analysis in M. truncatula; Birgit Schroeer (Max-Planck-Institute of Molecular Plant Physiology) for her technical support in the root hair promoter project at the Institut für Genbiologische Forschung (Berlin); and Sabine Klarer and Katalyn Konya for taking care of the greenhouse plants at ETH (Zurich).
Footnotes
This work was partially supported by the Swiss National Science Foundation, Biotechnology Priority Program (grant no. 5002–39814 to M.B.) and by ETH Zurich (to M.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010998.
LITERATURE CITED
- Ahn J, Choi Y, Kwon Y, Kim S, Choi Y, Lee J. A novel extensin gene encoding a hydroxyproline-rich glycoprotein requires sucrose for its wound-inducible expression in transgenic plants. Plant Cell. 1996;8:1477–1490. doi: 10.1105/tpc.8.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenijevic-Maksimovic I, Broughton W, Krause A. Rhizobia modulate root-hair-specific expression of extensin genes. Mol Plant-Microbe Interact. 1997;10:95–101. doi: 10.1094/MPMI.1997.10.1.95. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thalianaplants. CR Acad Sci Ser III Sci Vie. 1993;316:1194–1199. [Google Scholar]
- Bernhardt C, Tierney ML. Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol. 2000;122:705–714. doi: 10.1104/pp.122.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacteriumvectors for plant transformation. Nucleic Acids Res. 1984;12:8711. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 1999;17:657–665. doi: 10.1046/j.1365-313x.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000;24:591–599. doi: 10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- Brisson L, Tenhaken R, Lamb C. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell. 1994;6:1703–1712. doi: 10.1105/tpc.6.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M, Schroeer B, Willmitzer L, Riesmeier JW. Two genes encoding extensin-like proteins are predominantly expressed in tomato root hair cells. Plant Mol Biol. 1997;35:497–508. doi: 10.1023/a:1005869717158. [DOI] [PubMed] [Google Scholar]
- Cassab G. Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- Cormack RGH. Cell elongation and the development of root hairs in tomato roots. Am J Bot. 1945;32:190–196. [Google Scholar]
- Daram P, Brunner S, Persson BL, Amrhein N, Bucher M. Functional analysis and cell-specific expression of a phosphate transporter from tomato. Planta. 1998;206:225–233. doi: 10.1007/s004250050394. [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J. Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 1985;13:4777–4788. doi: 10.1093/nar/13.13.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depicker A, Stachel S, Dhaese P, Zambryski P, Goodman HM. Nopaline synthase: transcript mapping and DNA sequence. J Mol Appl Genet. 1982;1:561–573. [PubMed] [Google Scholar]
- Devreux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. The ArabidopsisAthb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organisation of the Arabidopsis thalianaroot. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Epstein J, Lamport D. An intramolecular linkage involving isodityrosine in extensin. Phytochemistry. 1984;23:1241–1246. [Google Scholar]
- Fillatti J, Kiser J, Rose R, Comai L. Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciensvector. Bio/Technology. 1987;5:726–730. [Google Scholar]
- Filpula DR, Lee SM, Link RP, Strausberg SL, Strausberg RL. Structural and functional repetition in a marine mussel adhesive protein. Biotechnol Prog. 1990;6:171–177. doi: 10.1021/bp00003a001. [DOI] [PubMed] [Google Scholar]
- Fleming AJ, Mandel T, Roth I, Kuhlemeier C. The patterns of gene expression in the tomato shoot apical meristem. Plant Cell. 1993;5:297–309. doi: 10.1105/tpc.5.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A, Guilley H, Jonard G, Richards K, Hirth L. Nucleotide sequence of Cauliflower Mosaic virus DNA. Cell. 1980;21:285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsisroot. Dev Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- Gielen J, De Beuckeleer M, Seurinck J, Deboeck F, De Greve H, Lemmers M, Van Montagu M, Schell J. The complete nucleotide sequence of the TL-DNA of the Agrobacterium tumefaciensplasmid pTiAch5. EMBO J. 1984;3:835–846. doi: 10.1002/j.1460-2075.1984.tb01894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie CS, Sherman DL, Blair GE, Brophy PJ. Periaxin, a novel protein of myelinating Schwann cells with a possible role in axonal ensheathment. Neuron. 1994;12:497–508. doi: 10.1016/0896-6273(94)90208-9. [DOI] [PubMed] [Google Scholar]
- Gilroy I, Jones DL. Through form to function: root hair development and nutrient uptake. Trends Plant Sci. 2000;5:56–60. doi: 10.1016/s1360-1385(99)01551-4. [DOI] [PubMed] [Google Scholar]
- Grierson C, Roberts K, Feldmann K, Dolan L. The COW1 locus of Arabidopsisacts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol. 1997;115:981–990. doi: 10.1104/pp.115.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions:β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 3901–3907 [DOI] [PMC free article] [PubMed]
- Katavic V, Haughn G, Reed D, Martin M, Kunst L. In planta transformation of Arabidopsis thaliana. Mol Gen Genet. 1994;245:363–370. doi: 10.1007/BF00290117. [DOI] [PubMed] [Google Scholar]
- Keller B, Lamb CJ. Specific expression of novel cell wall hydroxyproline-rich glycoprotein genes in lateral root initiation. Genes Dev. 1989;3:1639–1646. doi: 10.1101/gad.3.10.1639. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y. An improved assay for beta-glucuronidase in transformed cells: Methanol almost completely suppresses a putative endogenous beta-glucuronidase activity. Plant Sci. 1990;70:133–140. [Google Scholar]
- Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lawton MA, Dean SM, Dron M, Kooter JM, Kragh KM, Harrison MJ, Yu L, Tanguay L, Dixon RA, Lamb CJ. Silencer region of a chalcone synthase promoter contains multiple binding sites for a factor, SBF-1, closely related to GT-1. Plant Mol Biol. 1991;16:235–249. doi: 10.1007/BF00020555. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Locket TJ. A bacteriophage lambda DNA purification procedure suitable for the analysis of DNA from either large or multiple small lysates. Anal Biochem. 1990;185:230–234. doi: 10.1016/0003-2697(90)90284-g. [DOI] [PubMed] [Google Scholar]
- Masucci J, Schiefelbein J. The rhd6 mutation of Arabidopsis thalianaalters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 1994;106:1335–1346. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsisroot. Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R, Jacobson Y, Melamed S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A. A new model system for tomato genetics. Plant J. 1997;12:1465–1472. [Google Scholar]
- Memelink J, Swords KM, de Kam RJ, Schilperoort RA, Hoge JH, Staehelin LA. Structure and regulation of tobacco extensin. Plant J. 1993;4:1011–1022. doi: 10.1046/j.1365-313x.1993.04061011.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, Willmitzer L. Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J. 1989;8:23–29. doi: 10.1002/j.1460-2075.1989.tb03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosahl S, Schell J, Willmitzer L. Expression of a tuber-specific storage protein in transgenic tobacco plants: demonstration of an esterase activity. EMBO J. 1987;6:1155–1159. doi: 10.1002/j.1460-2075.1987.tb02348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan E, Grierson C, Cavell A, Steer M, Dolan L. TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol. 1998;138:49–58. [Google Scholar]
- Sambrook J, Frisch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- Schiefelbein J, Galway M, Masucci J, Ford S. Pollen tube and root-hair tip growth is disrupted in a mutant of Arabidopsis thaliana. Plant Physiol. 1993;103:979–985. doi: 10.1104/pp.103.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville CR. Genetic control of root hair development in Arabidopsis thaliana. Plant Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabelrauch LS, Kieliszewski M, Upham BL, Alizedeh H, Lamport DT. Isolation of pl 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 1996;9:477–489. doi: 10.1046/j.1365-313x.1996.09040477.x. [DOI] [PubMed] [Google Scholar]
- Schneider K, Mathur J, Boudonck K, Wells B, Dolan L, Roberts K. The ROOT HAIRLESS 1 gene encodes a nuclear protein required for root hair initiation in Arabidopsis. Genes Dev. 1998;12:2013–2021. doi: 10.1101/gad.12.13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Wells B, Dolan L, Roberts K. Structural and genetic analysis of epidermal cell differentiation in Arabidopsisprimary roots. Development. 1997;124:1789–1798. doi: 10.1242/dev.124.9.1789. [DOI] [PubMed] [Google Scholar]
- Sessa G, Morelli G, Ruberti I. The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 1993;12:3507–3517. doi: 10.1002/j.1460-2075.1993.tb06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers A, Schnepf E. Morphogenesis and polarity of tubular cells with tip growth. In: Kiermayer O, editor. Cell Biology Monographs. Vol. 8. New York: Springer-Verlag; 1981. pp. 265–299. [Google Scholar]
- Solano R, Nieto C, Avila J, Canas L, Diaz I, Paz-Ares J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 1995;14:1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiré C, De Rycke R, De Loose M, Inzé D, Van Montagu M, Engler G. Extensin gene expression is induced by mechanical stimuli leading to local cell wall strengthening in Nicotiana plumbaginifolia. Planta. 1994;195:175–181. doi: 10.1007/BF00199676. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. Epidermal cell differentiation in Arabidopsisdetermined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymer CL, Bibikova TN, Gilroy S. Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 1997;12:427–439. doi: 10.1046/j.1365-313x.1997.12020427.x. [DOI] [PubMed] [Google Scholar]
- Ye Z-H, Varner J. Tissue-specific expression of cell wall proteins in developing soybean tissues. Plant Cell. 1991;3:23–37. doi: 10.1105/tpc.3.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]