Abstract
The gl8 gene is required for the normal accumulation of cuticular waxes on maize (Zea mays) seedling leaves. The predicted GL8 protein exhibits significant sequence similarity to a class of enzymes that catalyze the reduction of a ketone group to a hydroxyl group. Polyclonal antibodies raised against the recombinant Escherichia coli-expressed GL8 protein were used to investigate the function of this protein in planta. Subcellular fractionation experiments indicate that the GL8 protein is associated with the endoplasmic reticulum membranes. Furthermore, polyclonal antibodies raised against the partially purified leek (Allium porrum) microsomal acyl-coenzyme A (CoA) elongase can react with the E. coli-expressed GL8 protein. In addition, anti-GL8 immunoglobulin G inhibited the in vitro elongation of stearoyl-CoA by leek and maize microsomal acyl-CoA elongase. In combination, these findings indicate that the GL8 protein is a component of the acyl-CoA elongase. In addition, the finding that anti-GL8 immunoglobulin G did not significantly inhibit the 3-ketoacyl-CoA synthase, 3-ketoacyl-CoA dehydrase, and (E) 2,3-enoyl-CoA reductase partial reactions of leek or maize acyl-CoA elongase lends further support to our previous hypothesis that the GL8 protein functions as a β-ketoacyl reductase during the elongation of very long-chain fatty acids required for the production of cuticular waxes.
Cuticular waxes are complex mixtures of very long-chain fatty acids (VLCFAs; >C18) and derivatives such as hydrocarbons, alcohols, aldehydes, ketones, and esters (Tulloch, 1976; Walton, 1990). These cuticle components are synthesized by plant epidermal cells (Kolattukudy, 1968; Kolattukudy and Buckner, 1972; Cassagne and Lessire, 1974, 1978). Although the processes by which VLCFAs are biosynthesized in higher plants are largely unknown, it has been proposed that the elongation of fatty acids occurs in a manner analogous to that involved in de novo fatty acid biosynthesis (Stumpf, 1984; von Wettstein-Knowles, 1995). De novo fatty acid biosynthesis is performed by fatty acid synthase, which, in plants, is a collection of four distinct enzymes that catalyze four sequential reactions: condensation, reduction, dehydration, and a second reduction. These reactions occur in the stroma of plastids and are catalyzed by soluble enzymes that, in a cyclic manner, add two carbons from malonyl-acyl carrier protein (ACP) to a growing acyl chains that are covalently bound to the prosthetic group of ACP (for review, see Ohlrogge and Jaworski, 1997).
In contrast to the involvement of ACP in de novo fatty acid synthesis, VLCFA biosynthesis involves the addition of two carbon atoms from malonyl-coenzyme A (CoA) to a growing acyl-CoA chain. In addition, whereas fatty acid synthase occurs in the plastid stroma and is catalyzed by soluble enzymes, VLCFA biosynthesis occurs in the cytoplasm and is catalyzed by enzymes associated with the endoplasmic reticulum (ER) membranes (Cassagne and Lessire, 1978; Agrawal et al., 1984; Post-Beittenmiller, 1996). These enzymes are collectively referred to as the acyl-CoA elongase. Acyl-CoA elongases have been partially purified from various plants, including leek (Allium porrum), Lunaria annua, Sinapis alba, Limnanthes alba, and oilseed rape (Brassica napus; for review, see Harwood, 1988). SDS-PAGE of partially purified leek acyl-CoA elongases reveals that they are composed of several protein components (Bessoule et al., 1989). Hence, it is thought that the acyl-CoA elongase is a complex of several enzymes that have distinct functions, but that collectively catalyze acyl elongation.
Experimental evidence has been accumulating over the past decade to support this hypothesis (for review, see von Wettstein-Knowles, 1987; Post-Beittenmiller, 1996). One line of evidence came from the identification of intermediates during acyl-CoA elongation in leek epidermal cells (Lessire et al., 1989, 1999). In addition, a 57-kD protein has been purified and cloned from jojoba (Simmondsia Chinensis) that has ketoacyl-CoA synthase activity and that can generate β-ketoacyl-CoA (Lassner et al., 1996). Additional evidence comes from the recent cloning of several Arabidopsis genes that encode ketoacyl-CoA synthases involved in the elongation of the fatty acids. These include FAE1, KCS1, and CUT1. Mutations in FAE1 specifically inhibit the accumulation of VLCFAs in seeds (James et al., 1995), whereas mutations in KCS1 (Todd et al., 1999) and CUT1 (Millar et al., 1999; Fiebig et al., 2000) affect the accumulation of VLCFAs associated with cuticular waxes.
Here, we report evidence that the maize (Zea mays) gl8 gene encodes the β-ketoacyl-CoA reductase of the acyl-CoA elongase. Sequence analysis of the cloned gl8 gene from maize reveals that it encodes a protein with significant amino acid similarity with a large family of keto reductases (Xu et al., 1997). Mutants of gl8 accumulate reduced amounts of cuticular waxes on seedling leaves. In addition, the chain lengths of those acyl derivatives that do accumulate are reduced relative to the wax of wild-type seedlings (Bianchi et al., 1979; Avato et al., 1987). Therefore, it was hypothesized that the maize gl8 gene encodes the β-ketoacyl-CoA reductase involved in the biosynthesis of the VLCFA required for the production of cuticular waxes (Xu et al., 1997). Here, subcellular fractionation studies have been performed, and they demonstrate that GL8 localizes to the ER. In addition, immunoinhibition of leek and maize acyl-CoA elongase activity by anti-GL8 immunoglobulin G (IgG) demonstrate that GL8 is a component of acyl-CoA elongase and provides strong evidence to support GL8 as the β-ketoacyl-CoA reductase of acyl-CoA elongase.
RESULTS
Characterization of the GL8 Protein in Maize
Antibodies are very convenient reagents for detecting and characterizing the function of a specific gene product. Therefore, to begin the characterization of the biochemical function of the gl8 gene, the GL8 protein was first expressed in Escherichia coli, and the resulting recombinant protein was used as the antigen for immunizing rabbits to generate GL8 antibodies. The 0.8-kb partial gl8 cDNA described by Xu et al. (1997) was cloned in-frame into the pET-30c expression vector (Novagen, Madison, WI) to generate pR8. A second construct containing only the C-terminal coding region from the 0.8-kb gl8 cDNA was cloned in-frame into the pET-30b expression vector (Novagen) to generate pCT. The proteins expressed by pR8 and pCT are expected to contain 167 amino acids and 77 amino acids, respectively, of GL8 protein fused at the N terminus to S- and His-tags from the expression vectors. The predicted sizes of the fusion proteins expressed by pR8 and pCT are 24 and 15 kD, respectively.
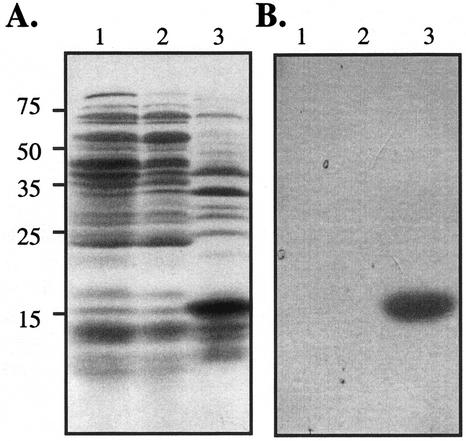
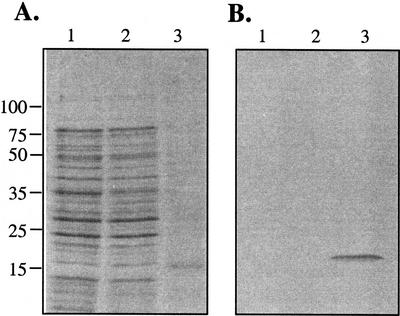
Figure 1A shows that E. coli BL21(DE3) that harbors pCT and has been induced with IPTG accumulates a prominent protein with the expected size of the GL8 fusion protein (15 kD). The identity of this protein was confirmed by western blotting using the S-protein alkaline phosphatase conjugate that detects the S-tag fused to the N terminus of the pCT protein (Fig. 1B). Similar analyses confirmed the size and identity of the pR8-expressed protein (data not shown).
Figure 1.
Expression of the His6-S-tag-GL8 fusion protein in E. coli. A, Protein extracts from E. coli strain BL21(DE3) (lane 1), BL21(DE3) harboring pET 30b (lane 2), and E. coli cell harboring pCT (lane 3). Following the induction of protein expression with isopropyl β-d-thiogalactoside (IPTG), proteins were extracted from the resulting cultures, fractionated by SDS-PAGE, and stained with Coomassie Brilliant Blue. Approximately 100 μg of protein was loaded in each lane. The positions of the molecular mass standards are indicated in kilodaltons. B, Proteins from a gel identical to that shown in A were transferred to a nitrocellulose filter. The expressed protein was detected with S-protein alkaline phosphatase conjugate as described in “Materials and Methods.”
The partial GL8 proteins expressed from pR8 and pCT were purified and injected into rabbits to generate polyclonal antibodies (see “Materials and Methods”). The serum from the rabbit challenged with the pCT-derived protein had a higher titer than that obtained with the pR8 construct and, therefore, was used for further experiments. The GL8 anti-serum was affinity purified as described in “Materials and Methods,” and was found to be able to immunologically detect the pCT- and pR8-derived GL8-expressed proteins (data not shown).
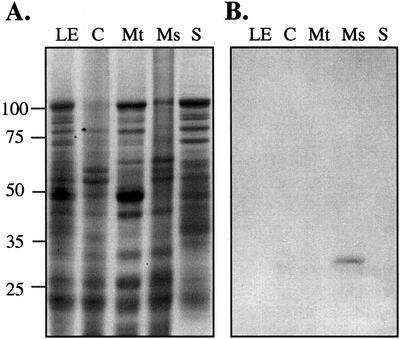
To assess the subcellular localization of the GL8 protein in maize leaf cells, cellular membranes were separated by differential centrifugation. Figure 2 reveals that the affinity-purified GL8 antibody detects a protein in the microsomal-enriched fraction, and that this protein is absent from chloroplast- and mitochondria-enriched membrane fractions and from the 100,000g supernatant. This immunologically detected protein is approximately 32 kD, the expected size of the mature GL8 protein after the cleavage of the predicted N-terminal signal peptide (Xu et al., 1997). The inability to immunologically detect the GL8 protein in unfractionated seedling leaf extract probably reflects its low abundance. The preimmune serum does not detect any proteins on western blots (data not shown).
Figure 2.
Subcellular localization of the GL8 protein. A, Proteins extracts from maize seedling leaves (LE), chloroplast membranes (C), mitochondria membranes (Mt), microsomal pellet (Ms), and 100,000g supernatant (S) fractions were fractionated by SDS-PAGE and stained with Coomassie Brilliant Blue. Each lane was loaded with 200 μg of protein. The positions of the molecular mass standards are indicated in kilodaltons. B, Protein samples from a gel identical to that shown in A were transferred to nitrocellulose and were incubated with affinity-purified GL8 antibodies as described in “Materials and Methods.”
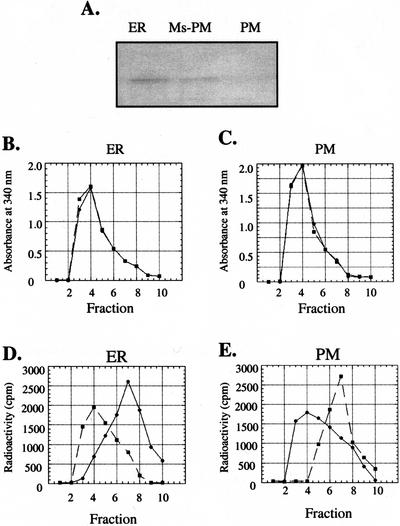
Because differential centrifugation of the postmitochondrial supernatant at 100,000g was used to isolate the microsomal fraction, this fraction is a mixture of small ER vesicles and other membranous material derived from the Golgi apparatus (GA), tonoplast (TN), and plasma membrane (PM; Lord, 1987). To localize the GL8 protein among these different membranes, each membrane fraction was purified and immunologically analyzed for the presence of the GL8 protein. The PM-enriched fraction was purified via a two-phase system as described in “Materials and Methods.” The ER membrane-enriched fraction was purified from the fraction remaining after the PM fraction was removed from the microsomal fraction. As shown in Figure 3A, the GL8 protein was immunologically detected in the ER-enriched fraction and the microsomal fraction following the removal of the PM, but was not detectable in the PM-enriched fraction. The identities and purities of these membrane fractions were determined by assaying for stereospecific NADH-ferricyanide reductase (Fredlund et al., 1996). The ER contains only the α-specific NADH-ferricyanide reductase, whereas the PM contains only the β-specific NADH-ferricyanide reductase. These enzyme activities were measured by monitoring the release of tritium (in separate reactions) from the α and β positions of the nicotinamide ring of NADH into the aqueous assay medium. Following each assay, the tritium associated with NADH was separated from that released into the assay medium by gel filtration chromatography. Fractions were collected, and the A340 and the radioactivity associated with each fraction was determined. As judged by the A340 profile, NADH was consistently eluted in fraction four from the ER- and PM-derived experiments (Fig. 3, B and C, respectively). When [4α-3H]NADH was incubated with the ER-enriched membrane fraction, tritium was recovered in fraction seven, which contains tritium released from NADH. In contrast, when [4β-3H]NADH was used as the substrate, the highest level of tritium was recovered in fraction four, coeluting with NADH (Fig. 3D). These results indicate that the ER-enriched fraction contains the α-specific but not the β-specific NADH-ferricyanide reductase activity. Hence, the ER-enriched fraction is not contaminated at a detectable level with the PM. In a similar manner, when [4α-3H]NADH or [4β-3H]NADH was incubated with the PM-enriched membrane fraction, tritium was only released from [4β-3H]NADH and not [4α−3H]NADH (Fig. 3E). Hence, the PM-enriched fraction contains the β-specific but not the α-specific NADH-ferricyanide reductase activity. These results establish that the PM-enriched fraction is not contaminated with the ER at a detectable level.
Figure 3.
Assay for the presence of the GL8 protein in ER- and PM-enriched fractions. A, Immunoblot analysis of the GL8 protein in the ER membrane-enriched fraction (ER), microsomal fraction after the removal of the PM (Ms-PM), and the PM-enriched fraction (PM). Eighty micrograms of protein was loaded per lane. Stereospecific NADH-ferricyanide reductase activity associated with the ER (B and D) and PM (C and E) fractions was determined by monitoring the release of tritium from [4α-3H]NADH (●) and [4β-3H]NADH (▪). The products of each assay were fractionated by gel filtration chromatography through a Sephadex G-10 column, and the A340 (B and C) and radioactivity (D and E) associated with each fraction were determined. The A340 identified the elution of NADH, which was recovered in fraction four. Tritium released from [4α-3H]NADH or [4β-3H]NADH was recovered in fraction seven. The ER fraction contains the α-specific NADH ferricyanide reductase, and the PM fraction contains the β-specific NADH ferricyanide reductase.
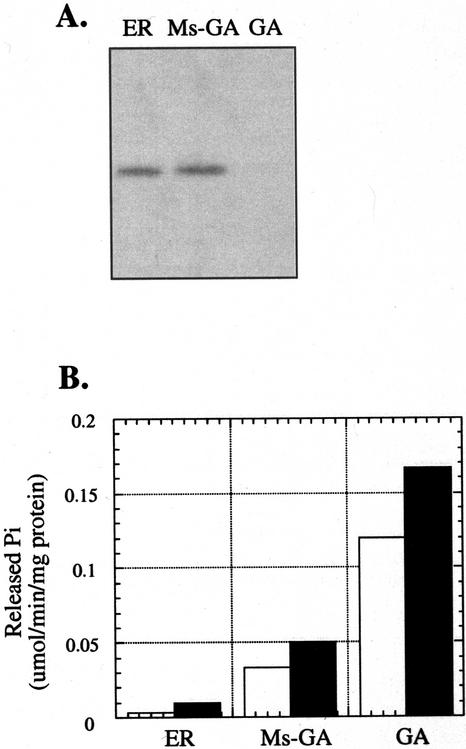
A GA-enriched fraction was purified from the microsomal fraction as described in “Materials and Methods.” A fraction containing the remaining intracellular membranes (including the ER) was also recovered. As shown in Figure 4A, the GL8 protein was immunologically detected in the ER-enriched fraction and the microsomal fraction after the removal of the GA, but not in the GA-enriched fraction. The enzyme marker IDPase was used to assay the identity and purity of the GA-enriched fraction. As shown in Figure 4B, the GA-enriched fraction has much higher IDPase activity as compared with the remaining membrane fractions, indicating that the ER fraction is relatively free of GA contamination.
Figure 4.
Assay for the presence of the GL8 protein in GA-enriched fraction. A, Immunoblot analysis of the GL8 protein in the ER-enriched fraction (ER), microsomal fraction after the removal of the GA (Ms-GA), and the GA-enriched fraction (GA). Eighty micrograms of protein was loaded per lane. B, IDPase activity associated with the membrane fractions described above. The open and filled bars represent IDPase activity in the absence and presence of Triton X-100, respectively.
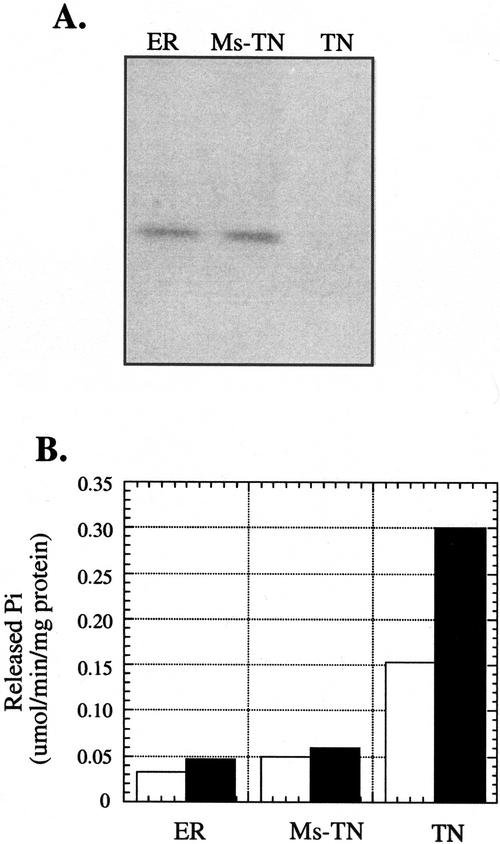
A TN-enriched fraction was purified from the microsomal fraction as described in “Materials and Methods.” The remaining intracellular membranes were also recovered. As shown in Figure 5A, the GL8 protein was immunologically detected in the ER-enriched fraction and the microsomal fraction after the removal of the TN, but not in the TN-enriched fraction. The nitrate-sensitive Mg2+-dependant ATPase was used as an enzyme marker to assay the identity and purity of the TN-enriched fraction. The TN-enriched fraction has substantially more ATPase activity than the other two membrane fractions. The addition of nitrate led to a substantially greater reduction in ATPase activity in the TN-enriched fraction than in the other membrane fractions. This result confirms the identity of the TN-enriched fraction and establishes that there is little TN contamination in the ER fraction.
Figure 5.
Assay for the presence of the GL8 protein in the TN-enriched fraction. A, Immunoblot analysis of the GL8 protein from the ER-enriched fraction (ER), microsomal fraction after the removal of the TN (Ms-TN), and the TN-enriched membrane fraction (TN). Eighty micrograms of protein was loaded per lane. B, Nitrate-sensitive Mg2+-dependent ATPase activity associated with the membrane fractions. Open and filled bars represent ATPase activities in the presence and absence of nitrate, respectively.
The GL8 Protein Is a Component of Acyl-CoA Elongase
Antibodies raised against the leek epidermis acyl-CoA elongase complex (Bessoule et al., 1989, 1992) detect the recombinant GL8 protein expressed from the pCT construct (Fig. 6). Similar results were obtained with the protein expressed from the pR8 construct (data not shown). The finding that the leek acyl-CoA elongase complex antibodies can interact with the GL8 protein provides a strong indication that this protein is one of the components of the acyl-CoA elongase complex.
Figure 6.
Immunoreaction of the GL8 protein with leek anti-acyl-CoA elongase antibodies. A, Protein extracts from E. coli strain BL21(DE3) (lane 1), BL21(DE3) harboring pET 30b (lane 2), and purified expressed protein from BL21 (DE3) cells harboring pCT and induced with IPTG (lane 3). Proteins were fractionated by SDS-PAGE and were stained with Coomassie Brilliant Blue. Approximately 140 μg of protein was loaded in lanes 1 and 2, and 5 μg was loaded in lane 3. B, Immunoblot analysis of protein samples as described in A with the leek anti-acyl-CoA elongase antibody at a dilution of 1:500.
This hypothesis was tested by determining the effects of GL8 antibodies on acyl-CoA elongase activity. The effects of the anti-GL8 IgG on stearoyl-CoA elongase activity in microsomal fractions isolated from etiolated leek and maize seedlings was assayed by comparing the stearoyl-CoA-dependent in vitro incorporation of [2-14C]malonyl-CoA into fatty acids after incubation of leek or maize microsomal fractions with the preimmune IgG or the anti-GL8 IgG fraction. Following incubation of the microsomal fraction with control preimmune IgG, stearoyl-CoA elongase activity was 3.4 ± 0.4 and 0.73 ± 0.05 nmol mg−1 h−1 for leek and maize, respectively. However, following incubation with anti-GL8 IgG, the stearoyl-CoA elongase activity was inhibited by at least 40% of the control levels in the leek and maize microsomal fractions (Tables I and II).
Table I.
Inhibition of leek acyl-CoA elongase activity by anti-GL8 IgG
| Enzyme | Preimmune | Anti-GL8 | Inhibition |
|---|---|---|---|
| % | |||
| Stearoyl-CoA elongase | 3.4 ± 0.4 (5) | 1.9 ± 0.1 (5) | 44 |
| 3-Ketoacyl-CoA synthase | 0.32 ± 0.08 (4) | 0.29 ± 0.1 (4) | 9.4 |
| 3-Hydroxyacyl-CoA dehydrase | 50 ± 1 (4) | 44 ± 2 (4) | 12 |
| (E) 2,3 Enoyl-CoA reductase | 54 ± 2 (4) | 46 ± 2 (4) | 14 |
All enzymatic activities were measured using linear standard conditions. The stearoyl-CoA elongase activity was expressed as nanomoles per milligram per hour, the synthase as nanomoles per milligrams per 15 min, and the dehydrase and reductase as nanomoles per milligram per 30 min. IgG and microsomal proteins were mixed in a ratio of 2:1. The numbers of independent experiments performed are indicated in parentheses; in each experiment, assays were performed in triplicate.
Table II.
Inhibition of maize acyl-CoA elongase activity by anti-GL8 IgG
| Enzyme | Preimmune | Anti-GL8 | Inhibition |
|---|---|---|---|
| % | |||
| Stearoyl-CoA elongase | 0.73 ± 0.05 (6) | 0.44 ± 0.08 (5) | 40 |
| 3-Ketoacyl-CoA synthase | 0.15 ± 0.02 (6) | 0.14 ± 0.03 (6) | 6.7 |
| 3-Hydroxyacyl-CoA dehydrase | 104 ± 6 (4) | 102 ± 4 (4) | 2.0 |
| (E) 2,3 Enoyl-CoA reductase | 107 ± 5 (4) | 103 ± 5 (4) | 4.0 |
All enzymatic activities were measured using linear standard conditions. The stearoyl-CoA elongase activity was expressed as nanomoles per milligram per hour, the synthase as nanomoles per milligram per 15 min, and the dehydrase and reductase as nanomoles per milligram per 30 min. IgG and microsomal proteins were mixed in a ratio of 2:1. The numbers of independent experiments performed are indicated in parentheses; in each experiment, assays were performed in triplicate.
Acyl-CoA elongation is thought to be achieved by a complex of proteins with four separate enzymatic activities. These include 3-ketoacyl-CoA synthase, 3-ketoacyl-CoA reductase, 3-hydroxyacyl-CoA dehydrase, and (E) 2,3 enoyl-CoA reductase activities. The effect of anti-GL8 antibodies on three of these activities [3-ketoacyl-CoA synthase, 3-hydroxyacyl-CoA dehydrase, and (E) 2,3 enoyl-CoA reductase] was determined individually. 3-Ketoacyl-CoA synthase activity was determined by an assay analogous to the stearoyl-CoA elongase assay, but in the absence of reducing reagents (NADH and NADPH). In the absence of these reducing agents, the elongation of stearoyl-CoA proceeds only to the 3-ketoacyl-CoA intermediate, and cannot proceed further. The product of this reaction, 3-ketoicosanyl-CoA, is chemically unstable, and was recovered after processing of the reaction products as the methylketone, nonadecanone. Thus, the 3-ketoacyl-CoA synthase activity was measured as the rate of stearoyl-CoA-dependent incorporation of radioactivity from [2-14C]malonyl-CoA into nonadecanone. When the reaction was conducted after incubation of the elongase-containing microsomes with preimmune IgG or anti-GL8 IgG, 3-ketoacyl-CoA synthase activity was inhibited by only 9.4% (Table I) and 6.7% (Table II) in leek and maize, respectively.
3-Hydroxyacyl-CoA dehydrase activity was determined as the conversion of [1-14C]3-hydroxyeicosanoyl-CoA to icosenoyl-CoA, which was recovered following saponification and separation by thinlayer chromatography (TLC) as the free unsaturated acid. Relative to the effect of incubating elongase-containing microsomes with control preimmune IgG, anti-GL8 IgG inhibited the leek and maize dehydrase activities by only 12% (Table I) and 2% (Table II), respectively. The effect of the anti-GL8 antibodies on (E) 2,3 enoyl-CoA reductase activity was also determined. This activity was measured as the conversion of [1-14C](E)-2,3-eicosenoyl-CoA to icosanoyl-CoA, which was recovered following saponification and separation by TLC as the free fatty acid. In these experiments, anti-GL8 IgG caused 14% inhibition of this activity in leek microsomes (Table I) and 4% inhibition in maize microsomes (Table II).
Because of the chemical instability of 3-ketoacyl derivatives, it was not possible to directly measure 3-ketoacyl-CoA reductase. However, based on the sequence similarity between the GL8 protein and ketoacyl reductases and the observation that anti-GL8 antibodies inhibited the overall elongase activity of leek and maize microsomal extracts by at least 40%, and yet inhibited the 3-ketoacyl-CoA synthase, 3-hydroxy acyl-CoA dehydrase, and (E) 2,3 enoyl-CoA reductase activities by 14% or less, it is reasonable to consider that the primary target of the anti-GL8 antibodies is the 3-ketoacyl-CoA reductase component of the acyl-CoA elongase.
DISCUSSION
Polyclonal antibodies generated against the maize GL8 protein were used to experimentally confirm the sequence-based prediction that this protein is membrane associated. Specifically, differential centrifugation of cellular membranes of maize seedlings indicates that the GL8 protein is entirely recovered in the microsomal membrane fraction. Further fractionation of the microsomal membranes clearly indicates that the GL8 protein is not associated with PM, GA, or TN fractions, but that it is associated with the ER membranes. This finding is consistent with the role of GL8 as a component of acyl-CoA elongase because studies in leek and other species have demonstrated that elongase activity is associated with the ER fraction (Agrawal et al., 1984; Lessire et al., 1985a, 1985b, 1985c, 1989).
The experimental finding that the GL8 protein is associated with the ER is in contradiction to earlier computational predictions generated by the PSORT algorithm (Nakai and Kanehisa, 1992), which suggested that the GL8 protein is associated with the PM. The PSORT algorithm predicts that GL8 contains an N-terminal 29-amino acid signal peptide, which targets this protein to the ER, but because the GL8 protein does not contain any of the known ER retention consensus signals, the protein was predicted to be associate with the PM. The discrepancy between the computational prediction and the experimental findings may be explained by reports that demonstrate that Lys residues at position −3 and −4 or −3 and −5 can serve as ER retention and retrieval signals for ER proteins (Jackson et al., 1990; Andersson et al., 1999). Therefore, the Lys-rich C terminus of GL8 (−KKKAL) is likely an effective signal for ER retention.
ER-associated fatty acid elongases from maize and leek utilized stearoyl-CoA as elongation primers (Cassagne and Lessire, 1978; Lessire, et al., 1982). The stearoyl-CoA elongase activity involves four separate reactions: 3-ketoacyl-CoA synthase, 3-ketoacyl-CoA reductase, 3-hydroxyacyl-CoA dehydrase, and (E) 2,3 enoyl-CoA reductase. It has been proposed that the gl8 gene encodes the 3-ketoacyl-CoA reductase associated with the stearoyl-CoA elongase activity. The ability of antibodies raised against a partially purified acyl-CoA elongase from leek to react with the E. coli-expressed GL8 protein indicates that GL8 is a component of acyl-CoA elongase. To further demonstrate this, antibodies raised against the maize GL8 protein were tested for their ability to inhibit the in vitro stearoyl-CoA elongase activity that has been previously characterized for leek microsomal fractions (Lessire et al., 1999). The maize GL8 antibody was able to inhibit the elongation of stearoyl-CoA by leek and maize microsomal extracts by at least 40%. These data provide strong evidence that GL8 is a component of the acyl-CoA elongase.
Support for the hypothesis that GL8 encodes the 3-ketoacyl reductase component of the elongase complex was obtained by measuring the immunoinhibitions of three component reactions from the elongase complex, 3-ketoacyl-CoA synthase, 3-hydroxy acyl-CoA dehydrase, and 2,3 enoyl-CoA reductase. It is unfortunate that it was not possible to directly measure the immunoinhibition of 3-ketoacyl-CoA reductase, the predicted GL8 enzymatic function, due to the extreme instability of the substrate (3-ketoacyl-CoA) for this enzyme. However, because anti-GL8 antibodies immunoinhibited elongase activity by at least 40%, whereas inhibition of the other three component activities was 14% or less, it is compelling to conclude that the 3-ketoacyl-CoA reductase is the primary target for the anti-GL8 IgG inhibition. Hence, these data, in combination with the sequence of the GL8 protein and its localization to the ER, provide convincing evidence to support the hypothesis that GL8 is the β-ketoacyl reductase component of acyl-CoA elongase.
MATERIALS AND METHODS
Plant Materials
Maize (Zea mays) seedling leaves (inbred line B73) were harvested at the two-leaf stage after being grown for 10 d in a greenhouse sand bench. Leek (Allium porrum) seeds, stored overnight at 4°C, were surface sterilized with sodium hypochlorite in the presence of Triton X-100 for 2 min and were then washed with distilled water. Sterilized seeds were germinated and grown for 7 d in the dark.
Preparation of Membrane Fractions
Maize membrane fractions were prepared as described by Walker et al. (1987) and Douce et al. (1987) with minor modifications. All steps were performed at 4°C. In brief, seedling leaves were pulverized in liquid N2 and were further homogenized in extraction buffer (50 mm Tris HCl, pH 8.0, 1 mm EDTA, 10% [w/v] Suc, 40 mm 2-mercaptoethanol, and 1 mm phenylmethylsulfonyl fluoride). The homogenate was filtered through four layers of cheesecloth and was centrifuged at 10,000g for 10 min to pellet chloroplast-derived membranes. The resulting supernatant was centrifuged at 20,000g for 30 min to pellet the mitochondrial membranes. The resulting supernatant was centrifuged at 100,000g for 1 h to pellet the microsomal fraction. The remaining supernatant was retained as the soluble fraction.
A high-purity PM fraction was prepared using an aqueous Dextran-polyethylene glycol two-phase system (Larsson et al., 1987). The microsomal fraction was suspended in 10 mL of suspension buffer (0.33 m Suc, 3 mm KCl, and 5 mm potassium phosphate, pH 7.8). Nine grams of this suspension was mixed with 27.0 g of the phase mixture (11.2 g of 20% [w/v] Dextran T-500, 5.58 g of 40% [w/v] polyethylene glycol 3350, 3.05 g of Suc, 0.675 mL of 0.2 m potassium phosphate, pH 7.8, and 0.041 mL of 2 m KCl, plus distilled water to a final weight of 27.0 g). This two-phase mixture was mixed thoroughly and was separated by centrifugation at 1,500g for 5 min. The upper phase was recovered and subjected to two additional phase partitions. The final upper phase, enriched for the PM fraction (Larsson et al., 1987), was diluted with two volumes of suspension buffer and was centrifuged at 100,000g for 30 min to pellet the PMs.
The procedure for the isolation of ER membranes was modified from that of Lord (1987). The lower phase recovered from the PM isolation procedure was diluted at least 10-fold with the suspension buffer and was subjected to centrifugation at 100,000g for 30 min. The pellet was resuspended in 150 mm Tricine, pH 7.5, 10 mm KCl, 1 mm EDTA, pH 7.5, 1 mm MgCl2, and 12% (w/w) Suc. This suspension was layered onto a continuous Suc density gradient. This gradient consisted of 25 mL of Suc solution increasing linearly in concentration from 30% to 60% (w/w) Suc, with a 5-mL layer of 20% (w/w) Suc on top and a 2-mL cushion of 60% (w/w) Suc on the bottom. All of the Suc solutions contained 1 mm EDTA, pH 7.5. The gradient was centrifuged at 83,000g for 3 h. The interface below the 20% (w/w) Suc layer was rescued as the ER-enriched fraction.
The procedure for the isolation of the GA was modified from that of Green (1983). The microsomal fraction was layered over a discontinuous Suc gradient consisting of 7 mL of 1.6 m Suc layered over 7 mL of 1.8 m Suc and was centrifuged at 30,000g for 30 min. Immediately after the centrifugation, the layer above the 1.6 m Suc pad was collected as the microsomal fraction without GA and was pelleted by centrifugation at 100,000g for 1 h. Meanwhile, 7-mL aliquots of the 1.5, 1.25, and 0.5 m Suc solutions were layered on top of the interface above the 1.6 m Suc layer, and the gradient was centrifuged at 100,000g for 3 h. The interface that formed between the 0.5 and 1.25 m Suc layers and between the 1.25 and 1.5 m Suc layers were collected and pooled, diluted with 0.2 m Suc, and pelleted by centrifugation at 100,000g for 1 h. The resulting pellet was recovered as the GA-enriched fraction.
A TN-enriched fraction was purified from the microsomal fraction via the method of Jacoby (1987). The microsomal pellet was washed with washing buffer (0.25 m Suc, 2 mm dithiothreitol [DTT], 5 mm Tris/MES [2-(N-morpholino)-ethanesulfonic acid], pH 6.5, and 500 mm KI), and the pellet was resuspended in 0.25 m Suc, 2 mm DTT, and 5 mm Tris/MES, pH 6.5, and loaded on a discontinuous Suc gradient with 10% and 23% (w/v) Suc layers. Following centrifugation at 100,000g for 2 h, the TN-enriched fraction was recovered from the interface between the 10% and 23% (w/v) Suc layers.
Leek microsomal fractions were prepared by homogenizing 5 to 8 g of 7-d-old etiolated leek seedlings in a mortar with 50 mL of 0.08 m HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] buffer (pH 7.2) containing 10 mm 2-mercaptoethanol and 0.32 m Suc. The homogenate was then filtered through two layers of cheesecloth and was centrifuged at 3,000g for 5 min. The supernatant was centrifuged at 12,000g for 20 min. The resulting pellet was discarded and the supernatant was centrifuged at 189,000g for 15 min. The microsomal pellet was resuspended in 2 mL of 0.08 m HEPES buffer (pH 6.8) containing 10 mm 2-mercaptoethanol.
Protein concentrations were determined via the method of Bradford (1976) by using the Bio-Rad Protein Assay kit (Hercules, CA).
Enzyme Assays
Two NADH-ferricyanide reductases with different stereospecificities for the α-hydrogen and the β-hydrogen atoms on the nicotinamide ring of NADH were used as enzyme markers for the ER and PM. The stereospecificities of the NADH-ferricyanide reductase activities obtained from the different membrane fractions were assayed as described by Fredlund et al. (1996). The reaction buffer included 180 nm [4α-3H]NADH or [4β-3H]NADH, 100 μM potassium ferricyanide, and 50 mm MOPS [3-(N-morpholino)-propanesulfonic acid]-KOH, pH 7.2. The reaction was started by the addition of equal amounts of protein (200 μg) from the indicated membrane fraction, followed by incubation at room temperature for 10 min, and the reaction was stopped by boiling for 2 min. After adding unlabeled NADH to a final concentration of 0.5 mm, an aliquot was subjected to gel filtration chromatography through a 9-mL Sephadex G-10 column and was eluted with 50 mm MOPS, pH 7.2. The radioactivity in each 1-mL elution fraction was determined by liquid scintillation counting. The presence of NADH in the fractions was monitored by A340.
[4α-3H]NADH and [4β-3H]NADH were synthesized by reducing [4-3H]NAD+ (Amersham Biosciences AB, Uppsala) with Glc-6-P dehydrogenase (Boehringer Molecular Biochemicals, Indianapolis) and alcohol dehydrogenase (Boehringer Molecular Biochemicals), respectively (Fredlund et al., 1996). The H+ donors for the two reactions were Glc-6-P and ethanol, respectively.
IDPase was used as an enzyme marker for the GA-enriched fraction (Green, 1983). The reaction was started by the addition of the membrane suspension to the reaction buffer (5 mm IDP, 1 mm MgCl2, and 50 mm Tris, pH 7.5, with or without 0.1% [v/v] Triton X-100), it incubated at 37°C for 60 min, and the reaction was stopped with the addition of cold 12.5% (w/v) trichloroacetic acid. Released phosphate was measured by the method of Taussky and Shorr (1953). In brief, the concentration of phosphate was determined by A720 in ferrous sulfate-ammonium molybdate reagent (100 mL of solution freshly prepared with 1 g of ammonium molybdate, 10 mL of 10 n sulfuric acid, and 5 g of FeSO4 7H2O) and was compared with concentration standards.
Nitrate-sensitive Mg2+-ATPase was used as an enzyme marker for the TN (Jacoby, 1987). The reaction was started by the addition of a membrane aliquot to the reaction buffer (30 mm Tris/MES, pH 8.0, 50 mm KCl, 3 mm Tris/ATP, 3 mm MgSO4, 0.1 mm vanadate, 0.5 mm azide, and 0.1 mm ammonium molybdate) in the presence or absence of 20 mm KNO3. Following incubation at 37°C for 60 min, the reaction was stopped by the addition of cold 12.5% (w/v) trichloroacetic acid. The amount of released phosphate was determined by the method of Taussky and Shorr (1953).
Stearoyl-CoA elongase activity was assayed by mixing 60 μg of microsomal proteins in a 0.08 m HEPES buffer (pH 7.2) with 0.5 mm NADPH, 0.5 mm NADH, 2 mm DTT, 1 mm MgCl2, 9 μm acyl-CoA, and 17 μm [2-14C] malonyl-CoA (Lessire et al., 1999). The reaction mixture (0.1 mL) was incubated at 30°C for 1 h. Then, 100 μL of 5 n KOH and 10% (w/v) methanol were added, and the lipids were saponified for 1 h at 70°C. After acidification with 0.1 mL of 10 n H2SO4 containing 10% (w/v) malonic acid, the fatty acids were extracted with 2 mL of chloroform, and the radioactivity associated with fatty acids was determined after evaporation of chloroform using a liquid scintillation counter.
To assay 3-ketoacyl-CoA synthase, 60 μg of microsomal proteins was incubated for 15 min at 30°C under the same conditions as the stearoyl-CoA elongase assay except that NADH and NADPH were omitted. In these conditions (i.e. the absence of reducing agents), the product of the condensation reaction, 3-ketoeicoanoyl-CoA, was recovered as the methylketone, nonadecanone, which was extracted into the chloroform fraction, and the radioactivity associated with that fraction was determined by liquid scintillation counting.
To assay for 3-hydroxyacyl-CoA dehydrase, 60 μg of microsomal proteins was incubated for 30 min at 30°C in the presence of 11.4 μM [1-14C]3-hydroxyeicosanoyl-CoA, 1 mm MgCl2, 2 mm DTT, and 3 mm Triton X-100. The reaction was stopped by the addition of 0.1 mL of 5 n KOH, and the reaction mixture was heated for 1 h at 70°C. After acidification, the fatty acids were extracted with chloroform and separated by TLC. The silica gel containing icosanoate was scraped from the TLC plate, and the radioactivity associated with this fraction was determined.
(E) 2,3 Enoyl-CoA reductase activity was measured by incubating 100 μm NADPH, 1 mm MgCl2, 11.4 μM [1-14C] (E)-2,3-eicosenoyl-CoA, 2 mm DTT, and 3 mm Triton X-100 in a 0.08 m HEPES (pH 7.2) buffer in a final volume of 0.1 mL for 30 min at 30°C. As with the elongase assay, the reaction was stopped by saponification, followed by acidification and extraction of the fatty acid product with chloroform. Following TLC fractionation, the radioactivity associated with icosanoate was determined.
Analysis of Elongation Products
Following the acyl-CoA elongase assay and the elongase component assays, the chloroform extracts that contained the products of the reactions were fractionated by TLC using 60F 254 plates (Merck, Whitehouse Station, NJ) developed with hexane:diethyl ether:acetic acid (75:25:1). The different components were identified by comparison with the RF of standards, and the radioactivity was quantified by autoradiography or by using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Generation of Antibodies
The pR8 expression construct was generated by cloning the 0.8-kb partial gl8 cDNA from p88 m into the EcoRI and XhoI sites of the pET-30c (Novagen, Madison WI) in-frame with the expression cassette. The pCT expression construct was generated by PCR-amplifying a portion of the p88 m cDNA insert with the universal (5′-GTAAAACGACGGCCAGT-3′) and gl8e3 (5′-GTGGCGACAAAGCTTGCATCTATCAGGAAGTCT-3′) primers. Primer gl8e3 contains a sequence mismatch relative to the gl8 sequence that generates a HindIII restriction site in the amplified product, which then allowed the product to be cloned in-frame with the expression cassette of the pCT-30b expression vector (Novagen). Both constructs were transformed into BL21(DE3). Expression was induced by the addition of IPTG to a final concentration of 0.4 mm to exponentially growing cultures. pR8 and pCT fusion proteins accumulated as inclusion bodies. The proteins were solubilized with 6 m urea and were purified using Novagen His-Bind resin columns. Eluted proteins were dialyzed against a series of 1× phosphate-buffered saline (170 mm NaCl, 6.2 mm KCl, 12.6 mm Na2HPO4, and 2.2 mm KH2PO4, pH 7.4) solutions containing decreasing concentrations of urea (4, 3, 2, 1, and 0.5 m), and finally against phosphate-buffered saline without urea (both proteins precipitated at 2 m urea). Protein precipitate suspensions were then injected into rabbits to generate polyclonal antibodies for each GL8 fusion protein. Rabbits were injected with approximately 0.5 mg of GL8 protein emulsified with Freund's Complete Adjuvant and were subsequently challenged with two additional injections of 0.5 mg of GL8 protein emulsified with Freund's Incomplete Adjuvant.
Affinity Purification of Antibodies
The resulting anti-GL8 sera were affinity purified (Sambrook et al., 1989). The expressed GL8 protein from the pR8 construct was fractionated by SDS-PAGE and was transferred to nitrocellulose membranes. Sections of nitrocellulose membranes containing the expressed GL8 protein were excised and incubated in blocking solution (5% [w/v] bovine serum albumin, 0.05% [w/v] Tween 20, 500 mm NaCl, and 20 mm Tris-HCl, pH 7.5) for 1 h. These blots were then incubated for 8 h at 4°C in a 1:50 dilution of the antiserum derived from the pCT construct. The strip was then washed three times with blocking solution for 30 min. The affinity-purified antibodies were then eluted with a low pH buffer (0.1 m Gly, pH 2.7) and were immediately neutralized with 1 m sodium phosphate buffer (pH 7.7) to a final concentration of 50 mm. They were stored at 4°C in the presence of 0.02% (w/v) sodium azide.
IgG Preparation
Preimmune and anti-GL8 sera (1 mL) were loaded onto 1-mL columns of Protein-A Sepharose pre-equilibrated with a 0.1 m phosphate (pH 8.0) buffer. The columns were then washed with the same buffer, and the IgG fractions were eluted using a 0.1 m citrate (pH 4.0) buffer. The IgG fractions were precipitated with 30% (w/v) polyethylene glycol and were resuspended in the phosphate buffer.
Immunoblot Analyses
SDS-PAGE was conducted according to standard methods (Sambrook et al., 1989). After SDS-PAGE, gels were stained with Coomassie Brilliant Blue R-250 or were electrophoretically transferred to nitrocellulose membranes in transfer buffer (48 mm Tris, pH 9.2, 39 mm Gly, and 20% [w/v] methanol) at 15 V for 40 min using a Trans-Blot Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). Immunoblotting procedures were based on the manufacture's protocol (Hames and Rickwood, 1981). In brief, blots were preincubated with blocking solution (5% [w/v] bovine serum albumin, 10 mm Tris, pH 8.0, and 150 mm NaCl) for 2 h at room temperature, followed by incubation with affinity-purified antibodies at the indicated dilutions for 1 to 3 h, washed with washing solution (10 mm Tris, pH 8.0, 150 mm NaCl, and 0.05% [w/v] Tween 20), and then incubated with anti-rabbit IgG alkaline phosphatase conjugate (Sigma, St. Louis) at a 1:30,000 dilution for 1 h. The blots were washed again with the washing solution and were incubated in the substrate mixture, 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium in alkaline phosphatase buffer (100 mm NaCl, 5 mm MgCl2, and 100 mm Tris, pH 9.5) until color developed (about 15 min). S-Tag western blotting was performed similarly. In brief, proteins were transferred from the SDS-PAGE gel to nitrocellulose membrane with transfer buffer (12 mm Tris, 96 mm Gly, pH 8.3, and 20% [w/v] methanol), incubated for 15 min at room temperature in Tris-buffered saline plus Tween 20 buffer (10 mm Tris, pH 8.0, 150 mm NaCl, and 0.1% [w/v] Tween 20) plus 1% (w/v) gelatin to block excess protein binding sites, S-protein alkaline phosphatase was added to a dilution of 1:5,000, and this was incubated for another 15 min at room temperature, washed with Tris-buffered saline plus Tween 20, and then treated as described above to allow color development.
ACKNOWLEDGMENTS
We thank Dr. Kristin R. Harkins for advice regarding the production of the GL8 antibodies, and Joel Hansen for assistance with the liquid scintillation counter.
Footnotes
This study was supported by the National Science Foundation (grant nos. IBM–9316832 and IBN–9808559 to P.S.S. and B.J.N.). C.R.D. was funded in part by a U.S. Department of Agriculture National Needs Fellowship in Plant Biotechnology. This is Journal Paper no. 19,396 of the Iowa Agricultural and Home Economics Experiment Station (Ames, IA). This is project no. 3,409 and was supported by Hatch Act and State of Iowa funds.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010621.
LITERATURE CITED
- Andersson H, Kappeler F, Hauri HP. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J Biol Chem. 1999;274:15080–15084. doi: 10.1074/jbc.274.21.15080. [DOI] [PubMed] [Google Scholar]
- Agrawal VP, Lessire R, Stumpf PK. Biosynthesis of very long chain fatty acids in microsomes from epidermal cells of Allium porrum L. Arch Biochem Biophys. 1984;230:580–589. doi: 10.1016/0003-9861(84)90438-7. [DOI] [PubMed] [Google Scholar]
- Avato P, Bianchi G, Nayak A, Salamini F, Gentinetta E. Epicuticular waxes of maize as affected by the interaction of mutant gl8 with gl3, gl4 and gl15. Lipids. 1987;22:11–16. doi: 10.1007/BF02534868. [DOI] [PubMed] [Google Scholar]
- Bessoule JJ, Creach A, Lessire R, Cassagne C. Evaluation of the amount of acyl-CoA elongase purified from leek (Allium porrum L.) epidermal cells. Biochem Biophys Acta. 1992;1117:78–82. doi: 10.1016/0304-4165(92)90165-q. [DOI] [PubMed] [Google Scholar]
- Bessoule JJ, Lessire R, Cassagne C. Partial purification of the acyl-CoA elongase of Allium porrum leaves. Arch Biochem Biophys. 1989;268:475–484. doi: 10.1016/0003-9861(89)90315-9. [DOI] [PubMed] [Google Scholar]
- Bianchi G, Avato P, Salamini F. Glossy mutants of maize. Heredity. 1979;42:391–395. [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cassagne C, Lessire R. Studies on alkane biosynthesis in epidermis of Allium porrum L. leaves. Arch Biochem Biophys. 1974;165:274–280. doi: 10.1016/0003-9861(74)90165-9. [DOI] [PubMed] [Google Scholar]
- Cassagne C, Lessire R. Biosynthesis of saturated very long chain fatty acids by purified membrane fractions from leek epidermal cells. Arch Biochem Biophys. 1978;191:146–152. doi: 10.1016/0003-9861(78)90076-0. [DOI] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Brouguisse R, Neuburger M. Isolation of intact mitochondria: general principles and criteria of integrity. Methods Enzymol. 1987;148:403–415. [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell. 2000;12:2001–2008. doi: 10.1105/tpc.12.10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredlund KM, Widell S, Moller IM. Stereospecificity of NADH-ferricyanide reductase is a convenient marker for the endoplasmic reticulum of plant cells. Plant J. 1996;10:925–933. [Google Scholar]
- Green JR. The Golgi Apparatus. In: Hall JL, Moore AL, editors. Isolation of Membranes and Organelles from Plant Cells. New York: Academic Press; 1983. pp. 135–152. [Google Scholar]
- Hames BD, Rickwood D. Gel Electrophoresis of Proteins: A Practical Approach. Oxford: IRL Press; 1981. [Google Scholar]
- Harwood JL. Fatty acid metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:101–138. [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;10:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby B. Characterization of tonoplast enzyme activities and transport. Methods Enzymol. 1987;148:105–115. [Google Scholar]
- James DW, Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy PE. Further evidence for an elongation decarboxylation mechanism in the biosynthesis of paraffins in leaves. Plant Physiol. 1968;43:375–383. doi: 10.1104/pp.43.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy PE, Buckner JS. Chain elongation of fatty acids by cell-free extract of epidermis from pea leaves (Pisum sativum) Biochem Biophys Res Commun. 1972;47:1306–1313. doi: 10.1016/s0006-291x(72)80212-2. [DOI] [PubMed] [Google Scholar]
- Larsson C, Widell S, Kjellbom P. Preparation of high-purity plasma membrane. Methods Enzymol. 1987;148:559–569. [Google Scholar]
- Lassner MW, Lardizabal K, Metz JG. A jojoba β-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell. 1996;8:281–292. doi: 10.1105/tpc.8.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessire R, Bessoule JJ, Cassagne C. Solubilization of C18-CoA and C20-CoA elongases from Allium porrum L. epidermal cell microsomes. FEBS Lett. 1985a;187:314–320. [Google Scholar]
- Lessire R, Bessoule JJ, Cassagne C. Involvement of a β-ketoacyl-CoA intermediate in acyl-CoA elongation by an acyl-CoA elongase purified from leek epidermal cells. Biochem Biophys Acta. 1989;1006:35–40. [Google Scholar]
- Lessire R, Chevalier S, Lucet-Levannier K, Lellouche JP, Mioskowski C, Cassagne C. Study of the 3-hydroxy eicosanoyl-coenzyme A dehydratase and (E)-2,3 enoyl-coenzyme A reductase involved in acyl-coenzyme A elongation in etiolated leek seedlings. Plant Physiol. 1999;119:1009–1015. doi: 10.1104/pp.119.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessire R, Hartmann-Bouillon MA, Cassagne C. Very long chain fatty acids: occurrence and biosynthesis in membrane fractions from etiolated maize coleoptiles. Phytochemistry. 1982;21:55–59. [Google Scholar]
- Lessire R, Juquelin H, Moreau P, Cassagne C. Elongation of acyl-CoA by microsomes from etiolated leek seedlings. Phytochemistry. 1985b;24:1187–1192. [Google Scholar]
- Lessire R, Juquelin H, Moreau P, Cassagne C. Nature of the reaction product of [1-14C] stearoyl-CoA elongation by etiolated leek seedling microsomes. Arch Biochem Biophys. 1985c;239:260–269. doi: 10.1016/0003-9861(85)90834-3. [DOI] [PubMed] [Google Scholar]
- Lord JM. Isolation of endoplasmic reticulum: general principles, enzymatic markers, and endoplasmic reticulum-bound polysomes. Methods Enzymol. 1987;148:576–584. [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;11:825–838. doi: 10.1105/tpc.11.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG. Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant and Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D. Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol and Mol Biol. 1996;47:405–430. doi: 10.1146/annurev.arplant.47.1.405. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Stumpf PK. Fatty acid biosynthesis in higher plants. In: Numa S, editor. Fatty Acid Metabolism and Its Regulation. Amsterdam: Elsevier; 1984. pp. 155–199. [Google Scholar]
- Taussky HH, Shorr E. A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem. 1953;202:675–685. [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;17:119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- Tulloch AP. Chemistry of waxes of higher plants. In: Kolattukudy PE, editor. Chemistry and Biochemistry of Natural Waxes. Amsterdam: Elsevier; 1976. pp. 235–287. [Google Scholar]
- von Wettstein-Knowles P. Genes, elongases and associated enzyme systems in epicuticular wax synthesis. In: Stumpf PK, Mudd JB, Nes WD, editors. The Metabolism, Structure, and Function of Plant Lipids. New York: Plenum Press; 1987. pp. 489–498. [Google Scholar]
- von Wettstein-Knowles P. Biosynthesis and genetics of waxes. In: Hamilton RJ, editor. Waxes: Chemistry, Molecular Biology and Functions. Allowry, Scotland: Oily Press; 1995. pp. 91–130. [Google Scholar]
- Walker DA, Cerovic ZG, Robinson SP. Isolation of intact chloroplasts: general principles and criteria of integrity. Methods Enzymol. 1987;148:145–157. [Google Scholar]
- Walton TJ. Waxes, cutin and suberin. In: Harwood JL, Bowyer JR, editors. Methods in Plant Biochemistry: Lipids, Membranes and aspects of Photobiology. San Diego: Academic Press; 1990. pp. 105–158. [Google Scholar]
- Xu XJ, Dietrich CR, Delledonne M, Xia YJ, Wen TJ, Robertson DS, Nikolau BJ, Schnable PS. Sequence analysis of the cloned glossy8 gene of maize suggests that it may code for a β-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol. 1997;115:501–510. doi: 10.1104/pp.115.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]