Abstract
The peptide snakin-2 (StSN2) has been isolated from potato (Solanum tuberosum cv Jaerla) tubers and found to be active (EC50 = 1–20 μm) against fungal and bacterial plant pathogens. It causes a rapid aggregation of both Gram-positive and Gram-negative bacteria. The corresponding StSN2 cDNA encodes a signal sequence followed by a 15-residue acidic sequence that precedes the mature StSN2 peptide, which is basic (isoelectric point = 9.16) and 66 amino acid residues long (molecular weight of 7,025). The StSN2 gene is developmentally expressed in tubers, stems, flowers, shoot apex, and leaves, but not in roots, or stolons, and is locally up-regulated by wounding and by abscisic acid treatment. Expression of this gene is also up-regulated after infection of potato tubers with the compatible fungus Botritys cinerea and down-regulated by the virulent bacteria Ralstonia solanacearum and Erwinia chrysanthemi. These observations are congruent with the hypothesis that the StSN2 is a component of both constitutive and inducible defense barriers.
An important component of plant defense is a diverse set of constitutive and pathogen-inducible antimicrobial compounds that includes the so-called pathogenesis-related proteins, several families of antimicrobial peptides, a variety of chemically diverse organic compounds classified as phytoalexins and phytoanticipins, and certain active oxygen and nitrogen species (Osbourn, 1996, 1999; Broekaert et al., 1997; Kombrink and Somssich, 1997; García-Olmedo et al., 1998, 2001). Accumulation of these compounds and the ability of a given pathogen to deal with them may be decisive for the outcome of the interaction (Titarenko et al., 1997a; López-Solanilla et al., 1998, 2001; Miguel et al., 2000; Alamillo and García-Olmedo, 2001; García-Olmedo et al., 2001). Thus, it has been shown that increased levels of certain antimicrobial peptides, either through overexpression of the corresponding genes or by appropriate exogenous treatments, result in enhanced tolerance to particular pathogens (Carmona et al., 1993; Terras et al., 1995; Epple et al., 1997; Molina and García-Olmedo, 1997; Holtorf et al., 1998; Thomma et al., 1998, 1999). Furthermore, pathogen mutants that are sensitive to antimicrobial peptides show decreased virulence when inoculated in the plant (Titarenko et al., 1997a; López-Solanilla et al., 1998).
Several families of antimicrobial peptides have been characterized in plants (García-Olmedo et al., 1992, 1995, 1998, 2001; Broekaert et al., 1997). The majority of them are Cys-rich and their globular structure is stabilized by disulphide bridges, although linear Gly-/His-rich and macrocyclic Cys-knot peptides have also been recently identified (Tam et al., 1999; Park et al., 2000). The peptides are generally encoded by multigenic families in which some genes are developmentally regulated in certain tissues, whereas others are pathogen inducible, and a number of them show both constitutive and pathogen-inducible expression (García-Olmedo et al., 1995, 1998, 2001; Broekaert et al., 1997).
In a previous report, a novel 12-Cys, antimicrobial peptide from potato (Solanum tuberosum cv Jaerla) tubers, snakin-1 (StSN1), was described (Segura et al., 1999). Its amino acid sequence was homologous to those deduced from a number of anonymous cDNAs reported in different plant species (Shi et al., 1992; Herzog et al., 1995; Ben-Nissan and Weiss, 1996; Kotilainen et al., 1999). Most of the corresponding genes are constitutively expressed, and some of them (GAST1 from tomato [Lycopersicon esculentum; GA-stimulated transcript] and GASA1 and GASA4 from Arabidopsis [GA stimulated in Arabidopsis]) have been shown to be up-regulated by gibberellic acid (GA) in GA-deficient mutants and, to a lesser degree, in wild-type plants (Shi et al., 1992; Herzog et al., 1995; Ben-Nissan and Weiss, 1996; Kotilainen et al., 1999).
The StSN1 gene from potato is constitutively expressed in different tissues during development and does not respond to GA and other abiotic or biotic treatments (Segura et al., 1999). Here, we report a second snakin peptide (StSN2) from potato that represents a quite divergent (38% conserved residues) snakin/GASA subfamily. Its spectrum of antimicrobial activity against the bacterial and fungal pathogens tested is quite similar to that of StSN1 and different from that of defensin peptides from the same tissues. However, expression of the StSN2 gene is locally induced by wounding and shows differential responses to pathogen infection, which is in contrast with that of the StSN1 gene. Expression patterns and antimicrobial activities of StSN2 are congruent with its possible participation in both constitutive and inducible defense barriers of potato.
RESULTS
Isolation and Characterization of StSN2
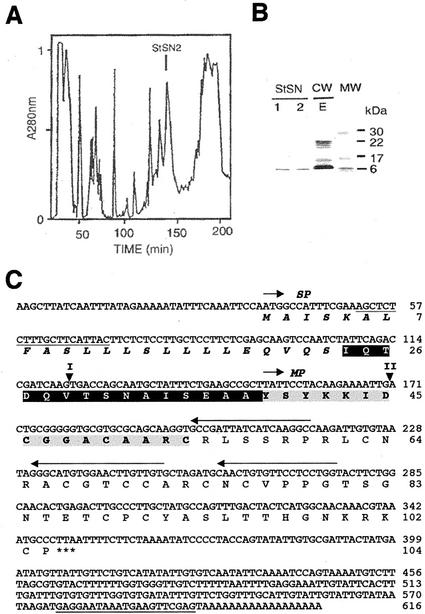
An HPLC fraction with antimicrobial activity (StSN2; Fig. 1A) was isolated from a crude cell wall extract that was obtained from potato tubers as previously described (Moreno et al., 1994; Segura et al., 1999). This fraction was homogeneous, as judged by SDS-PAGE, and comigrated with snakin-1 (StSN1; Segura et al., 1999; Fig. 1B). The homogeneity of this fraction was confirmed by matrix-assisted laser desorption ionization (MALDI)-time of flight mass spectrometry (MS), which detected a unique compound with a molecular mass of 7,024.93 D. The peptide was named snakin-2 (StSN2) because its N-terminal amino acid sequence, determined up to the 17th residue by Edman degradation, indicated homology to snakin-1, as well as to deduced amino acid sequences of the GASA family. The concentration of StSN2 in tubers was estimated to be in a range of 2 to 4 μmol kg−1 fresh weight
Figure 1.
Purification and characterization of StSN2. A, reverse phase-HPLC fractionation of the cell wall extract (CWE) from potato tubers. The linear gradient used was water (0.1% [v/v] trifluoroacetic acid)-2-propanol, 0% to 30% for 180 min and 30% to 50% for 15 min. Fraction corresponding to StSN2 is indicated. B, Separation by SDS-PAGE of the purified proteins StSN2 and StSN1 (Segura et al., 1999), and CWE from potato tuber. Molecular mass markers (MW) are indicated. C, Nucleotide sequence of StSN2 cDNA (AJ312904) and amino acid sequence of the corresponding protein. The gray-shaded amino acid sequence was obtained by direct N-terminal Edman degradation of the purified StSN2 and the rest of protein sequence was deduced from the cDNA sequence. Signal peptide (SP) is followed by a black-shaded amino acid sequence corresponding to the acidic sequence that preceded StSN2 mature peptide (MP). Predictions of SP were done by using the SignalP (http://www.cbs.dtu.dk/services/SignalP/) and Psort (http://psort.nibb.ac.jp/) program. Oligonucleotides sequences used for 5′-RACE are indicated by horizontal arrows, and those used for PCR amplification of the StSN2 gene are underlined. The position of the introns (I and II) in the StSN2 gene (AJ312424) are indicated by triangles.
To clone the StSN2 cDNA, 3′-/5′-RACE was carried out, using tuber cDNA as template. The full-length cDNA of StSN2 encoded a protein with a signal peptide sequence (residues 1–23), followed by a 15-residue-long acidic peptide (pI = 3.1) and then by a 66-residue sequence whose N-terminal was identical to that directly determined by Edman degradation of the purified peptide (Fig. 1C). The mature peptide was basic (pI = 9.6) and its calculated molecular mass was 7,025.14 D (assuming that the 12 Cys were in disulphide form), a figure that differs by less than 1 D from that directly determined by MALDI-time of flight MS.
Amino acid sequence alignments shown in Figure 2A indicate that StSN2 represents one of the three subfamilies into which snakin/GASA sequences can be classified. In addition to the 12 characteristic Cys, residues at nine positions are highly conserved throughout the family, whereas a number of motifs define each of the three subfamilies (Fig. 2A). StSN2 also shows some sequence similarity with Cys-rich domains from animal proteins, such as von Willebrand factors (vWF; Shelton-Inloes et al., 1986; Verweij et al., 1986), mucins (Li et al., 1998), and MDC (metalloproteinase-like desintegrin-like Cys-rich) proteins (Wolfsberg et al., 1995; Sagane et al., 1998; Fig. 2B). Although plant snakin peptides share certain motifs with disintegrin hemotoxic snake venoms, they lack the RGD motif that is responsible for disintegrin activity (Fig. 2B; Dennis et al., 1990).
Figure 2.
Alignment of snakins/GASAs amino acid sequences. A, Comparison of amino acid sequences of snakins/GASAs of subfamilies II, I, and III, respectively. Amino acids conserved across all the family members are black shaded and indicated in the consensus. Highly conserved residues that are relevant for subfamily classification (conserved in known, non-represented members of each group) are gray shaded. Sequences taken for the alignment are the following: StSN1 (Segura et al., 1999) and StSNIII (BG597515) from potato; AtGASA1 and AtGASA4 (Herzog et al., 1995), AtGASA7 (AC005396.2), and AtGASA8 (AC004218.2) from Arabidopsis; LsGAST1 (Shi et al., 1992), LsGASAI (BG130738), and LsGASAII (AI77478) from tomato; GhGEG from Gerbera hybrida (Kotilainen et al., 1999); FaGAST from Fragraria ananassa (AF039183); Rc153 from Ricinus communis (EMBL T24153); PhGIP1 from Petunia hybrida (Ben-Nissan and Weiss, 1996); and PmGAST1 from Picea marina (AF051227). The putative N-terminal sequences of the proteins have been determined by homology with those of StSN1 and StSN2, and by using the posttranslational predictions programms indicated in Figure 1C. B, Alignment of potato StSN2 sequence with amino acid sequence of Cys-rich domains of proteins from mammals and the hemotoxic snaken venom disintegrin. Identical residues are black shaded and similar, conservative positions (polar, apolar, and charged) are gray shaded. Proteins used in the alignment are the following: HsvWF (X04385), HsMuc (Q9ESP3), HsMDC, and HsMDC2a (Sagane et al., 1998) from human; BtvWF from Bos taurus (P80012); and disintegrin from agkistrodon halys blomhoffii snake (Gloydius blomhoffii; P21858). A stretch of 15 amino acids of HsvWF that has not been included in the alignment is indicated by an asterisk. The RGD motif of disintegrin essential for protein action is underlined. Similarities between StSN2 and the aligned proteins were found using the Jpred program for secondary structure predictions (http://barton.ebi.ac.uk/).
Activity against Bacterial and Fungal Pathogens
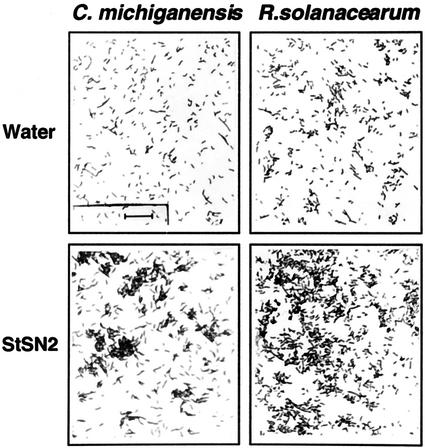
In Table I, the in vitro antimicrobial activity of purified StSN2 can be compared with that of StSN1 and defensin StPTH1, which also accumulate in potato tubers (Moreno et al., 1994; Segura et al., 1999). StSN2 was active (EC50 = 1–20 μm) against all the fungal species tested, the Gram-positive bacterium C. michiganenesis subsp. sepedonicus, and the Gram-negative bacterium R. meliloti, whereas it was inactive against the Gram-negative bacteria R. solanacearum and E. chrysanthemi (Table I). Despite their divergent amino acid sequences (38% identical residues), StSN2 and StSN1 antimicrobial activity spectra were essentially equivalent (Table I). In contrast, there were significant differences with respect to that of defensin StPTH1. For example, F. culmorun and R. meliloti were resistant to StPTH1, but sensitive to StSN2, and F. solani was 15 times more sensitive to StPTH1 than to StSN2 (Table I). Similar to StSN1 and in contrast with other types of antibiotic peptides, StSN2 caused rapid aggregation of both Gram-positive and Gram-negative bacteria (Fig. 3), although this property did not correlate with its inhibitory activity. Finally, as described for other plant antimicrobial peptides (De Samblanx et al., 1997; Segura et al., 1998, 1999), the in vitro activity of StSN2 was reverted when salt (50 mm KCl + 1 mm CaCl2) was added to the growth media (data not shown).
Table I.
Inhibition of bacterial and fungal pathogens by snakins StSN2 and StSN1 and defensin StPTH1 from potato
| Pathogen | Protein (EC50)a
|
||
|---|---|---|---|
| StSN2 | StSN1 | StPTH1 | |
| μm | |||
| Bacteria | |||
| Clavibacter michiganensis | 1 | 4 | 7 |
| Ralstonia solanacearum | NA | NA | 25 |
| R. solanacearum (rfa−) | 30 | 15 | 25 |
| Erwinia chrysanthemi | NA | NA | NA |
| Rhizobium meliloti | 8 | nt | NA |
| Fungi | |||
| Botrytis cinerea | 2 | 0.8 | 1 |
| Fusarium solani | 3 | 2 | 0.2 |
| Fusarium culmorum | 2 | 2 | NA |
| Fusarium oxysporum f. sp. conglutinans | 10 | 13 | nt |
| Fusarium oxysporum f. sp. lycopersici | 20 | 13 | nt |
| Plectosphaerella cucumerina | 10 | 10 | 10 |
| Colletotrichum graminicola | 10 | 20 | 2 |
| Colletotrichum lagenarium | 10 | 10 | nt |
| Bipolaris maydis | 20 | 20 | 10 |
| Aspergillus flavus | 20 | 20 | nt |
EC50, Effective concentration for 50% inhibition; NA, not active at concentration <20 μm; nt, not tested.
Figure 3.
Aggregation of bacteria caused by potato StSN2. A 5-μL suspension (105 colony forming units [cfu] mL−1) of the Gram-positive bacterium C. michiganensis subsp. sepedonicus or the Gram-negative bacterium R. solanacearum was deposited in a microscope slide, then 5 μL of a 20 μm solution of StSN2 or a 5-μL drop of water was added, and a photograph was taken immediately under a microscope. Bar represents 20 μm.
Developmental Expression of the StSN2 Gene
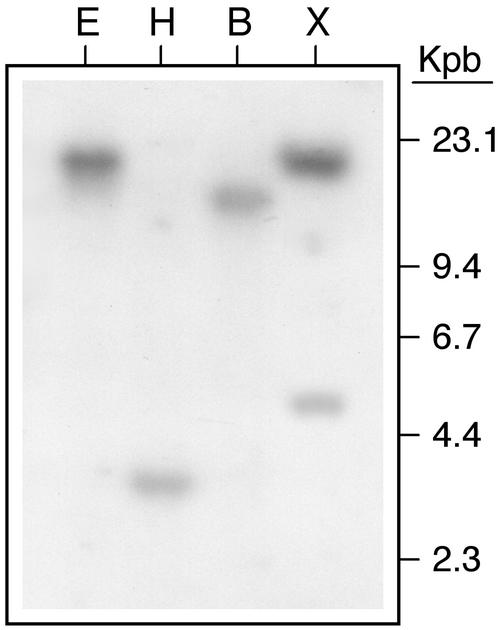
The StSN2 cDNA, which did not give any cross-hybridization with potato StSN1 DNA (data not shown), was used for Southern-blot hybridization of potato DNA and the observed patterns were compatible with the presence of one or two copies of the gene in the genome (Fig. 4). PCR amplification and sequencing of StSN2 demonstrated the presence of two introns that were 249 and 163 bp in length, respectively, and were in similar positions to those of other genes of the same subfamily (Fig. 1C).
Figure 4.
Southern-blot analysis of StSN2 gene. Potato genomic DNA (10 μg) was digested with the EcoRI (E), HindIII (H), BamHI (B), or XhoI (X) restriction endonucleases. The StSN2 cDNA fragment obtained by 5′-RACE (nucleotides 170–616) was [32P] labeled and used as probe.
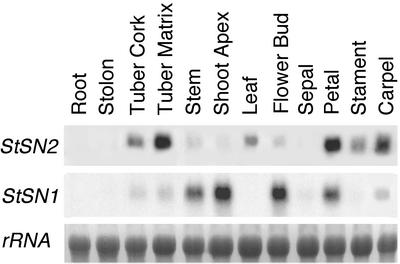
The expression of StSN2 gene in potato plants was analyzed and compared with that of the StSN1 by the northern-blot technique. Steady-state levels of StSN2 mRNA were high in tubers, as well as in petals, stamens, and carpels from fully developed flowers (Fig. 5). In all these organs except petals, StSN1 was not highly expressed (Fig. 5). Conversely, low levels of StSN2 mRNA were detected in stem, shoot apex, and flower buds where StSN1 was highly expressed (Fig. 5). StSN2, but not StSN1, was expressed in leaves, where it reached a peak at emergence in 4-week-old plants and decreased at the start of senescence (Fig. 5; data not shown). StSN2 mRNA was not detected in other tissues and organs analyzed, including roots, stolons, and sepals (Fig. 5).
Figure 5.
Expression of StSN2 gene in potato. Nothern-blot analysis of total RNAs (5 μg) extracted from the indicated parts of the potato plant. Blot was hybridized with the StSN2 probe and an StSN1 probe (Segura et al., 1999). Equal loading was confirmed by rehybridization of the blot with a potato 18S-ribosomal RNA probe.
Response of the StSN2 Gene to Abiotic Treatments
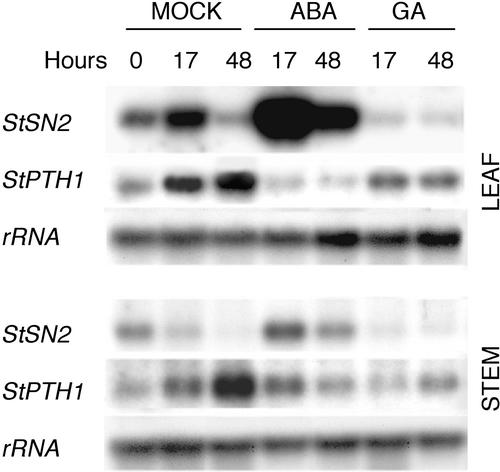
Excised potato plants were treated with GA (50 μm) and abscisic acid (ABA; 100 μm) by stem feeding. As shown in Figure 6, mock treatment slightly increased the basal StSN2 mRNA level. This effect was significantly enhanced in leaves and stems by ABA and prevented by GA (Fig. 6). Expression of the defensin StPTH1 gene from potato (Moreno et al., 1994), used as a negative control, was also slightly increased in the mock experiment, but unaffected by either ABA or GA (Fig. 6).
Figure 6.
Expression of StSN2 gene is induced by ABA. Nothern-blot analysis of total RNAs (7.5 μg) extracted from leaves and stems of mock-treated plants (M), or plants treated with 100 μm ABA or 50 μm GA3. Blot was hybridized with the StSN2 probe, and a potato defensin StPTH1 probe (Moreno et al., 1994). RNA equal loading was confirmed by rehybridization of the blot with a potato 18S-ribosomal RNA probe. This is one of three experiments carried out that gave similar results.
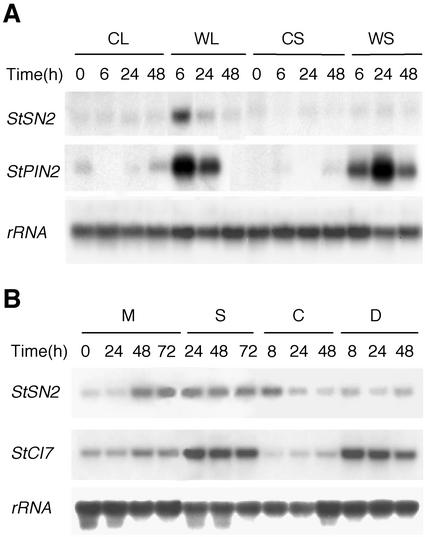
Because it is known that some defensin genes are wound inducible (Broekaert et al., 1997), the concomitant induction of the StSN2 gene in excised plants suggested that this gene could respond to mechanical damage. The enhanced expression of StSN2 gene after treatment with ABA, which plays a role in wound signaling in solanaceas (Peña-Cortés et al., 1989; León et al., 2001), was also in line with a putative regulation of StSN2 gene by wounding. To directly test this hypothesis, the experiment summarized in Figure 7 was done, using the wound-inducible StPIN2 gene as positive control (Peña-Cortés et al., 1989). Expression of the StSN2 was locally induced in the wounded leaves, but not in the upper systemic, non-damaged leaves (Fig. 7A). This induction was not observed in mechanically damaged tubers and stems of potato (data not shown). Chitosan and jasmonic acid (JA), which have also been implicated in wound response (Bishop et al., 1981; Farmer and Ryan, 1990), had no effect on the expression of StSN2 gene (data not shown).
Figure 7.
Response of StSN2 gene to wounding and water-stress. A, Northern blot (7.5 μg per sample) of total RNAs extracted from mechanically damaged leaves (WL) and upper systemic non-damaged leaves (WS) from wounded plants, and from same-age leaves of control, non-wounded plants (CL and CS). Blot was hybridized with the StSN2 probe and with a potato StPIN2 probe (Peña-Cortés et al., 1989). B, Northern-blot analysis of total RNAs (7.5 μg) extracted from leaves of excised plants mock treated (M) or incubated with salt (250 mm NaCl; S), intact control plants (C), and plants that were remove from soil and were left to dry (drought treatment; D). Blot was hybridized with the StSN2 probe, and a probe of the potato CI7 gene (Kirch et al., 1997). RNA equal loading was confirmed by rehybridization of the blot with a potato 18S-ribosomal RNA probe. This is one representative experiment of the two carried out that gave similar results.
Because it is known that ABA mediates adaptive responses to water stress (salinity or drought; Grill and Himmelbach, 1998), possible responses of the StSN2 gene to this type of stress were investigated, using as control the CI7 gene, which responds to salinity and drought (Kirch et al., 1997). Gene StSN2 was found to respond only weakly to the salinity treatment and was unaffected by drought (Fig. 7B).
Responses of the StSN2 Gene to Infection by Bacterial and Fungal Pathogens
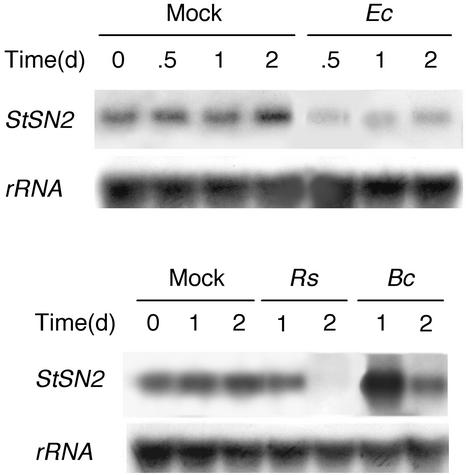
To investigate possible effects of pathogen infection on the expression of the StSN2 gene, potato tubers were inoculated with the bacterial pathogens R. solanacearum, which induces tuber necrosis 1 d after inoculation (Titarenko et al., 1997a), and E. chrysanthemi, which causes soft rot disease in potato and a wide range of plants (Boccara et al., 1991; López-Solanilla et al., 1998), or the fungus B. cinerea, which causes the gray mold disease in potato (Agrios, 1997). The gene was down-regulated by the two bacterial infections, whereas it was transiently induced after inoculation with the fungus (Fig. 8). In contrast, expression of the gene was unaffected when leaves were infected with the fungus or treated with ethylene (125 μL L−1), a hormone involved in activation of some defense responses (Solano and Ecker, 1998; Thomma et al., 1998, 1999), or with benzo[1,2,3]-thiadiazole-7-carbothioic acid S-methyl ester (0.3 mm), a compound that induces systemic acquired resistance (Ryals et al., 1996; data not shown).
Figure 8.
Response of StSN2 gene to infection of potato tubers with pathogens. Northern blot (7.5 μg per sample) of total RNAs extracted from tubers mock inoculated with 10 mm MgCl2 (M), infected with bacterial suspensions (50 μL of 107 cfu mL−1) of R. solanacearum (Rs) or E. chrysanthemi (Ec), or inoculated with a spore supension (50 μL of 2 × 105 spores mL−1) of the fungus B. cinerea (Bc). Blot was hybridized with the StSN2 probe and RNA equal loading was confirmed by rehybridization of the blot with a potato 18S-ribosomal RNA probe. This is one of two experiments carried out that gave similar results.
DISCUSSION
We have previously characterized the antimicrobial properties of the cell wall associated peptide snakin-1 (StSN1) from potato, the first purified member of what appears to be a widely distributed peptide type, the snakin/GASA family (Segura et al., 1999). This peptide was developmentally accumulated in different tissues of potato plants and the expression of its corresponding gene was unaffected by a variety of abiotic or biotic challenges (Segura et al., 1999). We have now described the isolation of a new peptide from potato (StSN2) that represents a widely divergent snakin subfamily. The StSN2 peptide is also active in vitro against bacterial and fungal plant pathogens and, in contrast with StSN1, expression of the corresponding gene is affected by certain external treatments including pathogen infection.
Like the majority of globular antimicrobial peptides from plants described so far (Broekaert et al., 1997; García-Olmedo et al., 1998, 2001), the snakin/GASA peptides are basic and rich in Cys residues, which may form six disulphide bridges that stabilize their structure. The amino acid sequences deduced from cloned snakin/GASA cDNAs and genes can be classified into three subfamilies (I–III). StSN1 and StSN2 are respectively the only purified members representing subfamilies I and II, whereas no member of subfamily III has been isolated so far.
Although StSN1 and StSN2 show only 38% sequence similarity, they have almost identical antimicrobial activity spectra. The concentration of StSN2 in tubers and in other tissues and organs of the potato plant seems to be well above that possibly required for in planta inhibition of some pathogens, based on in vitro inhibitory activity of this peptide. In potato tubers, other antimicrobial peptides, such as defensin StPTH1 (Moreno et al., 1994) and snakin StSN1 (Segura et al., 1999), accumulate at high concentrations together with StSN2. The complementarity of the antibiotic activity spectra of defensin StPTH1 and the snakins, as well as the synergism between the two peptide types (Moreno et al., 1994; Segura et al., 1999; M. Berrocal-Lobo, G. López, F. García-Olmedo, and A. Molina, unpublished data), suggests that the concomitant accumulation of snakins and defensins in a given tissue (i.e. in tubers or in flowers) may represent a constitutive, broad-spectrum, effective barrier against bacterial and fungal pathogens.
StSN2 causes a rapid aggregation of Gram-positive and Gram-negative bacteria in vitro, as previously reported for StSN1 (Segura et al., 1999). The aggregation did not correlate with antimicrobial activity, but could play a role in the control of the pathogen in vivo because snakin peptides show sequence similarity with Cys-rich domains of proteins from animals that are involved in protein-protein interactions, such as the vWFs (Shelton-Inloes et al., 1986; Verweij et al., 1986), mucins (Li et al., 1998), and MDC proteins (Wolfsberg et al., 1995; Sagane et al., 1998). In particular, mucins coat the epithelia of intestines, airways, and other mucus membrane-containing organs from animals, providing a protective, lubricating barrier against particles and infectious agents (Pérez-Vilar and Hill, 1999). The mechanism of action of snakins remains unknown, but, in contrast with other antibiotic peptides from plants, they do not interact with artificial lipid membranes (Caaveiro et al., 1997).
The StSN2 mRNA encodes a precursor with a signal peptide, which is congruent with the cell wall location of the mature peptide, followed by an N-terminal acidic pro-peptide, 15 amino acid residues in length. Acidic peptides at the N terminus can be also predicted for other members of the family (e.g. AtGASA1, AtGASA4, GhGEG, and LsGAST1), but not for StSN1. It has been suggested that the acidic amino acid sequence that sometimes precedes or follows different antibiotic peptides may act as an inhibitor of endogenous toxicity during transport (see García-Olmedo et al., 2001).
Though constitutively expressed, the StSN2 gene is up-regulated by ABA and down-regulated by GA, just the opposite of what has been described for other GASA genes, such as GASA1 from Arabidopsis and GAST1 gene from tomato, which are up-regulated by GA, and down-regulated by ABA (Shi et al., 1992; Herzog et al., 1995; Raventos et al., 2000). GA has been shown to down- or up-regulate the expression of the Arbidopsis GASA4 gene in a tissue-specific manner (Aubert et al., 1998). ABA is known to mediate some plant responses to environmental stress, including osmotic stress produced by cold, high salinity, or drought, as well as to mechanical injury produced by wound-causing agents (Peña-Cortés et al., 1989, 1995; Grill and Himmelbach, 1998; León et al., 2001). However, the StSN2 gene is not a typical water stress-inducible gene such as tomato TAS14 (Godoy et al., 1990) or potato CI7 (Kirch et al., 1997) because it did not respond clearly to salinity or drought.
StSN2 expression was locally induced in wounded leaves, but not in systemic non-damaged leaves, and it did not respond to JA treatment. Both JA-dependent and -independent wound signal transduction pathways have been described in plants (Farmer and Ryan, 1990; Peña-Cortés et al., 1995; O'Donnell et al., 1996; Dammann et al., 1997; Reymond et al., 2000; León et al., 2001), and wound-inducible genes that do not respond to JA have been identified (e.g. WR3 and Twi1; Nishiuchi et al., 1997; Titarenko et al., 1997b; O'Donnell et al., 1998; Reymond et al., 2000). The StSN2 gene belongs to the latter class, but its expression differs from that of tomato Twi1, which shows systemic activation upon wounding and is induced by salicylic acid (O'Donnell et al., 1998), whereas StSN2 did not. Expression pattern of StSN2 also differs from that of Arabidopsis WR3 gene, which is up-regulated by oligosacharides (Titarenko et al., 1997b; Rojo et al., 1999), whereas StSN2 was unaffected by chitosan treatment.
Responses to different pathogens and wound-inducible expression of the StSN2 gene are in line with its putative defense role. The gene was induced in tubers infected with the StSN2-sensitive fungus B. cinerea, and repressed after inoculation with R. solancearum and E. chrysanthemi. Furthermore, an StSN2 expressed sequence tag (BG591412; Potato Genomic Project) has been recently associated with leaves infected with Phytophthora infestans. Up-regulation by fungal pathogens and down-regulation by bacterial ones have been described for other genes encoding plant antimicrobial peptides, such as HvLTP4.3 from barley (Hordeum vulgare) and defensin StPTH1 from potato (Moreno et al., 1994; Molina et al., 1996). Down-regulation of defense genes by some pathogens has been suggested as a mechanism to overcome plant defense (Jakobek et al., 1993; Wada et al., 1995). The antimicrobial properties of StSN2 and its developmental and wound/pathogen-responsive expression patterns are congruent with a putative defense role. This function is also supported by the observed reduced virulence in planta of R. solanacearum and E. chysanthemi mutants, which are sensitive to potato snakins (Titarenko et al., 1997a; Solanilla et al., 1998; rfa− mutant in Table I). It should be noted that a defense role is not incompatible with the involvement of snakin/GASA genes in other plant processes and mechanisms (Herzog et al., 1995; Aubert et al., 1998; Kotilainen et al., 1999; Raventos et al., 2000).
MATERIALS AND METHODS
Biological Materials
Potato (Solanum tuberosum cv Jaerla) was cultivated in a growth chamber at 60% humidity at 28°C day and 22°C night, with a photoperiod of 14 h light/10 h dark. Bacterial pathogens Clavibacter michiganensis subsp. sepedonicus, strain C5, Ralstonia solanacearum, strain K60 and rfa− mutant (Titarenko et al., 1997a), Erwinia chrysanthemi, strain AC4150 (López-Solanilla et al., 1998), and Rhizobium meliloti, as well as the fungal pathogens Botrytis cinerea (strain 1) and Fusarium solani (strain 1), were from the Escuela Técnica Superior Ingenieros Agronomos collection (Madrid). The fungal species Aspergillus flavus, Bipolaris maydis, Fusarium culmorum, Colletotrichum lagenarium, and Colletotrichum graminicola were from the Novartis Corp. collection (Research Triangle Park, NC). The fungus Plectosphaerella cucumerina was the gift of Dr. Brigitte Mauch-Mani (University of Fribourg, Switzerland), and the fungi Fusarium oxysporum f. sp. lycopersici and F. oxysporum f. sp. conglutinans were kindly provided by Dr. María I.G. Roncero (Universidad de Cordoba, Spain).
Purification and Characterization of StSN2
A crude cell wall extract was obtained from frozen potato tuber material as previously described (Moreno et al., 1994; Segura et al., 1999) and subjected to reverse phase-HPLC on an Ultrapore C3 column (1 × 25 cm; 5-μm particle; 300-Å pore; Beckman, San Ramon, CA). The linear gradient used was water (0.1% [v/v] trifluoroacetic acid)-2-propanol, 0% to 30% for 180 min and 30% to 50% for 15 min, at 0.5 mL min−1. Fractions were collected by hand and freeze dried. SDS-PAGE of the proteins was carried out in 12% (w/v) acrylamide gels following standard procedures. MALDI-MS was performed in the MALDI-II system from Kratos (Michigan, UK), using α-cyano (Sigma, St. Louis) as matrix and angiotensin I (MH, Mr 1,297.5) and cytochrome C (MH, Mr 12,361.5; M2H, Mr 6,182.2) as standards for mass calibration. Amino acid sequencing was carried out by automated Edman degradation of the intact protein.
Pathogen Inhibition Tests
Bacterial inhibition tests were carried out in sterile microtiter plates by mixing 50-μL aliquots of bacterial suspension (final concentration 104 cfu mL−1) in nutrient broth (Oxoid, Basingstoke, UK) with different amounts of the protein dissolved in 100 μL of sterile water. After 16 to 24 h of incubation at 28°C, growth was recorded by measuring A490 in an ELISA plate reader. Fungal spores from 8-d-old cultures grown at 28°C on potato dextrose agar plates (Difco, Detroit, MI) were collected in sterile water and stored at −80°C in 20% (v/v) glycerol. Twenty-five microliters of a spore suspension (final concentration 104 spores mL−1) in potato dextrose (Difco) was mixed in microtiter plates with different amounts of the protein dissolved in 50 μL of sterile water. Plates were incubated at 28°C for 26 to 44 h and growth was recorded as above. Potato defensin StPTH1 and snakin StSN1 used in the inhibition experiments were purified as previously described (Moreno et al., 1994; Segura et al., 1999).
Characterization of StSN2
Total RNA extracted from potato tubers was used to obtain cDNA with the First-Strand Synthesis Kit (Amersham-Pharmacia, Rainham, UK) and an oligo(dT) primer. StSN2 cDNA was cloned by 3′-RACE using cDNA from potato tubers, an oligo(dT)-anchor primer from the kit indicated above, and two overlapping degenerate oligonucleotides corresponding to the amino acids sequences YKKIDCG and DCGGACA of the N-terminal region of the mature StSN2 peptide (PCR-annealing temperatures of 51°C and 56°C, respectively). The 5′ region of the SN2 cDNA was cloned with the 5′-/3′-RACE Kit from Boheringer (Mannheim, Germany) using cDNA from potato tubers and the primers indicated in Figure 1C (PCR-annealing temperatures of 51°C and 54°C, respectively). Genomic sequences of StSN2 was amplified by PCR using specific primers deduced from their correspoding cDNAs (Fig. 1C) and potato genomic DNA obtained as decribed (Dellaporta et al., 1983). The PCR-amplified cDNAs and genomic fragments were cloned in the pGEM vector (Promega, Madison, WI), and the corresponding clones were sequenced using the ABIPrism Kit (Perkin-Elmer, Norwalk, CT).
Northern and Southern Blots
A StSN2 cDNA fragment (nucleotides 170–616) was random labeled with [32P]ATP following standard procedures (Sambrook et al., 1989) and used as a probe for northern and Southern experiments. Potato genomic DNA used in the Southern experiments was obtained as decribed (Dellaporta et al., 1983), digested with different endonucleases, subjected to electrophoresis on 0.8% (w/v) agarose gels, and transferred to Hybond N+ membranes (Amersham-Pharmacia) using standard procedures (Sambrook et al., 1989). RNAs were purified from frozen tissues by phenol/chloroform extraction, followed by precipitation with 3 m LiCl (Lagrimini et al., 1987), and subjected to electrophoresis on 5% (v/v) formaldehyde/agarose gels. The gels were blotted to Hybond N+ membranes following standard procedures (Sambrook et al., 1989). RNA equal sample loads were checked by rehybridizing the blots with an 18S ribosomal cDNA probe of potato (X67238, nucleotides 361–959) obtained by PCR. Hybridization and washing of northern and Southern blots were carried out at 65°C according to Church and Gilbert (1984). Potato CI7 probe was obtained by PCR amplification (nucleotides 851–1,216; Kirch et al., 1997) using potato genomic DNA as template. StPIN2 probe was kindly provided for Dr. Jose J. Sánchez-Serrano (Centro Nacional Biotecnología-Consejo Superior de Investigaciones Científicas, Madrid).
External Treatments
Four-week-old plants (five- to six-leaf stage) were used in all these experiments. ABA (100 μm; Sigma) and GA (50 μm GA3; Sigma) were supplied in a phosphate solution (pH = 6.3) to excised potato plants, which were kept under the same growth conditions until the end of the experiments. For wounding experiment, expanded leaves were wounded perpendicular to the main vein with metallic forceps and material from the wounded leaves and non-damaged, upper leaves was harvested. Same-age leaves of non-wounded plants were used for controls. Salinity treatment was done by incubating excised potato plants in 250 mm NaCl, whereas control plants were incubated in a phosphate buffer (pH = 6.3). For drought treatment potato plants were removed from the pot and left to dry in the growth chamber. Chitosan treatment was done as described (Walker-Simmons and Ryan, 1984). Treatments with methyl jasmonate (50 μm; Apex Organics, Leicester, UK; >90% pure), and the systemic acquired resistance activator benzo[1,2,3]-thiadiazole-7-carbothioic acid S-methyl ester (0.3 mm; Novartis, Basel) or its corresponding wettable powder alone, were done by spraying the whole plants with these compounds. Ethylene treatment was carried out by injecting 125 μL L−1 of the ethylene gas in a chamber containing excised potato plants incubated in water. In all these experiments, samples were collected at different times after treatment and frozen in liquid nitrogen.
Inoculation of Potato with Pathogens
Potato tubers were inoculated at three points with a suspension of B. cinerea spores (50 μL of 2 × 105 spores mL−1) by using a plastic tip containing the spores, which was inserted at a constant depth of 1.5 cm in the tubers. Potato leaves of 4-week-old plants were inoculated in the upper side with four 20-μL drops of a B. cinerea spore suspension (2 × 105 spores mL−1). Inoculated plants and tubers were incubated at 28°C and 80% humidity. Tuber samples were collected by slicing the tuber and harvesting a disc (1-cm diameter and 2-cm heigth) of tissue around the inoculation points. Infection of potato tubers with E. chrysanthemi AC4150 strain (López-Solanilla et al., 1998) and R. solanacearum K60 strain (Titarenko et al., 1997a) were carried out as described above using 50 μL of 107 cfu mL−1 in 10 mm MgCl2. Mock inoculations of tubers and leaves were done with 10 mm MgCl2 alone. Samples were harvested as indicated above at different times after inoculation and frozen in liquid nitrogen.
ACKNOWLEDGMENTS
Technical help from Dolores Lamoneda and Joaquín García is gratefully acknowledged.
Footnotes
This work was supported by the Dirección General de Investigacion Cientifica y Technica (grant no. PB92–0325 to F.G.-O.), by the Comunidad de Madrid (grant no. 07B/0002/1999 to A.M.), by the Ministerio de Ciencia y Tecnología from Spain (grant no. BIO2000–1308 to A.M), and by the European Project BIOCT97–2120 (DGXII–SSMI to M.B.-L.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010685.
LITERATURE CITED
- Agrios GN. Plant Pathology. Ed. 4. San Diego: Academic Press; 1997. [Google Scholar]
- Alamillo JM, García-Olmedo F. Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J. 2001;25:529–541. doi: 10.1046/j.1365-313x.2001.00984.x. [DOI] [PubMed] [Google Scholar]
- Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol. 1998;36:871–883. doi: 10.1023/a:1005938624418. [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G, Weiss D. The petunia homologue of tomato gast1: Transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol Biol. 1996;32:1067–1074. doi: 10.1007/BF00041390. [DOI] [PubMed] [Google Scholar]
- Bishop PD, Makus DJ, Pearce G, Ryan CA. Proteinase inhibitor inducing factor activity in tomato leaves resides in oligosaccharides enzimically released from cell walls. Proc Natl Acad Sci USA. 1981;78:3536–3540. doi: 10.1073/pnas.78.6.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccara M, Vedel R, Lalo D, Lebrun MH, Lafay JF. Genetic diversity and host range in strains of Erwinia chrysanthemi. Mol Plant-Microbe Interact. 1991;4:293–299. [Google Scholar]
- Broekaert WF, Cammue BPA, De Bolle MFC, Thevissen K, De Samblanx GW, Osborn RW. Antimicrobial peptides from plants. Crit Rev Plant Sci. 1997;16:297–323. [Google Scholar]
- Caaveiro JMM, Molina A, González-Mañas JM, Rodríguez-Palenzuela P, García-Olmedo F, Goñi FM. Differential effect of five types of antipathogenic plant peptides on model membranes. FEBS Lett. 1997;410:338–342. doi: 10.1016/s0014-5793(97)00613-3. [DOI] [PubMed] [Google Scholar]
- Carmona MJ, Molina A, Fernández JA, López-Fando JJ, García-Olmedo F. Expression of the α-thionin gene from barley in tobacco confers enhanced resistance to bacterial pathogens. Plant J. 1993;3:457–462. doi: 10.1111/j.1365-313x.1993.tb00165.x. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann C, Rojo E, Sanchez-Serrano JJ. Abscisic acid and jasmonic acid activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J. 1997;11:773–782. doi: 10.1046/j.1365-313x.1997.11040773.x. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–22. [Google Scholar]
- Dennis MS, Henzel WJ, Pitti RM, Lipari MT, Napier MA, Deisher TA, Bunting S, Lazarus RA. Platelet glycoprotein IIb-IIIa protein antagonists from snake venoms: evidence for a family of platelet-aggregation inhibitors. Proc Natl Acad Sci USA. 1990;87:2471–2475. doi: 10.1073/pnas.87.7.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Samblanx GW, Goderis IJ, Thevissen K, Raemaekers R, Fant F, Borremans F, Acland DP, Osborn RW, Patel S, Broekaert WF. Mutational analysis of a plant defensin from radish (Raphanus sativus L.) reveals two adjacent sites important for antifungal activity. J Biol Chem. 1997;272:1171–1179. doi: 10.1074/jbc.272.2.1171. [DOI] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H. Overexpression of an endogenous thionin gives enhanced resistance of Arabidopsis thaliana against Fusarium oxysporium. Plant Cell. 1997;9:509–520. doi: 10.1105/tpc.9.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Interplant communication: Airbone methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci USA. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Olmedo F, Carmona MJ, Lopez-Fando JJ, Fernandez JA, Castagnaro A, Molina A, Hernandez-Lucas C, Carbonero P. Characterization and analysis of thionin genes. In: Boller T, Meins F, editors. Genes Involved in Plant Defense. Wien, Austria: Springer-Verlag; 1992. pp. 283–302. [Google Scholar]
- García-Olmedo F, Molina A, Alamillo JM, Rodríguez-Palenzuela P. Plant defense peptides. Biopolymers. 1998;47:479–491. doi: 10.1002/(SICI)1097-0282(1998)47:6<479::AID-BIP6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- García-Olmedo F, Molina A, Segura A, Moreno M. The defensive role of nonspecific lipid-transfer proteins in plants. Trends Microbiol. 1995;3:72–74. doi: 10.1016/s0966-842x(00)88879-4. [DOI] [PubMed] [Google Scholar]
- García-Olmedo F, Rodriguez-Palenzuela P, Molina A, Alamillo JM, Lopez-Solanilla E, Berrocal-Lobo M, Poza-Carrion C. Antibiotic activities of peptides, hydrogen peroxide and peroxynitrite in plant defense. FEBS Lett. 2001;498:219–222. doi: 10.1016/s0014-5793(01)02456-5. [DOI] [PubMed] [Google Scholar]
- Grill E, Himmelbach A. ABA signal transduction. Curr Opin Plant Biol. 1998;1:412–418. doi: 10.1016/s1369-5266(98)80265-3. [DOI] [PubMed] [Google Scholar]
- Godoy JA, Pardo JM, Pintor-Toro JA. A tomato cDNA inducible by salt stress and abscisic acid: nucleotide sequence and expression pattern. Plant Mol Biol. 1990;15:695–705. doi: 10.1007/BF00016120. [DOI] [PubMed] [Google Scholar]
- Herzog M, Dorne AM, Grellet F. GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol Biol. 1995;27:743–752. doi: 10.1007/BF00020227. [DOI] [PubMed] [Google Scholar]
- Holtorf S, Ludwig-Muller J, Apel K, Bohlmann H. High-level expression of a viscotoxin in Arabidopsis thaliana gives enhanced resistance against Plasmodiophora brassicae. Plant Mol Biol. 1998;36:673–680. doi: 10.1023/a:1005947904830. [DOI] [PubMed] [Google Scholar]
- Jakobek JL, Smith JA, Lindgren PB. Supression of bean defense responses by Pseudomonas syringae. Plant Cell. 1993;5:57–63. doi: 10.1105/tpc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch HH, van Berkel J, Glaczinski H, Salamini F, Gebhardt C. Structural organization, expression and promoter activity of a cold-stress-inducible gene of potato (Solanum tuberosum L.) Plant Mol Biol. 1997;33:897–909. doi: 10.1023/a:1005759925962. [DOI] [PubMed] [Google Scholar]
- Kombrink E, Somssich IE. Pathogenesis related proteins in plant defense. In: Carrol G, Tudzynski P, editors. The Mycota V Part A, Plant Relationships. Berlin: Spring-Verlag; 1997. pp. 107–128. [Google Scholar]
- Kotilainen M, Helariutta Y, Mehto M, Pollanen E, Albert VA, Elomaa P, Teeri TH. GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell. 1999;11:1093–1104. doi: 10.1105/tpc.11.6.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini LM, Burkhart W, Moyer M, Rothstein S. Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc Natl Acad Sci USA. 1987;84:7542–7546. doi: 10.1073/pnas.84.21.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J, Rojo E, Sanchez-Serrano JJ. Wound signaling in plants. J Exp Bot. 2001;52:1–9. doi: 10.1093/jexbot/52.354.1. [DOI] [PubMed] [Google Scholar]
- Li D, Gallup M, Fan N, Szymkowski DE, Basbaum CB. Cloning of the amino-terminal and 5′-flanking region of the human MUC5AC mucin gene and transcriptional up-regulation by bacterial exoproducts. J Biol Chem. 1998;273:6812–6820. doi: 10.1074/jbc.273.12.6812. [DOI] [PubMed] [Google Scholar]
- López-Solanilla E, Garcia-Olmedo F, Rodriguez-Palenzuela P. Inactivation of the sapA to sapF locus of Erwinia chrysanthemi reveals common features in plant and animal bacterial pathogens. Plant Cell. 1998;10:917–924. [PMC free article] [PubMed] [Google Scholar]
- López-Solanilla E, Llama-Palacio A, García-Olmedo F, Rodríguez-Palenzuela P. Relative effects on virulence of mutations in the sap, pel, and hrp loci of Erwinia chrysanthemi. Mol Plant-Microbe Interact. 2001;14:386–393. doi: 10.1094/MPMI.2001.14.3.386. [DOI] [PubMed] [Google Scholar]
- Miguel E, Poza-Carrión C, López-Solanilla E, Aguilar I, Llama-Palacios A, García-Olmedo F, Rodríguez-Palenzuela P. Evidence against a direct antimicrobial role of H2O2 in the infection of plants by Erwinia chrysanthemi. Mol Plant-Microbe Interact. 2000;13:421–429. doi: 10.1094/MPMI.2000.13.4.421. [DOI] [PubMed] [Google Scholar]
- Molina A, Diaz I, Vasil IK, Carbonero P, Garcia-Olmedo F. Two cold-inducible genes encoding lipid transfer protein LTP4 from barley show differential responses to bacterial pathogens. Mol Gen Genet. 1996;252:162–168. doi: 10.1007/BF02173216. [DOI] [PubMed] [Google Scholar]
- Molina A, García-Olmedo F. Enhanced tolerance to bacterial pathogens caused by transgenic expression of barley lipid transfer protein LTP2. Plant J. 1997;12:669–675. doi: 10.1046/j.1365-313x.1997.00669.x. [DOI] [PubMed] [Google Scholar]
- Moreno M, Segura A, García-Olmedo F. Pseudothionin-PTH1, a potato peptide active against potato pathogens. Eur J Biochem. 1994;223:135–139. doi: 10.1111/j.1432-1033.1994.tb18974.x. [DOI] [PubMed] [Google Scholar]
- Nishiuchi T, Hamada T, Kodama H, Iba K. Wounding changes the spatial expression pattern of the arabidopsis plastid omega-3 fatty acid desaturase gene (FAD7) through different signal transduction pathways. Plant Cell. 1997;9:1701–1712. doi: 10.1105/tpc.9.10.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell JP, Truesdale MR, Calvert C, Dorans A, Roberts MR, Bowles DJ. A novel tomato gene that rapidly responds to wound- and pathogen-related signals. Plant J. 1998;14:137–142. doi: 10.1046/j.1365-313X.1998.00110.x. [DOI] [PubMed] [Google Scholar]
- O'Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ. Ethylene as a signal mediating the wound response of tomato plants. Science. 1996;274:1914–1917. doi: 10.1126/science.274.5294.1914. [DOI] [PubMed] [Google Scholar]
- Osbourn AE. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell. 1996;8:1821–1831. doi: 10.1105/tpc.8.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn AE. Antimicrobial phytoprotectants and fungal pathogens: a commentary. Fungal Genet Biol. 1999;26:163–168. doi: 10.1006/fgbi.1999.1133. [DOI] [PubMed] [Google Scholar]
- Park CJ, Park CB, Hong SS, Lee HS, Lee SY, Kim C. Characterization and cDNA cloning of two glycine- and histidine-rich antimicrobial peptides from the roots of shepherd's purse, Capsella bursa-pastoris. Plant Mol Biol. 2000;44:187–197. doi: 10.1023/a:1006431320677. [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Fisahn J, Willmitzer L. Signals involved in the wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc Natl Acad Sci USA. 1995;92:4106–4113. doi: 10.1073/pnas.92.10.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Sánchez-Serrano JJ, Mertens R, Willmitzer L, Prat S. Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proc Natl Acad Sci USA. 1989;86:9851–9855. doi: 10.1073/pnas.86.24.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Vilar J, Hill RL. The structure and assembly of secreted mucins. J Biol Chem. 1999;274:31751–31754. doi: 10.1074/jbc.274.45.31751. [DOI] [PubMed] [Google Scholar]
- Raventos D, Meier C, Mattsson O, Jensen AB, Mundy J. Fusion genetic analysis of gibberellin signaling mutants. Plant J. 2000;22:427–438. doi: 10.1046/j.1365-313x.2000.00759.x. [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–720. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Leon J, Sanchez-Serrano JJ. Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 1999;20:135–142. doi: 10.1046/j.1365-313x.1999.00570.x. [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagane K, Ohya Y, Hasegawa Y, Tanaka I. Metalloproteinase-like, disintegrin-like, cysteine-rich proteins MDC2 and MDC3: novel human cellular disintegrins highly expressed in the brain. Biochem J. 1998;334:93–98. doi: 10.1042/bj3340093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis R. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Segura A, Moreno M, Madueno F, Molina A, Garcia-Olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant-Microbe Interact. 1999;12:16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- Segura A, Moreno M, Molina A, Garcia-Olmedo F. Novel defensin subfamily from spinach (Spinacia oleracea) FEBS Lett. 1998;435:159–162. doi: 10.1016/s0014-5793(98)01060-6. [DOI] [PubMed] [Google Scholar]
- Shelton-Inloes BB, Titani K, Sadler JE. cDNA sequences for human von Willebrand factor reveal five types of repeated domains and five possible protein sequence polymorphisms. Biochemistry. 1986;25:3164–3171. doi: 10.1021/bi00359a014. [DOI] [PubMed] [Google Scholar]
- Shi L, Gast RT, Gopalraj M, Olszewski NE. Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 1992;2:153–159. [PubMed] [Google Scholar]
- Solano R, Ecker JR. Ethylene gas: perception, signaling and response. Curr Opin Plant Biol. 1998;1:393–398. doi: 10.1016/s1369-5266(98)80262-8. [DOI] [PubMed] [Google Scholar]
- Tam JP, Yi-An L, Jin-Long Y, Koiu-Wei C. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci USA. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras FRG, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Vanderleyden J, Cammue BPA, Broekaert WF. Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell. 1995;7:573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Pennicckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Tierens KFMJ, Broekaert WF. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 1999;121:1093–1101. doi: 10.1104/pp.121.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Lopez-Solanilla E, García-Olmedo F, Rodriguez-Palenzuela P. Mutants of Ralstonia (Pseudomonas) solanacearum sensitive to antimicrobial peptides are altered in their lipopolysaccharide structure and are avirulent in tobacco. J Bacteriol. 1997a;179:6699–6704. doi: 10.1128/jb.179.21.6699-6704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Rojo E, Leon J, Sanchez-Serrano JJ. Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 1997b;115:817–826. doi: 10.1104/pp.115.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij CL, Diergaarde PJ, Hart M, Pannekoek H. Full-length von Willebrand factor (vWF) cDNA encodes a highly repetitive protein considerably larger than the mature vWF subunit. EMBO J. 1986;5:1839–1847. doi: 10.1002/j.1460-2075.1986.tb04435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Kato H, Malik K, Sriprasertsak P, Ichinose Y, Shiraishi T, Yamada T. A supprescin from a phytopathogenic fungus deactivates transcription of a plant defense gene encoding phenylalanine ammonia-lyase. J Mol Biol. 1995;249:513–519. doi: 10.1006/jmbi.1995.0313. [DOI] [PubMed] [Google Scholar]
- Walker-Simmons M, Ryan CA. Proteinase inhibitor synthesis in tomato leaves. Plant Physiol. 1984;76:787–790. doi: 10.1104/pp.76.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing a disintegrin and metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995;131:275–278. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]