Abstract
We discovered that sequences essential for replication origin function are frequently conserved in sensu stricto Saccharomyces species. Here we use analysis of phylogenetic conservation to identify replication origin sequences throughout the Saccharomyces cerevisiae genome at base pair resolution. Origin activity was confirmed for each of 228 predicted sites—representing 86% of apparent origin regions. This is the first study to determine the genome-wide location of replication origins at a resolution sufficient to identify the sequence elements bound by replication proteins. Our results demonstrate that phylogenetic conservation can be used to identify the origin sequences responsible for replicating a eukaryotic genome.
Keywords: ACS, ARS, ORC, Saccharomyces, sensu stricto, yeast
Eukaryotic chromosomes are replicated from multiple discrete sites called replication origins, each of which initiates two diverging replication forks. Replication origins are best understood in the budding yeast Saccharomyces cerevisiae, where the ability of origin sequences to support plasmid maintenance provided a convenient assay for origin function, leading to their original designation as ARS elements or autonomously replicating sequences.
The few S. cerevisiae origins that have been investigated at the sequence level are almost all intergenic and consist of ∼200-base-pair (bp) sequences containing an essential ARS consensus sequence (ACS) and several nonessential secondary “B” elements (Shirahige et al. 1993; Weinreich et al. 2004). The ACS has been determined by alignment of known essential elements to consist of an 11-bp motif (T/A)TTTAT(A/G)TTT(T/A), sometimes represented as an extended 17-bp motif based on a larger number of origins (Theis and Newlon 1997). Most origins contain multiple imperfect matches to this motif with the best match not necessarily corresponding to the essential ACS. Since there are >12,000 potential ACS matches in the genome but ∼400 origins, the motif cannot be used to predict origin location. A match to the ACS is essential but not sufficient for origin function, indicating that there are additional sequences and/or chromatin requirements that are at present not understood.
Microarray-based studies have used two approaches to map the approximate location of replication origins. First, by determining the replication time of all genomic sequences and taking advantage of the fact that replication origins are locally the earliest replicating sequences, it has been possible to identify regions with origin activity in S. cerevisiae, Drosophila, and human cultured cells (Raghuraman et al. 2001; Yabuki et al. 2002; MacAlpine et al. 2004; Jeon et al. 2005). Second, chromatin immunoprecipitation (ChIP) of origin-binding factors (ORC [origin recognition complex] and Mcm2-7) facilitated identification of replication origin regions in S. cerevisiae and Drosophila (Wyrick et al. 2001; MacAlpine et al. 2004). However, the fairly low resolution of these studies combined with the degeneracy (in S. cerevisiae) or absence (in Drosophila) of known origin motifs has so far precluded the precise identification of replication origin sequences genome-wide (MacAlpine and Bell 2005). A computational study attempted to assign precise S. cerevisiae origin locations by searching for ACS matches that conform to an extended matrix (Breier et al. 2004). This approach worked better than previous computational attempts; however, the algorithm used made multiple assignments at some locations but none at all at the majority of experimentally determined origin regions (MacAlpine and Bell 2005), and therefore this study was not used for genome annotation.
Here we demonstrate that most replication origin sequences are phylogenetically conserved among closely related Saccharomyces species, and we use this conservation to permit genome-wide identification of the DNA sequences responsible for replicating S. cerevisiae chromosomes.
Results and Discussion
Evolutionary conservation of replication origin sequences
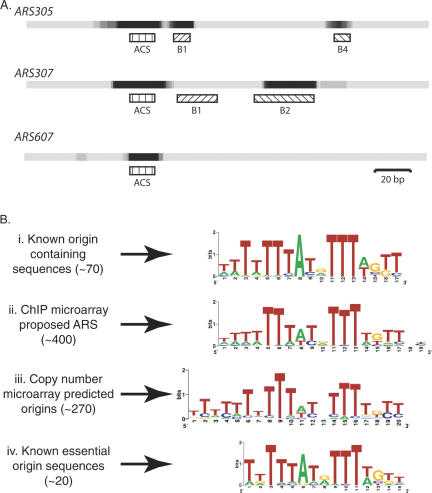
We examined evolutionary conservation at ∼20 previously characterized S. cerevisiae replication origins, comparing them with the corresponding sequences from four closely related sensu stricto Saccharomyces species (Saccharomyces paradoxus, Saccharomyces mikatae, Saccharomyces kudriavzevii, and Saccharomyces bayanus) (Cliften et al. 2003; Kellis et al. 2003). Sequences important for origin function were frequently conserved in the related species, particularly the essential ACS element that is bound by ORC (Fig. 1A; Theis et al. 1999; Breier et al. 2004; Nieduszynski et al. 2005). Transcription factor-binding sites show similar phylogenetic conservation, an observation that allowed the discovery or rediscovery of the motifs bound by each factor (Harbison et al. 2004; Xie et al. 2005). We combined origin location data with intergenic phylogenetic conservation data (Bejerano et al. 2005) to identify replication origin motifs. Three different sources of origin location data were used: (i) the location of 70 known ARS-containing restriction fragments, (ii) ∼400 regions that bind origin proteins (ORC and Mcm2-7) as identified by ChIP (Wyrick et al. 2001), and (iii) the location of ∼270 origins identified as early replicating regions (Yabuki et al. 2002). All three sources of origin location data revealed essentially the same motif (Fig. 1B; Supplementary Fig. S2). Comparison of these motifs to a reference motif generated by aligning 20 known essential replication origin elements (iv) showed that we had rediscovered the ACS. Therefore, like transcription factor-binding sites, replication factor-binding sites are evolutionarily conserved.
Figure 1.
Evolutionary conservation of functionally important origin sequence elements allows the rediscovery of the essential ORC-binding motif. (A) Density plots representing phylogenetic sequence conservation at S. cerevisiae replication origins ARS305, ARS307, and ARS607. Darker shading indicates higher levels of phylogenetic conservation. Functional sequence elements are shown by hatched bars (see also Supplementary Fig. S1); the essential ACS element is recognized by ORC. The B4 at ARS305 structurally and functionally resembles B2 elements at other origins (Lin and Kowalski 1997). (B) Phylogenetically conserved sequences (Bejerano et al. 2005) from intergenes close to origins, (as assigned by various sources: i, Hirschman et al. 2006; ii, Wyrick et al. 2001; and iii, Yabuki et al. 2002) were analyzed by the program MEME (Bailey and Elkan 1994) to determine the most significant motif. (iv) An ACS motif determined from 20 known ACS sequences. Motifs are drawn as LOGOs (Crooks et al. 2004).
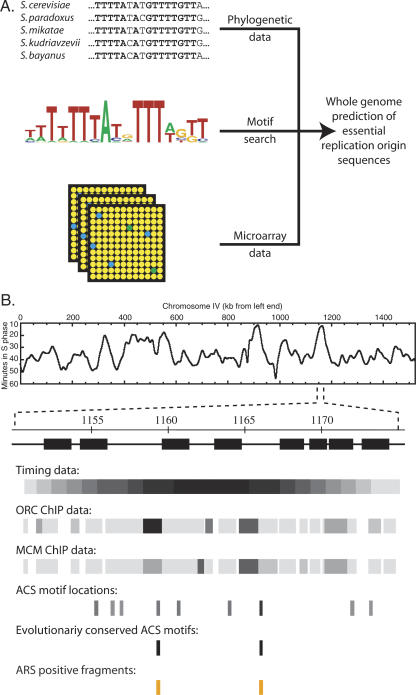
We used this evolutionary conservation to identify the critical origin consensus elements throughout the S. cerevisiae genome. ACS matches were scored on three criteria: phylogenetic conservation across the five Saccharomyces species (see Materials and Methods), proximity to an origin region (as identified in array-based studies) (Raghuraman et al. 2001; Wyrick et al. 2001; Yabuki et al. 2002), and similarity to our ACS motif (Fig. 2A). In analyzing the ChIP data set and ACS similarity values, we found it useful to adopt lower thresholds than the previous studies (Wyrick et al. 2001; Breier et al. 2004), where stringent thresholds had been employed to control the number of false positives. To test our combinatorial approach to origin identification, we examined a region of chromosome IV suggested by microarray studies to contain an origin (Fig. 2B). The ChIP study proposed an origin (called proARS432) at ∼1159 kb; however, a second region at ∼1165 kb fell just below the thresholds to be designated an origin. We identified nine high-scoring matches to our ACS motif across this region, but only two were intergenic and evolutionarily conserved. One of these fell within the region identified as proARS432, while the other lay close to the second weaker ChIP data point. We cloned 250-bp fragments containing the two conserved ACS elements and showed that both had ARS (origin) activity (Fig. 2B) that depended upon the ACS (see below). Therefore, our combined criteria precisely identified proposed origin ARS432 (which we call ARSIV-1159) and a novel replication origin (ARSIV-1166), and phylogenetic conservation correctly predicted both essential origin sequence elements.
Figure 2.
Combining motif searches, phylogenetic conservation, and microarray data allows the prediction of essential replication origin sequences throughout the genome. (A) We combined the motif from Figure 1B, section i, with phylogenetic sequence conservation data (ACS from ARS305 shown) and microarray data to predict the location of ACS elements throughout the genome. (B) An example from chromosome IV shows how combining data sources allows precise localization of replication origin sequences. From top to bottom are shown the following: chromosome IV replication timing plot (Raghuraman et al. 2001) identifies a peak corresponding to an early replication origin at ∼1160 kb; genes (black boxes) and intergenes (thin lines) in this region; replication timing data (early replicating sequences shown darker) (Yabuki et al. 2002); ORC and MCM-binding data (darker shading indicates stronger signal) (Wyrick et al. 2001); ACS motifs (high scoring shown darker, not to scale); conserved ACS motifs; and regions found to be ARS-positive on cloning.
Confirmation of origin activity
To confirm our identified origin sites and extend this analysis genome-wide, we developed a high-throughput transformation assay for ARS activity (Fig. 3; Supplementary Fig. S3). This assay distinguishes between sequences with and without ARS activity, allowing us to test proposed ACS (proACS) sites in vivo. We identified 228 ARS-positive fragments, each between 230 and 315 bp in length and containing one proACS (Supplementary Table S1; Supplementary Fig. S4). (In many but not all cases, proACS sequences correspond to one of the proARS calls made by Wyrick et al. [2001.) The previous genome-wide studies inevitably include a number of false positives, but we can have greatest confidence in the 151 locations they agree upon and we have identified origin sequences at 130, or 86%, of these (Supplementary Table S2; Supplementary Fig. S5). For example, on chromosome VII we identified and confirmed ARS activity for 19 proACS sites (Fig. 3A,B), including all of the chromosomally active origins identified by Raghuraman et al. (2001) and Yabuki et al. (2002). Since we tested the ability of each of the sites listed in Supplementary Tables S1 and S2 to behave as an origin, these lists contain no false positives.
Figure 3.
Confirmation of proACS activity across chromosome VII. (A) Schematic representation of chromosome VII showing the locations of proACS elements with ARS activity that we have verified (colored vertical bars). The origins in the 14 regions predicted by all three previous studies (Raghuraman et al. 2001; Wyrick et al. 2001; Yabuki et al. 2002) are shown as red bars. Origins at locations identified by only one or two of the previous studies are shown as orange bars. For representations of other chromosomes, see Supplementary Figure S4. The centromere is indicated by a black box and the telomeres are indicated by zigzags. (B) Representative ARS assay plates for a subset of the chromosome VII origins tested. (C) Results of ARS assays when the proACS is mutated.
ACS elements of previously identified origins were not used in determining proACS locations and therefore can be used to assess the accuracy of our predictions. At 12 of the 13 origins for which the essential ACS has previously been assigned, our predictions were in precise agreement with experimental data. The exception is ARS121 (ARSX-684), where our proACS overlaps but is distinct from the reported ACS (Walker et al. 1990). We therefore performed linker scan analysis at ARS121 and showed that the conserved proACS is essential whereas the previously identified ACS is not (Supplementary Fig. S6). We mutated a further 20 proACS elements (examples in Fig. 3C), including those at most chromosome VII origins. In 19 cases mutating the proACS abolished or dramatically reduced ARS activity; in one case the mutation did not affect ARS activity (Supplementary Table S3). Some origins have multiple redundant ACS elements (Theis and Newlon 2001), and it is likely that the mutated proACS that did not abolish ARS activity falls into this category. Therefore, phylogenetic conservation allows the location at base pair resolution of most of the essential sequence elements required for origin activity.
Properties of replication origin sequences
An ACS is essential but not sufficient for origin activity, and therefore, the sequence context of the ACS is important for its function. Reported contextual features such as flanking B sequence elements also contribute to origin activity. Several origins contain a B2 element of unknown molecular function that, intriguingly, shows sequence resemblance to the ACS. At ARS305 (Lin and Kowalski 1997) and ARS307 (Rao et al. 1994; Theis and Newlon 1994), the B2 elements are phylogenetically conserved (Fig. 1A). However we identified phylogenetically conserved sequences similar to B2 elements at only a small number of additional origins (Supplementary Fig. S7), perhaps indicating that only a subset of replication origins contains a B2 element. Alternatively, the nonessential nature of origin B2 elements may result in quicker phylogenetic drift, for example, variation in the spacing of the ACS and B2 elements among sensu stricto species. We did not identify any sequence motifs other than the ACS that are shared by all S. cerevisiae origin regions.
A second reported property of replication origins is a region of helical instability, thought to facilitate DNA unwinding (Natale et al. 1993). Sequences from the various sensu stricto species that flank a phylogenetically conserved ACS element share the property of helical instability despite lacking primary sequence conservation (data not shown). Therefore, origins show evolutionary conservation of both primary sequence elements and secondary sequence characteristics.
This study represents the first precise location of replication origin sequences across a eukaryotic genome. It therefore provides the first opportunity to examine the chromosomal context of replication origins genome-wide. We looked at the relationship between replication origins and transcription units. We observed a bias toward occurrence of replication origins in large intergenic spaces and between convergent transcription units (P = 0.00003) (Supplementary Table S4). Thirty-six percent of origins identified lie in convergent intergenic spaces, although only 22% of intergenic spaces are convergent. Where replication origins lie between tandem transcription units, we observed a bias for origins to lie closer to transcriptional terminators than promoters (69% of origins identified between tandem transcription units; P = 0.0003). Together these data suggest either exclusion of origins from promoters (perhaps due to the presence of transcription factors) and/or enrichment in terminator regions, possibly because termination zones tend to have greater helical instability than promoters (Benham 1996).
Next we examined nucleosome occupancy across replication origins. It had been reported that nucleosomes are excluded from the ACS and B elements of ARS1 and ARS307 (Lipford and Bell 2001). Accurate nucleosome positioning data are available at eight additional origins for which we have ACS (or proACS) locations (Supplementary Fig. S8; Yuan et al. 2005). At each origin a nucleosome-free region is bounded on one side by the ACS; the probability of 10 ACS elements lying in a nucleosome-free region by chance is 9.8 × 10−4.
Finally, we examined the modification status of nucleosomes close to replication origins. To investigate whether nucleosomes surrounding origins show specific modification patterns, we analyzed the results of a whole-genome study of histone modification (Pokholok et al. 2005). These data confirmed that origins are located in intergenic regions with low levels of H4 N-terminal acetylation (Vogelauer et al. 2002); in addition, these data suggest an even stronger tendency for the nucleosomes surrounding origins to have low levels of H3K79 trimethylation (Supplementary Fig. S9). Not all histone covalent modifications are reduced close to origins, since origins were randomly distributed among intergenic sequences ranked according to H3K4 monomethylation. It has been suggested that H3 or H4 acetylation state affects the time of origin initiation (Vogelauer et al. 2002); surprisingly, we observed no correlation between origin initiation time and any of the chromatin modifications examined by Pokholok et al. (2005) (Supplementary Fig. S10); neither did any modification relate to whether an origin initiates prior to the hydroxyurea-induced S-phase checkpoint.
To summarize, we have precisely assigned the location of the majority of S. cerevisiae replication origins and shown that sequences identified have ARS activity. For each origin we have proposed the essential 15-bp sequence element proACS. This analysis represents a increase in resolution over previous studies of approximately three orders of magnitude. Our data set unambiguously assigns the intergenic space occupied by each origin and reveals that replication origin sequences tend to fall close to transcriptional terminators. Knowledge of the precise location of replication origin sequences throughout a eukaryotic genome provides a valuable resource, making available an increased repertoire of origins for reductionist studies, and facilitating systems biology approaches to improve our understanding of DNA replication.
All of the origins identified in this study lie in intergenic regions. The very high levels of phylogenetic conservation in ORFs mask the lower levels of conservation observed at ACS elements and therefore prevent the identification of origin sequence elements that may lie within genes. To date only two origins have been identified within ORFs. These are ARS604, which lies within a transcribed gene but is chromosomally inactive for replication initiation, and ARS605, which is chromosomally active during mitotic growth but lies within a gene that is expressed only in meiosis (Hirschman et al. 2006). The fact that no cases are known where active origins coincide with active transcription, combined with the fact we have identified intergenic origin sequences for the majority of active origins, implies that very few origins lie within transcription units.
ACS elements show lower levels of phylogenetic conservation than genes, presumably because origins have higher levels of functional redundancy; several origins can be deleted from a chromosome without deleterious consequences (Dershowitz and Newlon 1993). The tendency for ACS elements and helical instability to be more evolutionarily conserved than the intergenic average implies that some selective pressure to retain origin sites does exist. We fail to observe phylogenetic conservation at certain replication origins (e.g., ARS1) (Supplementary Fig. S1). This observation may reflect genuine differences in some replication initiation sites between sensu stricto species; however in some cases, our analysis was limited by incomplete sequence data in the related yeast species. In other cases, failure to identify phylogenetic conservation may result from technical difficulties in aligning the genomic sequences.
The yeast genome contains ∼12,000 matches to the ACS motif, yet only ∼400 are functional. Whether a particular match to the ACS behaves as an origin may be determined by several other contributing factors—specifically, the ease of unwinding of DNA in the B region 3′ to the ACS, the presence of a nucleosome-excluding element flanking the B region (possibly a transcription factor-binding site or a run of T or A residues), and a surrounding chromatin conformation that is favorable for origin function. While no single one of these contributing properties may be essential for origin activity, together these features may determine whether a particular ACS motif can function as a replication origin.
We have shown that comparative genomics can be combined with origin location data to discover essential origin sequences. Using this approach, we have precisely identified replication origin sites and confirmed their activity in vivo throughout the S. cerevisiae genome. Our list of precise origin locations permitted the first whole-genome analysis of chromatin conformation close to replication origins revealing global properties of replication initiation sites.
Determining the sequence requirements for DNA replication origins in metazoans has proven elusive, in part hampered by the lack of assays analogous to the yeast ARS assay. Recent studies have greatly increased our knowledge of the approximate location of origins in several metazoans (MacAlpine et al. 2004; Jeon et al. 2005). Combining these results with comparative genomics may offer the first possibility to identify sequence elements or characteristics that regulate metazoan origin selection.
Materials and methods
Comparative genomics
Evolutionary conservation was assessed and scored using the University of California at Santa Cruz (UCSC) Genome Browser (http://genome. ucsc.edu; Bejerano et al. 2005). The custom track facility was used to annotate the Saccharomyces genomes with origin location data and extract phylogenetically conserved intergenic sequences. These sequences were assessed for motifs using MEME (http://meme.sdsc.edu; Bailey and Elkan 1994) and were visualized using WebLogo (Crooks et al. 2004).
ACS identification
Proposed ACS sites were identified on the basis of (1) proximity to an origin identified by microarray studies, (2) similarity to our ACS motif, and (3) phylogenetic sequence conservation. ACS matches in the vicinity of microarray-identified origins (<1.5 kb from a proARS (Wyrick et al. 2001); <3 kb from a copy number-derived origin) (Yabuki et al. 2002) were scored as potentially positive. The data from the ChIP microarray study (Wyrick et al. 2001) were reassessed using lower thresholds to allow identification of origins (e.g., ARS306) that fell just below the published thresholds. Occurrences of the ACS motif in the vicinity of microarray-identified origins were scored using MAST (http://meme.sdsc.edu; Supplementary Table S1; Bailey and Elkan 1994), and the highest-scoring occurrences that also shared phylogenetic conservation were tested for ARS activity. Phylogenetic conservation was assessed using alignments from either the UCSC Genome Browser or the Saccharomyces Genome Database and scored as potentially positive if either 12 out of 15 ACS bases were identical between S. cerevisiae and at least one other sensu stricto species, or the UCSC Genome Browser (phastCons table) annotated the ACS with a phylogenetic conservation score >0.01 (Supplementary Table S1). Because phylogenetic sequence data are unavailable at some locations and a minority of ACS may not be phylogenetically conserved, we tested 21 ACS occurrences for which there was no observed conservation but that scored highly on the other two criteria; these had ARS activity and are included in Supplementary Tables S1 and S2.
Yeast techniques
All ARS assays were performed in the S. cerevisiae strain Y00000 (MATa his3− leu2− met15− ura3−). Candidate ARS fragments were PCR-amplified using oligonucleotides designed to include 50–100 bp of sequence 5′ of the proACS and 150–200 bp of sequence 3′ of the proACS. PCR fragments were ligated into the vector pGEM-T (Promega). The high-throughput recombination-based ARS assay is summarized in Supplementary Figure S3 and will be described in detail elsewhere. Mutagenesis of proACS sites (shown in Fig. 2; Supplementary Table S3) was performed using PCR-mediated site-directed mutagenesis. Oligonucleotide sequences are available on request.
Data access
Our list of origins is being deposited with the Saccharomyces Genome Database and the UCSC Genome Browser to allow genome annotation with origin elements. The custom track facility of the UCSC Genome Browser allows comparison of our data set (available at http://www.oridb.org) with any other S. cerevisiae genome-wide data set.
Acknowledgments
We thank John Diffley for the initial suggestion of examining phylogenetic conservation at replication origins, and Shin-ichiro Hiraga for helpful discussion and advice. A.D.D. is a Royal Society University Research Fellow.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.385306
References
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bejerano G., Siepel A.C., Kent W.J., Haussler D. Computational screening of conserved genomic DNA in search of functional noncoding elements. Nat. Methods. 2005;2:535–545. doi: 10.1038/nmeth0705-535. [DOI] [PubMed] [Google Scholar]
- Benham C.J. Duplex destabilization in superhelical DNA is predicted to occur at specific transcriptional regulatory regions. J. Mol. Biol. 1996;255:425–434. doi: 10.1006/jmbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- Breier A.M., Chatterji S., Cozzarelli N.R. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 2004;5:R22. doi: 10.1186/gb-2004-5-4-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dershowitz A., Newlon C.S. The effect on chromosome stability of deleting replication origins. Mol. Cell. Biol. 1993;13:391–398. doi: 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J.E., Balakrishnan R., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., Fisk D.G., Hong E.L., Livstone M.S., Nash R., et al. Genome Snapshot: A new resource at the Saccharomyces Genome Database (SGD) presenting an overview of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2006;34:D442–D445. doi: 10.1093/nar/gkj117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y., Bekiranov S., Karnani N., Kapranov P., Ghosh S., Macalpine D., Lee C., Hwang D.S., Gingeras T.R., Dutta A. Temporal profile of replication of human chromosomes. Proc. Natl. Acad. Sci. 2005;102:6419–6424. doi: 10.1073/pnas.0405088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Lin S., Kowalski D. Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol. Cell. Biol. 1997;17:5473–5484. doi: 10.1128/mcb.17.9.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford J.R., Bell S.P. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Bell S.P. A genomic view of eukaryotic DNA replication. Chromosome Res. 2005;13:309–326. doi: 10.1007/s10577-005-1508-1. [DOI] [PubMed] [Google Scholar]
- MacAlpine D.M., Rodriguez H.K., Bell S.P. Coordination of replication and transcription along a Drosophila chromosome. Genes & Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale D.A., Umek R.M., Kowalski D. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 1993;21:555–560. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski C.A., Blow J.J., Donaldson A.D. The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic Acids Res. 2005;33:2410–2420. doi: 10.1093/nar/gki539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler E.A., Collingwood D., Hunt S., Wodicka L., Conway A., Lockhart D.J., Davis R.W., Brewer B.J., Fangman W.L. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Rao H., Marahrens Y., Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Mol. Cell. Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K., Iwasaki T., Rashid M.B., Ogasawara N., Yoshikawa H. Location and characterization of autonomously replicating sequences from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:5043–5056. doi: 10.1128/mcb.13.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J.F., Newlon C.S. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol. Cell. Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J.F., Newlon C.S. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl. Acad. Sci. 1997;94:10786–10791. doi: 10.1073/pnas.94.20.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J.F., Newlon C.S. Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites. Mol. Cell. Biol. 2001;21:2790–2801. doi: 10.1128/MCB.21.8.2790-2801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J.F., Yang C., Schaefer C.B., Newlon C.S. DNA sequence and functional analysis of homologous ARS elements of Saccharomyces cerevisiae and S. carlsbergensis. Genetics. 1999;152:943–952. doi: 10.1093/genetics/152.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Rubbi L., Lucas I., Brewer B.J., Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol. Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Walker S.S., Francesconi S.C., Eisenberg S. A DNA replication enhancer in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 1990;87:4665–4669. doi: 10.1073/pnas.87.12.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., Palacios DeBeer M.A., Fox C.A. The activities of eukaryotic replication origins in chromatin. Biochim. Biophys. Acta. 2004;1677:142–157. doi: 10.1016/j.bbaexp.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Wyrick J.J., Aparicio J.G., Chen T., Barnett J.D., Jennings E.G., Young R.A., Bell S.P., Aparicio O.M. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N., Terashima H., Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]