Abstract
Stable transformation of tomato (Lycopersicon esculentum cv Ailsa Craig) plants with a construct containing the antisense sequence for the receiver domain and 3′-untranslated portion of the tomato ethylene receptor (LeETR1) under the control of an enhanced cauliflower mosaic virus 35S promoter resulted in some expected and unexpected phenotypes. In addition to reduced LeETR1 transcript levels, the two most consistently observed phenotypes in the transgenic lines were delayed abscission and reduced plant size. Fruit coloration and softening were essentially unaffected, and all the seedlings from first generation seed displayed a normal triple response to ethylene. Two independent lines with a single copy of the transgene and reduced LeETR1 transcript accumulation were selected for detailed phenotypic analysis of second generation (R1) plants. Delayed abscission, shorter internode length, and reduced auxin movement all correlated with the presence of the transgene and the degree of reduced LeETR1 transcript accumulation. No significant differences were noted for fruit coloration or fruit softening on R1 plants and all seedlings from R1 and R2 seed displayed a normal triple response. LeETR2 transcript accumulation was only slightly reduced in the R1 plants compared with azygous plants, and LeETR3 (NR) transcript levels appeared to be unaffected by the transgene. We propose that ethylene signal transduction occurs through parallel paths that partially intersect to regulate shared ethylene responses.
Chang et al. (1993) described the cloning and characterization of the gene, AtETR1, responsible for a dominant ethylene-insensitive mutant in Arabidopsis and discovered that it shared many similarities with two-component regulators in yeast (Saccharomyces cerevisiae) and bacteria. Expression of the AtETR1 gene in yeast generated ethylene-binding sites with similar affinity for ethylene to that estimated from the dose response curve for ethylene inhibition of hypocotyl growth of Arabidopsis seedlings (Schaller and Bleecker, 1995). The AtETR1 ethylene receptor has three domains: a sensor, a His kinase, and a receiver domain (response regulator). Ethylene is bound within the N-terminal sensor domain that includes three membrane-spanning helices (Schaller and Bleecker, 1995). The predicted His kinase domain of the AtETR1 gene product is autophosphorylated (Gamble et al., 1998). Mutation of the His or other putative catabolic residues in the His domain-abolished autophosphorylation of AtETR1 (Gamble et al., 1998). However, a function for the His kinase has not been determined. Moreover, a function for the putative receiver domain is also unknown. Based on similarity with other two component systems, phosphorylation of the conserved Asp group in the receiver domain may serve to propagate a signal received by the sensor domain; however, an essential role for the Asp in ethylene signal transduction has not been established (Chang and Shockey, 1999).
There are currently five characterized genes that make up a family of ethylene receptors in Arabidopsis (Bleecker, 1999). Dominant mutations in all five have been demonstrated to affect ethylene action (Hua and Meyerowitz, 1998). All five gene products have the three transmembrane domains required for ethylene binding and a putative, GAF-like, cyclic nucleotide-binding domain (Bleecker, 1999). However, two of the five, AtERS1 and AtERS2, do not include a receiver domain, and three of the five gene products, AtETR2, AtEIN4, and AtERS2, do not possess amino acids deemed necessary for the His kinase activity found in AtETR1 (Bleecker, 1999). Although the proteins are structurally different, Hua and Meyerowitz (1998) proposed that at least four of the ethylene receptors serve redundant functions in Arabidopsis. They arrived at this conclusion based on their analysis of single and multiple loss-of-function mutants of four of the five ethylene receptor genes. In addition, because the double, triple, and quadruple loss-of-function mutants resulted in a progressively greater constitutive ethylene-like response, it was concluded that the ethylene response pathway is negatively regulated by the ethylene receptors in Arabidopsis. Their findings supported an earlier proposal of negative regulation based on the observation that loss-of-function mutations in the AtCTR1 gene, a gene product that functions downstream from the ethylene receptors, cause a constitutive ethylene-like response in the absence of ethylene (Kieber et al., 1993).
Using the Arabidopsis ETR1 cDNA as a probe, we identified two orthologues of the Arabidopsis cDNA in tomato (Lycopersicon esculentum cv Ailsa Craig), eTAE1 (Zhou et al., 1996a) and TFE27 (Zhou et al., 1996b). These genes have been named LeETR1 and LeETR2, respectively. In addition to these, the mutant gene causing the NR (Never-Ripe) phenotype in tomato was demonstrated to be closely related to the AtERS1 ethylene receptor in Arabidopsis (Wilkinson et al., 1995; Payton et al., 1996). For simplicity, the NR gene will be referred to here as LeETR3. The gene products for both LeETR1 and LeETR2 possess the three domains of the AtETR1 protein (sensor, His kinase, and receiver domains), whereasLeETR3, like AtERS1, does not contain a receiver domain. Two additional genes encoding tomato ethylene receptors were identified more recently: LeETR4 and LeETR5 (Tieman and Klee, 1999). LeETR4 and LeETR5 both contain sequence for a putative receiver domain but do not have the necessary amino acids for His kinase activity (Tieman and Klee, 1999). As with Arabidopsis (Schaller, 2000), the two tomato receptors lacking a functional His kinase domain include an additional 5′ membrane-spanning domain that may serve as a signal peptide for translation on the endoplasmic reticulum (ER; predicted using the Genetics Computer Group [Madison, WI] software, SPScan).
In addition to the structural differences described above for the five tomato ethylene receptor genes, their patterns of gene expression also differ. The LeETR1 and LeETR2 genes are constitutively expressed in all the tomato tissues examined to date (Zhou et al., 1996a; Lashbrook et al., 1998). LeETR3 is also expressed in all the tissues examined but is considerably enhanced in ripening fruit and pedicel abscission zones (Wilkinson et al., 1995; Payton et al., 1996; Lashbrook et al., 1998). LeETR4 and LeETR5 expression is high in reproductive tissues and lower in vegetative tissues (Tieman and Klee, 1999).
Differences in structure and expression patterns for the tomato ethylene receptors constitutes reason to believe that, in addition to partially redundant functions (Hua and Meyerowitz, 1998; Tieman et al., 2000), the ethylene receptors might also serve specialized functions in the plant. To test this hypothesis, a construct was prepared that contained an antisense copy of the receiver domain and 3′-untranslated portion of the LeETR1 cDNA fused to an enhanced cauliflower mosaic virus (CaMV) 35S promoter (Kay et al., 1987). Because of the high nucleotide sequence identity (78%) between the receiver domains of LeETR1 and LeETR2, it was anticipated that both might be inhibited by the one construct. Although both the LeETR4 and LeETR5 transcripts include sequence for a receiver domain, they share only 52% nucleotide identity with the receiver domain of LeETR1; therefore, inclusion of the LeETR1 antisense construct in the tomato genome was not expected to directly affect the expression level of the LeETR4 or LeETR5 transcripts.
RESULTS
Transformation, Transgene Copy Number, and Phenotypic Characteristics of Primary Transformants (R0)
A construct was prepared in pCGN1547 (McBride and Summerfelt, 1990) that included an enhanced CaMV 35S promoter (Kay et al., 1987) fused to the receiver domain and the 3′-untranslated region of the LeETR1 cDNA clone eTAE1 (accession no. u41103) in the antisense orientation. Several primary transformants (R0) were selected for kanamycin resistance and screened by PCR to ascertain the presence of the nptII gene. Seven transformants produced a PCR product of the expected size. Genomic DNA from these seven transformants, a wild-type plant, and two additional transformants that did not produce a PCR product was used for Southern-blot analysis to confirm the PCR assay and approximate transgene copy number (Table I). The PCR-positive transformants were determined to include between one and three copies of the transgene and the PCR-negatives were determined to include none (Table I).
Table I.
Estimate of transgene copy no. and percent abscission of flower pedicels on explants from primary transformants (R0) incubated for 0 to 80 h in ethylene (25 μL L−1) at 25°C
| Transformant No. | Gene Copy No. | Flower Pedicel Abscission in 25 μL L−1 Ethylene
|
||||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 32 h | 40 h | 48 h | 56 h | 72 h | 80 h | ||
| % | ||||||||
| 13 | 1 | 0 | 0 | – | 11 | 17 | 33 | 44 |
| 34 | 1 | 50 | 70 | – | 100 | |||
| 37 | 2 | 75 | 92 | – | 100 | |||
| 38 | 1 | 0 | – | 25 | 42* | |||
| 39 | 3 | 30 | 80 | – | 100 | |||
| 42 | 3 | 0 | 13 | – | 50 | 100 | ||
| 14 | 0 | 42 | – | 100 | ||||
| 53 | 0 | 0 | – | 100 | ||||
| WT | 0 | 26 | 55 | 96 | 100 | |||
| 23 | 1 | ND | ||||||
Transgene copy no. was estimated from the no. of bands hybridizing to an npt II gene probe on a genomic Southern blot. WT, Wild type. A dash indicates that no data were collected for that time point. ND, Not determined. *, Data collection was terminated after 48 h in this set of measurements at which time all the control (non-transgenic) flowers had already fully abscised.
Notable observations among the R0 plants included several shorter plants that retained infertile flowers longer and in some instances displayed a slightly greater amount of epinastic curvature of the petioles than did the non-transgenic plants. To quantify the effect on abscission, fully open flowers with the pedicels attached were collected and placed in air plus 25 μL L−1 ethylene. Three of the seven transgenic lines had a marked delay in abscission compared with flowers from wild-type or nontransgenic plants that went through the transformation and tissue culture process (Table I). Among the R0 plants, plant number 13 (line 13) had the greatest delay in abscission (Table I). In a similar experiment, leaf petiole explants were exposed to ethylene and the time required for abscission monitored. Petiole abscission was similarly delayed in the same three primary transformants.
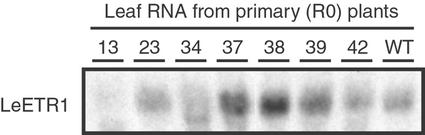
To approximate the levels of LeETR1 transcript in young leaves of wild-type and transgenic plants, RNA blots were hybridized with double-stranded full-length eTAE1 (LeETR1) probe at moderate stringency (for details, see Fig. 1). Multiple RNA extracts were prepared from the transgenic plant lines and several blots prepared. Although the relative signal strength for hybridization fluctuated among the samples on different blots, transgenic plant lines 13 and 34 consistently displayed reduced levels of LeETR1 transcript relative to other transgenic lines and wild-type plants. Of all the transgenic lines examined, line 13 had the least amount of LeETR1 transcript (Fig. 1).
Figure 1.
LeETR1 transcript levels in young fully expanded leaves from primary transformants (R0) of LeETR1 antisense plants and a wild-type plant. Numbers above each lane refer to the plant transformation number as described in Table I. Fifteen micrograms of total RNA was loaded per lane and blotted. Blots were probed with full-length nick-translated eTAE1 cDNA at 42°C in 60% (v/v) formamide, 5× SSPE, 2× Denhardt's, 0.1% (w/v) SDS, and 100 μg mL−1 sheared salmon sperm DNA.
LeETR1 Transcript Level in Leaves of Second Generation (R1) Plants
Although seven R1 transgenic lines were initially cultivated in the greenhouse, only transgenic lines 13 and 34 were selected for detailed study and continued propagation. Lines 13 and 34 displayed reduced LeETR1 transcript levels in R0 plants. In addition, line 13 exhibited the greatest delay in abscission and smallest stature compared with wild-type plants and line 34, which did not display a marked delay in abscission in the preliminary examination of primary transformants (Table I), did however display a somewhat shorter stature and an exaggerated epinastic response.
Ten plants from line 13 and five plants from line 34 were cultivated in the greenhouse. These plants were screened by PCR to determine the presence of the nptII gene. Six plants from line 13 tested positive for the transgene and four negative (azygous). All five plants for line 34 tested positive for the transgene. Southern-blot results on the R0 plants indicated that both transgenic lines might be single-copy genes (Table I). Segregation of the transgene in line 13 confirmed a single locus for the transgene integration into the genome. It was not possible to confirm that line 34 also contained a single locus of transgene integration because no azygous plants were obtained for this line. Six line 13 plants (two azygous and four transgenic) and two line 34 plants were selected for detailed study and propagated vegetatively to increase the amount of material available for analysis.
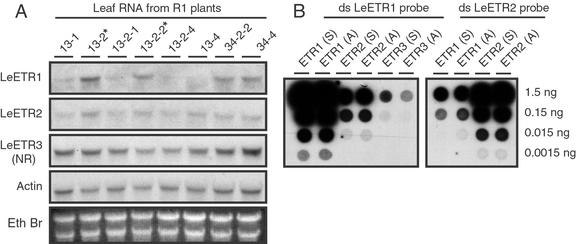
To ascertain transcript levels in young leaves of these plants, northern blots were hybridized with double-stranded LeETR1, LeETR2, LeETR3, and actin DNA probes (Fig. 2A). To reduce probe cross hybridization with the highly conserved sensor domain, DNA probes were prepared from PCR-amplified fragments of the respective His kinase domains. The LeETR1 His kinase domain has 83% and 68% nucleotide sequence identity with the His kinase domains of LeETR2 and LeETR3, respectively. A value of 83% sequence identity is high enough to be concerned about possible cross hybridization on RNA blots. Therefore, cross hybridization of the His kinase probes to the ETR transcripts was assayed by hybridizing the double-stranded probes to dot blots containing in vitro-transcribed sense and antisense RNA transcripts for each of the ETR cDNAs (Fig. 2B). The strength of the hybridization signal for the LeETR1 probe to the LeETR1 transcript was at least 20- and 100-fold greater than the signal strength for hybridization to the heterologous LeETR2 and LeETR3 transcripts, respectively (Fig. 2B). In addition, hybridization of the LeETR2 probe was approximately 20-fold greater to itself than the LeETR1 transcript; thus, under the conditions used for hybridization, cross hybridization between the ETR transcripts was largely restricted (Fig. 2B).
Figure 2.
A, Transcript levels for LeETR1, LeETR2, LeETR3 (NR), and actin in leaf RNA from second generation (R1) transgenic plants. Numbers above lanes refer to the primary transformant number followed by the seedling number. Lanes marked with an asterisk are azygous for the transgene. Each lane was loaded with 40 μg of total RNA from young fully expanded leaves. B, Dot blot of sense (S) and antisense (A) RNA transcripts from LeETR1, LeETR2, and LeETR3 cDNA clones. Probes, hybridization, and washing conditions were the same for A and B. The His kinase domains for LeETR1, LeETR2, and LeETR3 were amplified by PCR and approximately 100 ng of double-stranded DNA labeled by nick translation. Hybridization conditions were at 50°C in 50% (v/v) formamide, 5× SSPE, 5× Denhardt's, 1% (w/v) SDS, and 100 μg mL−1 sheared salmon sperm DNA. The final wash of the blots was in 0.1× SSPE and 0.1% (w/v) SDS at 55°C. Blots were exposed to x-ray film with intensifying screen at −70°C overnight for actin and the dot blots and from 4 to 7 d for the ethylene receptor blots for R1 transgenic plants.
At the time these experiments were begun, sequence information for LeETR4 and LeETR5 was unavailable (Tieman and Klee, 1999). Once these sequences became available, it was determined that the His kinase domains of LeETR1, LeETR2, and LeETR3 range from 56% to 65% identity with LeETR4 and LeETR5. Cross hybridization to the LeETR4 and LeETR5 transcript is negligible at the conditions used for the above hybridizations.
Transcript accumulation for LeETR2, LeETR3, and actin in young mature leaves was relatively constant among all the transgenic and azygous lines examined (Fig. 2A). Transcript levels for LeETR1 were considerably more variable. The two azygous plants (marked with asterisks in Fig. 1A) had the highest amount of LeETR1 transcript relative to actin transcript, whereas transgenic lines 13-2-4 and 13-4 had the least amount of LeETR1 transcript (Fig. 2A). The relative level of LeETR1 transcript to actin for the line 34 series of plants is slightly less than the azygous line 13 series plants. As a guide to general abundance of the mRNAs, the blot probed with actin was exposed to film overnight, whereas the blots for the ethylene receptor probes were exposed from 4 to 7 d.
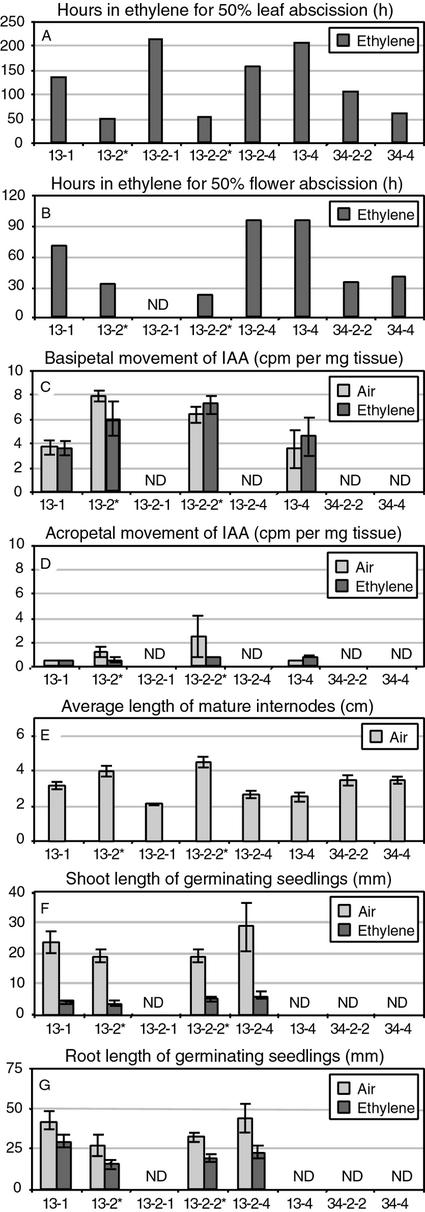
Rate of Abscission and LeETR1 Transcript Levels in Abscission Zones of R1 Plants
To assess the rate of leaf abscission, eight petiole explants were prepared from each of the plant lines in Figure 2A and exposed to 25 μL L−1 ethylene for 200 h. The time required for full abscission of each petiole was recorded. Figure 3A depicts the average time required for 50% of the petioles to abscise. A level of 50% abscission rather than 100% abscission was used because even after 200 h of exposure to ethylene, some of the petioles for the transgenic line 13 plants still had not abscised. A level of 50% abscission in the azygous plants after 50 h of exposure to 25 μL L−1 ethylene is typical of wild-type petioles. A 4-fold delay (from 50 to 200 h) as observed in 13-2-1 and 13-4 is quite remarkable and a 2-fold delay in line 34-2-2 is still nonetheless a very significant delay. A similar experiment was performed with flower explants that included between 15 to 19 flowers. The delay in flower abscission closely reflects the results for petiole abscission except that the time required for 50% of the pedicels to abscise is approximately one-half that of the petiole abscission rate (Fig. 3B). Akin to the results with the petiole abscission, many of the flowers from both transgenic line 13 and line 34 plants did not abscise after 96 h exposure to ethylene. Flowers on wild-type tomato cv Ailsa Craig explants typically will be 90% or greater abscised after only 48 h of exposure to ethylene. After 96 h of exposure to ethylene, an average of 45% of the flowers had abscised from the transgenic line 13 explants and 79% of the line 34 flowers. This is compared with 100% flower abscission after 96 h in the azygous line 13 plants. The reduction in the rate of abscission correlates closely with the reduction in LeETR1 transcript levels (Fig. 2A).
Figure 3.
Quantification of abscission, indole-3-acetic acid (IAA) movement, internode length, and seedling shoot and root length in second-generation R1 plants. The asterisks mark azygous plant lines. Plant tissues were exposed to air or 25 μL L−1 ethylene in air at 25°C. Numbers under bars denote transformant and seedling number. ND, Not determined. A and B, Leaf (stem and petiole) and flower explants were exposed to ethylene for 216 and 96 h, respectively, and the hours for 50% abscission estimated by interpolation between 24-h data points. C and D, 3H-labeled IAA in lanolin (approximately 10−7 m) was applied to distal or proximal ends, respectively, of 3-cm stem internodes and placed in air or ethylene for 20 h, after which 0.5 cm of the untreated ends were collected and soluble radioactivity measured. E, Fifty internodes were measured on three vegetatively propagated plants for each of the R1 plant lines. F and G, Seeds were germinated in the dark for 3 d on moist filter paper and then one-half of the seedlings were transferred to ethylene and one-half remained in air for another 5 d at which time shoot or root length were measured.
In a separate set of explants, the ends of the leaf petioles were dipped into a lanolin paste containing 10−5 m IAA and 4 h later exposed to 25 μL L−1 ethylene. In general, the IAA treatment delayed abscission severalfold in line 13 azygous plants but had very little or no affect on further extending the length of time before abscission occurred in the line 13 transgenic plants (data not shown).
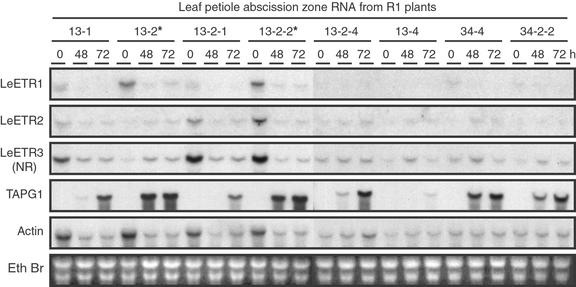
To ascertain transcript levels in abscission zones of lines 13 and 34, RNA was collected from each of the plant lines after 0, 48, and 72 h of exposure to ethylene (Fig. 4). Northern blots were hybridized with double-stranded DNA probes of LeETR1, LeETR2, LeETR3, TAPG1, and actin. Although loading of total RNA was equal between the samples, the levels of actin transcript varied more than in the leaf samples (compare Fig. 4 with 2A). Nevertheless, reduction in LeETR1 expression in transgenic plants relative to azygous plants (13-2 and 13-2-2) closely correlates with the presence of the transgene in abscission zone tissue when compared with actin. Changes in LeETR2 and LeETR3 did not always correlate with actin or the presence of the antisense transgene (e.g. compare 13-2 with 13-2-2 in Fig. 4). Differences in ethylene receptor may also be affected by age or stress conditions of the plant. Earlier studies demonstrated that TAPG1 (tomato abscission polygalacturonase 1) gene expression correlates with the onset of tomato flower and leaf abscission (Kalaitzis et al., 1997). Delayed accumulation of the TAPG1 transcript in the transgenic plants (Fig. 4) correlates precisely with the delayed abscission phenotype (Fig. 3A) and the suppression of the LeETR1 transcript (Fig. 4).
Figure 4.
Relative transcript levels for LeETR1, LeETR2, LeETR3 (NR), TAPG1 (tomato abscission polygalacturonase 1), and actin in flower abscission zones from second generation (R1) plants. Leaf petiole explants were exposed to 25 μL L−1 ethylene in air at 25°C for 0, 48, and 72 h and RNA extracted from the 2-mm abscission zone tissue at the base of the petiole. Fifteen micrograms of total RNA was loaded per lane. Probe preparation, hybridization and washing conditions, and exposure times were as described in Figure 2. The blot hybridized to the TAPG1 probe was exposed to x-ray film overnight with intensifying screen at −70°C.
Stem Internode Length and IAA Movement in R1 Plants
The short stature that was observed in the R0 transgenic plants segregated with the transgene in the R1 line 13 plants. The average internode length of 50 internodes from three vegetatively propagated plants for each of the different R1 line 13 plants was shorter for transgenic plants compared with azygous plants (Fig. 3E). The average internode length of line 34 plants was also shorter than azygous plants but to a lesser extent than line 13 transgenic plants (Fig. 3E). Auxin influences both abscission and cell elongation (Sexton and Roberts, 1982; Estelle, 1992). To determine if auxin movement or metabolism may be altered in the transgenic plants, the basipetal and acropetal movement and accumulation of 3H-labeled IAA were measured in 3-cm stem sections. The concentration of IAA in the lanolin paste was 10−7 m. The mean basipetal movement of label in stem sections exposed to air or ethylene at 25 μL L−1 was less in the transgenic plants than the azygous plants, which suggests that auxin movement is affected by the reduction in LeETR1 transcript levels (Fig. 3C). Acropetal movement of IAA was low in all cases (Fig. 3D).
Other Growth Responses Examined in R1 Plants
Seeds were collected from the seven first generation transgenic plants and wild-type plants, and 30 seeds (two plates of 15 seeds) from each line germinated in the dark in air. After 3 d, one plate of 15 seedlings was transferred to 25 μL L−1 ethylene for an additional 5 d, whereas the other plate remained in air 5 d. Hypocotyl length, stem circumference, apical hook, and gravitropic responses of the transgenic seedlings were identical to wild-type seedlings. None of the seedlings displayed a constitutive ethylene-like triple response in air and all the seedlings had an essentially normal triple response when exposed to ethylene. In addition, seeds from line 13 R1 plants were germinated in the dark for 3 d and then incubated for 5 d in either air or air plus 25 μL L−1 ethylene. The shoot and root length of etiolated seedlings were equally affected in azygous and transgenic plants when transferred to ethylene (Fig. 3, G and H). Moreover, no notable differences were observed in the apical hooks between seedlings from transgenic and azygous plants, indicating a normal response to ethylene. In general, roots and shoots of transgenic plants were slightly longer than azygous plants when germinated in air (Fig. 3, F and G); however, this may reflect differences in germination rates rather than cell elongation rates.
Epinastic curvature of petioles for transgenic plants growing in the greenhouse was only occasionally observed. Line 34 was included in detailed studies of the R1 plants partly because of its strong epinastic phenotype. Both line 34 R1 plants (34-2-2 and 34-4) included the transgene and both retained the strong epinastic phenotype when exposed to ethylene. Although the amount of curvature varied considerably among explants from the same plant, some line 34 petioles curved enough to form a shallow loop in the petiole. The epinastic curvature in ethylene-treated transgenic line 13 petioles was much less than line 34 petioles and not clearly different from wild-type or azygous explants. Based on the results for these two transgenic lines, it was not possible to correlate increased epinasty with reduced LeETR1 transcript (data not shown).
In general, fruit from azygous plants produced more normal-sized fertile seeds than fruit from transgenic plants. Most of the transgenic fruit contained very small underdeveloped seed and several R1 transgenic plants produced very few or no seeds. For example, only one normal-sized seed was obtained from several vegetatively propagated plants from the 13-4 series of transgenic plants and none from the 13-2-1 series. Although the trend for a low number of normal-sized viable seeds was observed in most of the primary (R0) transgenic lines, fruit from the azygous R1 plants also had fewer normal-sized seeds compared with wild-type plants that did not go through the transformation process. The difference in seed number in azygous and wild-type plants suggests that this phenotype might not be completely linked to the reduction in LeETR1 transcript levels and will require further study. Enough seeds were collected from 13-1 and 13-2-4 that a third generation (R2) of plants could be grown. The transgene in these R2 plants segregated at an approximate 3:1 ratio, indicating that the R1 plants from which the seeds were taken were all hemizygous for the transgene. No seeds were obtained from a homozygous R1 plant.
DISCUSSION
The ethylene receptors in Arabidopsis and tomato have several distinctive features that set the family members apart from each other. These include the presence or absence of a putative ER signal peptide, active or inactive His kinase domain, or the lack of a receiver domain (Bleecker, 1999). The absence of a receiver domain in AtERS1, AtERS2 and LeETR3 gene products is particularly distinctive. X-ray crystallography shows that the three-dimensional structure of the receiver domain for AtETR1 has many similarities with bacterial receiver domains where dimerization is important in the regulation of the C-terminal output domains (Müller-Dieckmann et al., 1999). Moreover, the unphosphorylated form of the AtETR1 receiver domain (residues 604–738) forms a homodimer in crystals and in solution as assayed by PAGE in nonreducing sample buffer without detergent (SDS; Müller-Dieckmann et al., 1999). Müller-Dieckmann et al. (1999) proposed that monomerization of AtETR1 protein may be phosphorylation dependent and may play a role in the interaction between native AtETR1 protein and the AtCTR1 protein.
The presence of the receiver domain and its dimerization may have a separate modifying effect on ethylene signal transduction that is not utilized in all ethylene responses. Also of interest in this regard is that, if the receiver domain was deleted from the AtETR1 protein, binding affinity between the AtETR1 and AtCTR1 proteins was much weakened (Clark et al., 1998). Moreover, the ability of the receiver domain to independently form dimers raises the possibility of interaction with other independent response regulators. A family of genes encoding individual response regulators exists in Arabidopsis that are induced by various stimuli (Brandstatter and Kieber, 1998; Imamura et al., 1999). Interactions of this nature could further enhance or inhibit ethylene signal transduction.
The LeETR4 protein includes a putative receiver domain. Tieman et al. (2000) demonstrated that reduction of LeETR4 transcript in transgenic tomato elicited several phenotypes typically associated with exposure to ethylene: severe epinastic curvature of petioles, enhanced senescence of flowers, and accelerated ripening of fruit. In addition, they demonstrated that crossing plant lines with reduced LeETR4 transcript with plants that constitutively overexpress the LeETR3 transcript eliminated the ethylene-sensitive phenotype of the former (Tieman and Klee, 1999). It would appear that the LeETR3 protein, although lacking the receiver domain, can compensate for the loss of the LeETR4 protein function.
To further investigate the hypothesis that different ethylene receptors might serve different signaling functions, we produced transgenic tomato plants with reduced LeETR1 transcript and normal LeETR3 transcript levels by introducing a transgene containing an antisense copy of the receiver domain and 3′-untranslated region of LeETR1 fused to a constitutively expressed enhanced CaMV 35S promoter. Here, we demonstrated that down-regulation of LeETR1 transcript in tomato resulted in some unexpected ethylene-related phenotypes that reinforce the contention of different roles for the different receptors in ethylene signaling and also emphasizes the complexity of interactions between multiple hormone response pathways.
Transgenic primary and secondary transformants displayed delayed abscission and a shorter stature (reduced internode length) that correlated with reduced expression of the LeETR1 transcript. However, although observed in several transformants, enhanced epinastic curvature of petioles and reduced seed set could not be conclusively correlated with LeETR1 transcript suppression. Moreover, there was no apparent effect on the triple response in seedlings or on fruit ripening, which appeared to be independent of the fact that transgenic fruit produced an abundance of small non-germinatable seed. Exaggerated epinasty and shorter internode length are phenotypes consistent with the negative regulation of ethylene responses by LeETR1. Nevertheless, these are complex phenotypes that are responsive to multiple stimuli (Ursin and Bradford, 1989; Estelle, 1992; Lehman et al., 1996). However, based on a negative regulation model, delayed abscission is perhaps the opposite of what might be expected in plants with reduced ethylene receptor because ethylene typically accelerates abscission (Sexton and Roberts, 1982).
In addition to ethylene, the other most important factor that regulates abscission is auxin. Application of auxin distal to the abscission zone strongly inhibits the induction of abscission by ethylene (Sexton and Roberts, 1982). Ethylene has been shown to affect the polar transport and conjugation of auxin in plants (Riov and Goren, 1979). Measurement of the movement of 3H-labeled IAA in 3-cm stem sections indicated a 30% reduction in the amount of label that accumulated at the basipetal end of plants exhibiting reduced LeETR1 transcript (Fig. 3C). As expected, acropetal movement and accumulation at the distal end was low in all stem sections (Fig. 3D). In these basic experiments, it is not possible to discriminate between reduction in transport of IAA and a change in the metabolism or conjugation of IAA. It is interesting that exposure of the stem sections to 25 μL L−1 ethylene compared with air did not further decrease the basipetal movement of labeled IAA in either the azygous or transgenic stem sections (Fig. 3C). This may indicate that the endogenous accumulation of wound- and stress-induced ethylene in the stem sections was sufficient to fully inhibit auxin movement or enhance its conjugation, and therefore the addition of exogenous ethylene had no additional effect.
If auxin movement is reduced in the antisense LeETR1 tomato plants and synthesis remains unchanged in source tissues, it is conceivable that the concentration of active auxin close to the source of auxin would be higher in transgenic plants than wild-type or azygous plants. This could result in an increase in the total amount of auxin in the pedicels and petioles of transgenic plants and lead to a delayed abscission phenotype. Detailed quantification of active auxin in the different plant tissues was beyond the scope of this project. Nevertheless, we performed a simple experiment that may relate to the auxin concentration in the abscission zones of antisense LeETR1 plants. When 10−5 m IAA was applied to the petiolar stump of the line 13 explants before exposure to 25 μL L−1 ethylene, separation occurred at approximately the same rate in azygous and transgenic plants (data not shown). This may indicate that the auxin concentration in the abscission zones of transgenic plants was already at the necessary threshold to inhibit abscission and that the added distal auxin did not adequately move and accumulate in the abscission zones to further delay abscission in the transgenic plants.
In addition to delayed abscission, tomato plants with reduced LeETR1 transcript also displayed reduced plant size (shorter internodes). Reduced plant size may be related to the reduced cell size phenotype observed for the AtETR1 loss-of-function mutant in Arabidopsis (Hua and Meyerowitz, 1998). Moreover, the reduced cell size phenotype in the AtETR1 loss-of-function mutant was not displayed in any of the other ethylene receptor loss-of-function mutants (Hua and Meyerowitz, 1998).
Examples of mutants having both ethylene and auxin phenotypes have been previously identified. Two examples that relate to the present study are the eir1 (ethylene insensitive root 1) mutant that plays a root-specific role in the transport of auxin (Luschnig et al., 1998) and the hls1 (hookless1) mutant that is defective in the response of the apical hook to ethylene, which is also dependent upon the level and distribution of auxin in the hook region (Lehman et al., 1996). Although these mutants are not ethylene receptor mutants, they further accentuate the complex nature of ethylene and auxin interaction in plant growth and development.
Our hypothesis at the outset of this research project was that the receiver domain of LeETR1 and LeETR2 might have a special function in ethylene signal transduction that could not be replaced by the LeETR3 protein, which lacks this domain. The results with the antisense LeETR1 plants suggest this is a viable possibility. The receiver domain may be an additional input or output domain that can modulate ethylene responses in parallel or intersecting ethylene signal transduction pathways. Phosphorylation of the Asp in the receiver domain and proposed phosphorylation-dependent dimerization of the receiver domain (Müller-Dieckmann et al., 1999) may be developmentally and/or cell specifically regulated or it may be a point for interaction with other hormone signaling pathways (Schaller, 2000). Abscission may be particularly sensitive to this control point, whereas the triple response, senescence, or fruit ripening are not. The discovery of two more ethylene receptor genes in tomato, LeETR4 and LeETR5, which also include sequence for receiver domains, complicates interpretation of the results for the antisense LeETR1. Nevertheless, the LeETR4 and LeETR5 genes include an additional membrane-spanning domain that might serve as a signal peptide (predicted using the Genetics Computer Group software, SPScan) for translation on the ER and transport to a different cellular location than LeETR1, LeETR2, and LeETR3 receptors. Moreover, the LeETR4 and LeETR5 receptors do not appear to have functional His kinase domains (Tieman and Klee, 1999). The lack of a functional His kinase domain might change their role in ethylene signal transduction. Although current technology allows the quantification of transcript levels for the ethylene receptors, it is not currently possible to determine the relative concentration of each of the different receptor proteins, nor has the exact location in the cell been determined for the different receptors. Moreover, the cell-specific expression patterns for the different ethylene receptors within a tissue or organ have not been determined. Further studies on tomato plants with reduced expression of genes encoding ethylene receptors will provide insight into the roles of the individual receptors and their interaction with other hormone signaling cascades.
MATERIALS AND METHODS
Construct Preparation and Transformation
The region from 1,969 to 2,665 of eTAE1 (accession no. u41103) was used for the antisense construct described. The region from 1,969 to 2,353 includes coding sequence for the receiver domain and that from 2,354 to 2,665 untranslated-3′ sequence of the LeETR1 ethylene receptor gene. The 3′-eTAE1 sequence was fused in the reverse orientation to an enhanced CaMV 35S promoter (Kay et al., 1987) and nopaline synthase 3′-termination sequence. This construct was then cloned into the BamHI site at the right border of the pCGN1547 binary vector. The binary vector was transformed into Agrobacterium tumefaciens (strain EH105) by the freeze-thaw method of Holsters et al. (1978). Transgenic tomato (Lycopersicon esculentum cv Ailsa Craig) plants were generated by the method of McCormick et al. (1986). Transformed shoots were selected on kanamycin (100 μg mL−1). After rooting of shoots, the plantlets were transferred to sterile potting soil and gradually acclimated before transfer to the greenhouse. PCR and Southern-blot analysis was used to check the integrity and copy number of the introduced genes.
Plant Material and Experimental Treatments
To increase the amount of plant material for experimentation and the number of seeds harvested, transgenic plants were vegetatively propagated by rooting 20-cm apical cuttings. Leaf abscission zone explants used for experimentation consisted of a stem segment with one petiole still attached and leaflets removed. A flower abscission zone explant consisted of a single fully opened flower removed from a panicle of flowers by cutting each at the base of the pedicel. Treatments with nonradioactive IAA were done by thoroughly mixing an aqueous solution of IAA with warm lanolin (approximately 50°C) at a ratio of 1:10 to obtain a final concentration of 10−5 m IAA in the lanolin paste. The ends of stems or petioles were dipped into the still warm lanolin paste (approximately 37°C). Radioactive IAA was applied similarly except that the final concentration of IAA in the lanolin paste was approximately 10−7 m. 3H-labeled IAA was measured in the ends of stems by cutting a 0.5-cm segment from the untreated ends of five stems that were combined and ground in 50 mm NaH2PO4 (pH 7.0), 10 mm EDTA, 0.1% (v/v) Triton X-100, 0.1% (w/v) SDS, and 10 mm β-mercaptoethanol at a ratio of 1:3 (w/v). Radioactivity was measured in a 100-μL aliquot with a liquid scintillation counter. Epinastic curvature of petioles on explants exposed to 25 μL L−1 ethylene in air at 25°C was determined by recording the angle between the stem and the end of the petiole (approximately 4 cm) at 0, 16, 22, and 38 h of exposure to ethylene.
RNA Blots
Total RNA was isolated using a phenol extraction procedure as described by (Sambrook et al., 1989). Fifteen or 40 μg, as indicated in figure legends, of total RNA from abscission zones or young leaves were loaded per lane and fractionated in a 3% (v/v) formaldehyde gel of 1% (w/v) agarose (Whitelaw et al., 1999). RNA was transferred by capillary action onto Hybond N membrane as described by the manufacturer (Amersham, Buckinghamshire, UK). Probe preparation, hybridization conditions, and exposure times are described in the figure legends.
ACKNOWLEDGMENTS
Special thanks to Brad Chapman for collection of tissue samples and preparation of RNA blots, and to Vanessa Thai for maintenance of plants in the greenhouse.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010782.
LITERATURE CITED
- Bleecker AB. Ethylene perception and signaling: an evolutionary perspective. Trends Plant Sci. 1999;4:269–274. doi: 10.1016/s1360-1385(99)01427-2. [DOI] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell. 1998;10:1009–1019. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–358. doi: 10.1016/s1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle M. The plant hormone auxin: insight in sight. Bioassays. 1992;14:439–444. doi: 10.1002/bies.950140703. [DOI] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters MB, Dewacle D, Depicker A, Mosseno E, van Montagu M, Schell J. Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet. 1978;163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999;40:733–742. doi: 10.1093/oxfordjournals.pcp.a029600. [DOI] [PubMed] [Google Scholar]
- Kalaitzis P, Solomos T, Tucker ML. Three different polygalacturonases are expressed in tomato leaf and flower abscission, each with a different temporal expression pattern. Plant Physiol. 1997;113:1303–1308. doi: 10.1104/pp.113.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J. Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann FA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Lashbrook CC, Tieman DM, Klee HJ. Differential regulation of the tomato ETR gene family throughout plant development. Plant J. 1998;15:243–252. doi: 10.1046/j.1365-313x.1998.00202.x. [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KE, Summerfelt KR. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
- Müller-Dieckmann H-J, Grantz AA, Kim S-H. The structure of the signal receiver domain of the Arabidopsis thaliana ethylene receptor ETR1. Structure. 1999;7:1547–1556. doi: 10.1016/s0969-2126(00)88345-8. [DOI] [PubMed] [Google Scholar]
- Payton S, Fray RG, Brown S, Grierson D. Ethylene receptor expression is regulated during fruit ripening, flower senescence and abscission. Plant Mol Biol. 1996;31:1227–1231. doi: 10.1007/BF00040839. [DOI] [PubMed] [Google Scholar]
- Riov J, Goren R. Effect of ethylene on auxin transport and metabolism in midrib sections in relation to leaf abscission of woody plants. Plant Cell Environ. 1979;2:83–89. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaller GE. Histidine kinases and the role of two-component systems in plants. In: Kreis M, Walker JC, editors. Advances in Botanical Research. Vol. 32. London: Academic Press; 2000. pp. 109–148. [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Sexton R, Roberts JA. Cell biology of abscission. Annu Rev Plant Physiol. 1982;33:133–162. [Google Scholar]
- Tieman DM, Klee HJ. Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiol. 1999;120:165–172. doi: 10.1104/pp.120.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Taylor MG, Ciardi JA, Klee HJ. The tomato ethylene receptors NR and LeETR4 are negative regulators of ethylene response and exhibit functional compensation within a multigene family. Proc Natl Acad Sci USA. 2000;97:5663–5668. doi: 10.1073/pnas.090550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursin VM, Bradford KJ. Auxin and ethylene regulation of petiole epinasty in two developmental mutants of tomato diageotropica and Epinastic. Plant Physiol. 1989;90:1341–1346. doi: 10.1104/pp.90.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw CA, Paul W, Jenkins ES, Taylor VM, Roberts JA. An mRNA encoding a response regulator protein from Brassica napus is up-regulated during pod development. J Exp Bot. 1999;50:335–341. [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Zhou D, Kalaitzis P, Mattoo AK, Tucker ML. The mRNA for an ETR1 homologue is constitutively expressed vegetative and reproductive tissue. Plant Mol Biol. 1996a;30:1331–1338. doi: 10.1007/BF00019564. [DOI] [PubMed] [Google Scholar]
- Zhou D, Mattoo AK, Tucker ML. Molecular cloning of a tomato cDNA (accession no. U47279) encoding an ethylene receptor. Plant Physiol. 1996b;110:1435. [Google Scholar]