Abstract
The soluble fraction of immature peanut (Arachis hypogaea) was capable of dephosphorylating [3H]lysophosphatidic acid (LPA) to generate monoacylglycerol (MAG). The enzyme responsible for the generation of MAG, LPA phosphatase, has been identified in plants and purified by successive chromatography separations on octyl-Sepharose, Blue Sepharose, Superdex-75, and heparin-agarose to apparent homogeneity from developing peanuts. This enzyme was purified 5,048-fold to a final specific activity of 858 nmol min−1 mg−1. The enzyme has a native molecular mass of approximately 39 kD determined by gel filtration and migrates as a single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis with a subunit molecular mass of 39 ± 1.5 kD. The Km values for oleoyl-, stearoyl-, and palmitoyl-sn-glycerol-3-phosphate were determined to be 28.6, 39.3, and 47.9 μm, respectively. The LPA phosphatase was specific to LPA and did not utilize any other substrate such as glycerol-3-phosphate, phosphatidic acid, or p-nitrophenylphosphate. The enzyme activity was stimulated by the low concentrations of detergents such as Triton X-100 and octylglucoside. Cations had no effect on the enzyme activity. Fatty acids, sphingosine, and sphingomyelin at low concentrations stimulated the enzyme activity. The identification of LPA phosphatase in plants demonstrates the existence of MAG biosynthetic machinery in plants.
Biosynthesis of lysophosphatidic acid (LPA) occurs by the acylation of glycerol-3-phosphate (G3P) by G3P acyltransferase, which is present in both the soluble and membrane fractions (Bell and Coleman, 1980; Browse and Somerville, 1991). There are three alternate ways of biosynthesis of LPA. LPA is synthesized by the acylation followed by the reduction of dihydroxyacetone phosphate that is catalyzed by dihydroxyacetone phosphate acyltransferase (Webber and Hajra, 1992) and the NADPH-dependent acyl-dihydroxyacetone phosphate reductase (Athenstaedt et al., 1999), respectively. LPA is also derived from phosphatidic acid (PA) by the action of phospholipase A1 or A2 (Thompson and Clark, 1995; Gait et al., 1997) and by the phosphorylation of monoacylglycerol (MAG) by the MAG kinase (Kanoh et al., 1986). LPA is the simplest phospholipid that has been implicated in signal transduction processes. It induces a wide range of activities in animal systems (van Crovan et al., 1989) and elimination of LPA is important for the termination of the cellular signals.
There are three ways of metabolism of LPA. LPA is (a) metabolized by LPA phosphatase to form MAG, (b) acylated to PA by LPA acyltransferase, and (c) hydrolyzed by LPA phospholipase to G3P. LPA phosphatase has been identified, purified from bovine brain, and cloned from animal systems (Xie and Low, 1994; Hiroyama and Takenawa, 1998, 1999). This enzyme has been implicated in the regulation of mitochondrial membrane lipid biosynthesis. Earlier reports on labeling studies with lipid biosynthetic precursors indicated that MAG was labeled significantly. However, the labeled MAG has been attributed to the hydrolytic breakdown of storage lipids or to the artifact in lipid isolation (Gurr, 1980). So far, there is no direct evidence for the formation of MAG and its role as a precursor for triacylglycerol (TAG) biosynthesis in plants. It has been shown that the purified membrane-bound PA phosphatase of avocado mesocarp has the ability to dephosphorylate PA, LPA, and ceramide 1-phosphate (Pearce and Slabas, 1998). To our knowledge, the LPA-specific phosphatase has not been identified and purified from any plant source. Here, we describe the identification, purification to apparent homogeneity, and characterization of LPA-specific and NaF-insensitive phosphatase from the soluble fraction of developing oilseed (Arachis hypogaea).
RESULTS
Examination of Different Tissues for LPA Phosphatase Activity
Hydrolysis of [32P]LPA was studied in the soluble and the particulate fractions from the tissues of leaf and immature seeds of peanut (Arachis hypogaea) and castor (Ricinus communis; Table I). LPA phosphatase was found mostly in the soluble fraction in all of the plant tissues studied. The immature seed soluble fraction showed a 144- to 198-fold higher total activity than the corresponding leaf tissues. These results suggest the presence of LPA phosphatase in plant tissues. Among the plant tissues examined, peanut immature seed showed the highest specific activity. Immature peanut cotyledons were used for further studies.
Table I.
Analysis of different plant tissues for LPA phosphatase activity
| Fraction | Protein | Activity |
|---|---|---|
| mg | pmol min−1 mg−1 | |
| Peanut leaf | ||
| Soluble | 704 | 0.23 ± 0.03 |
| Membranes | 52 | 0.06 ± 0.01 |
| Peanut immature seed | ||
| Soluble | 164 | 188.31 ± 13.8 |
| Membranes | 68 | 27.16 ± 2.6 |
| Castor leaf | ||
| Soluble | 685 | 0.15 ± 0.02 |
| Membranes | 49 | 0.04 ± 0.01 |
| Castor immature seed | ||
| Soluble | 198 | 72.74 ± 4.9 |
| Membranes | 74 | 10.11 ± 0.8 |
Fresh tissue (10 g) was used for preparing the soluble and the membrane fractions by differential centrifugation and 45 μg of protein used for assay. LPA phosphatase was measured for 10 min at 30°C by the release of [32P]Pi from [32P]LPA. Values are means ± sd of two independent experiments, each performed in duplicate.
Formation of MAGs, Diacylglycerols (DAGs), and TAGs from [3H]LPA and [14C]PA
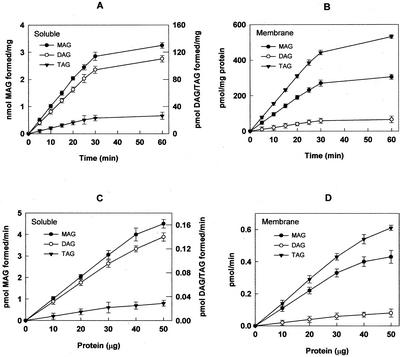
TAG biosynthetic activity was higher in 150,000g pellet than in the supernatant, whereas no significant formation of MAG was observed from PA. Incubation of the soluble fraction with [3H]LPA resulted in the formation of MAGs, DAGs, and TAGs (Table II) and the incorporation was linear with time (Fig. 1A) and protein amounts (Fig. 1C). MAG formation from LPA was about 10-fold higher in the soluble fraction as compared with the membranes. The incorporation of [3H]LPA into MAGs, DAGs, and TAGs in the membrane fraction was also linear with time (Fig. 1B) and protein amounts (Fig. 1D). When the soluble fraction was incubated with [14C]PA, the amount of label in DAG and TAG was less compared with the membranes. These results on the incorporation of [3H]LPA and [14C]PA revealed that only LPA is involved in generation of MAG. When the soluble fraction was incubated with either [3H]trioleoyl-sn-glycerol or [3H]phosphatidylcholine, there was no formation of labeled MAG ruling out the hydrolysis and transacylation of fatty acid (data not shown).
Table II.
Formation of MAGs, DAGs, and TAGs from [3H]LPA and [14C]PA by the soluble and the membrane fractions
| [3H]LPA
|
[14C]PA
|
|||
|---|---|---|---|---|
| Soluble | Membranes | Soluble | Membranes | |
| pmol min−1 mg−1 | ||||
| MAG | 92.05 ± 10.6 | 9.46 ± 0.7 | n.d. | 0.11 ± 0.01 |
| DAG | 3.18 ± 0.2 | 2.05 ± 0.1 | 0.66 ± 0.1 | 6.92 ± 0.6 |
| TAG | 0.73 ± 0.1 | 13.87 ± 1.8 | 0.08 ± 0.01 | 19.74 ± 1.3 |
Fresh immature peanut seed (50 g) was used for preparing the soluble (924 mg) and the membrane (384 mg) fractions. The incorporation of [3H]LPA or [14C]PA (50 μM) into MAG, DAG, and TAG were monitored for 15 min at 30°C in the presence of 20 μM palmitoyl-CoA and 35 μg of protein. Values are mean ± se of four separate experiments. n.d., Not detected.
Figure 1.
Formation of MAGs, DAGs, and TAGs from [3H]LPA. Incorporation of [3H]LPA into MAG, DAG, and TAG was carried out in a time-dependent manner in soluble (A) and membrane (B) fractions. Effect of increasing amounts of soluble proteins (C) and membrane proteins (D) on incorporation of [3H]LPA into MAG, DAG, and TAG. Values are the mean ± sd of four separate experiments, each performed in duplicate.
Generation of MAG
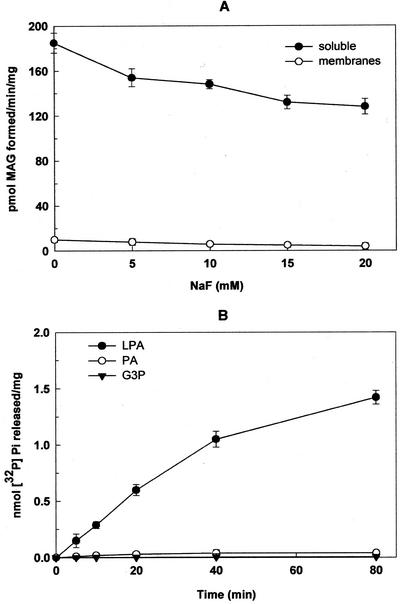
MAG could be formed either by the direct dephosphorylation of LPA or by the deacylation followed by dephosphorylation of PA. To find out the route by which MAG was synthesized, the soluble fraction was treated with various concentrations of NaF to inhibit phosphatase activity and the formation of MAG from [3H]LPA was measured. As shown in Figure 2A, there was about a 33% reduction in the formation of MAG in the presence of 20 mm NaF, but lower concentrations of NaF altered the formation of MAG to a lesser extent (17%) suggesting the presence of NaF-insensitive phosphatase in the soluble fraction. On the contrary, the membrane-bound PA phosphatase was sensitive to NaF (Tumaney et al., 2001). To confirm further, NaF-treated (20 mm) soluble fraction was incubated with [32P]LPA, and it was found that the [32P]Pi was released; whereas [32P]PA and [32P]G3P were not hydrolyzed under the same conditions (Fig. 2B). These results suggested that the phosphatase in the soluble fraction preferentially dephosphorylates LPA.
Figure 2.
Generation of MAG from LPA. A, Effect of NaF on the hydrolysis of [3H]LPA to form MAG was monitored for 10 min at 30°C in the soluble (25–35 μg) and the membrane (40–45 μg) fractions. Values are the mean ± sd of five separate experiments and each performed in duplicate. B, Time-dependent hydrolysis of [32P]LPA was carried out in the soluble fraction (25–35 μg of protein) in comparison with [32P]PA and [32P]G3P. Values are the mean ± sd of three separate experiments, each performed in triplicate.
Purification of LPA Phosphatase
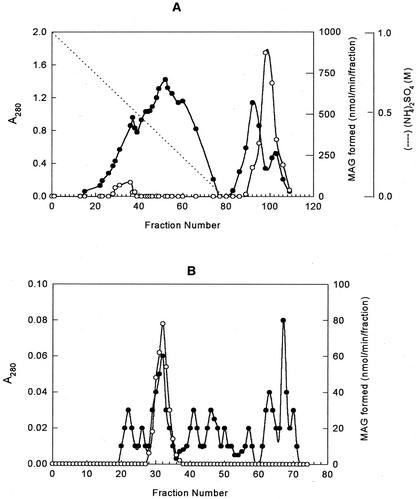
LPA phosphatase activity was found to be high in the soluble fraction and attempts were made to purify the LPA phosphatase using [3H]LPA as a substrate. Solid ammonium sulfate was added to the supernatant obtained after ultracentrifugation to bring to 1 m and then loaded onto an octyl-Sepharose column. After application to the matrix, the column was washed with buffer A containing 1 m ammonium sulfate, and the enzyme was eluted in the absence of salt. Fractions were collected, and the A280 and the LPA phosphatase activity were measured (Fig. 3A). This step was effective and resulted in a 33-fold purification (Table III). The active fractions from the octyl-Sepharose column were loaded onto a Blue Sepharose column. The enzyme did not bind to the matrix, but this step gave a 3.5-fold purification with 90% recovery of activity. The active unbound fraction from the Blue Sepharose was concentrated, the salt concentration was adjusted to 250 mm KCl and applied onto a preparative Superdex 75 FPLC column. The analysis of column fractions revealed that the most of the LPA phosphatase activity eluted between 31 and 33 fractions (Fig. 3B). The active fractions were pooled and dialyzed against buffer A. The dialyzed sample was applied onto a heparin-agarose column as the final step and the enzyme eluted with 0.5 m NaCl.
Figure 3.
Elution profile of LPA phosphatase on various column chromatography. A, Soluble fraction from immature peanuts was loaded onto an octyl-Sepharose column, which was preequilibrated with 1 m ammonium sulfate and the enzyme was eluted from the octyl-Sepharose column. Fractions (5 mL) were collected and assayed for the LPA phosphatase activity (○) and the protein (●). B, Unbound active fractions from Blue Sepharose were pooled, concentrated, and loaded onto a preparative gel filtration column (Superdex 75). Fractions (5 mL) were collected and assayed for the LPA phosphatase activity (○) and the protein (●).
Table III.
Purification of LPA phosphatase from developing peanut cotyledons
| Fraction | Protein | Total Activity | Specific Activity | Purification |
|---|---|---|---|---|
| mg | nmol min−1 | nmol min−1 mg−1 | −fold | |
| Cytosol | 947 | 16 | 0.2 | 1 |
| Octyl-Sepharose | 17.2 | 97.2 | 5.7 | 33 |
| Blue Sepharose | 4.6 | 87.4 | 19.1 | 112 |
| Superdex 75 | 0.6 | 46.6 | 75.1 | 442 |
| Heparin-agarose | 0.03 | 25.7 | 858 | 5048 |
The results are the summary of a typical purification of the LPA phosphatase. Frozen immature seed (50 g) was used for preparing the soluble fraction. The enzyme activities were measured using 3H-LPA as a substrate. The purification steps are described under “Materials and Methods.”
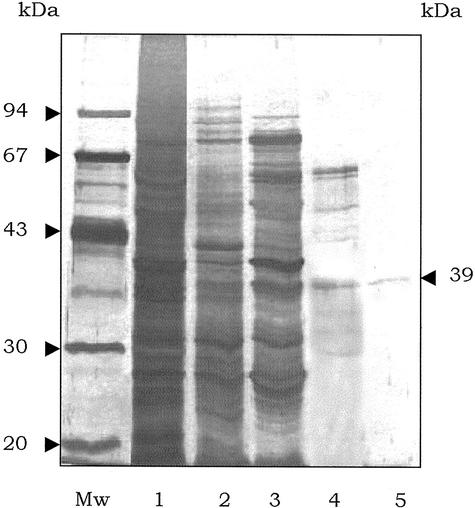
The overall purification of LPA phosphatase is summarized in Table III. The enzyme was purified to 5,048-fold with a 16% yield and the specific activity was 0.86 μmol MAG formed min−1 mg−1. The molecular mass of the enzyme under nondenaturing conditions was determined by gel filtration chromatography on a calibrated Superdex-75 FPLC column and the active LPA phosphatase eluted with an apparent molecular mass of 39 kD. SDS-PAGE of the purified protein yielded a major band upon silver staining, which had an apparent molecular mass of 39 ± 1.5 kD (Fig. 4).
Figure 4.
SDS-PAGE profile of LPA phosphatase purification. Aliquots from each purification step were analyzed on 12% (w/v) SDS-PAGE and visualized by silver staining. Lane Mw shows the molecular mass standards. Lane 1, 150,000g supernatant; lane 2, octyl-Sepharose pool; lane 3, Blue Sepharose pool; lane 4, Superdex 75 peak fraction; and lane 5, heparin-agarose pool.
To confirm the 39 kD is LPA phosphatase, the final preparation was subjected to 12% (w/v) denaturing PAGE without boiling the sample. The gel was cut into 0.5-cm segments, and the protein was eluted from each slice. The formation of MAG from [3H]LPA was measured, and it was found that the activity associated with the area of the gel corresponding to the 39-kD polypeptide (data not shown). The recovery of LPA phosphatase activity from SDS-PAGE was 9% ± 2% of that applied. These results demonstrated that the purified protein is indeed LPA phosphatase.
Characteristics of Peanut LPA Phosphatase
LPA phosphatase activity was linear with time and protein, and the pH optimum of the enzyme was found to be 7.0. To investigate the substrate specificity of LPA phosphatase, phosphate-containing compounds such as PA, G3P, and p-nitrophenyl phosphate were used. The purified enzyme hydrolyzed LPA specifically and did not hydrolyze other substrates (data not shown).
Substrate Specificity of LPA Phosphatase
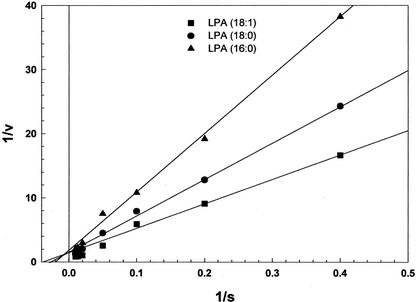
The rate of reaction increased linearly with increasing concentration of LPA (oleoyl) and reached the maximum at 50 μm. The initial rate data showed that the enzyme followed Michaelis-Menten kinetics. The specificity of LPA phosphatase was studied by providing LPA with varying chain lengths and unsaturation as substrates. The LPA phosphatase activity was the highest for oleoyl-LPA and relatively lower for palmitoyl-LPA. Based on the Lineweaver-Burk plot, apparent Vmax and Km values for LPA were obtained under the standard assay conditions (Table IV). The apparent Km values for LPA (oleoyl), LPA (stearoyl), and LPA (palmitoyl) were 28.6, 39.3, and 47.9 μm, respectively (Fig. 5). The saturated fatty acid containing LPA had lower Vmax and higher Km values as compared with oleoyl containing LPA.
Table IV.
Kinetic parameters of LPA phosphatase
| LPA | Km | Vmax | Vmax/Km |
|---|---|---|---|
| μm | μmol min−1 mg−1 | ||
| Palmitoyl | 47.9 ± 4.7 | 0.49 | 0.010 |
| Stearoyl | 39.3 ± 5.1 | 0.54 | 0.014 |
| Oleoyl | 28.6 ± 2.2 | 0.78 | 0.027 |
Apparent Km and Vmax values were calculated from data in Fig. 4.
Figure 5.
Lineweaver-Burk plot of LPA phosphatase activity toward 1-acyl G3P. The LPA phosphatase activity was measured with [3H]LPA (oleoyl), [32P]LPA (palmitoyl), or [32P]LPA (stearoyl) in 0.1% (w/v) Triton X-100 as a substrate. The enzyme activity in the purified enzyme (3 μg) was monitored for 10 min at 30°C as the function of LPA concentrations. The values are mean ± sd of two determinants, each performed in triplicate.
Effect of Detergents and Cations on LPA Phosphatase Activity
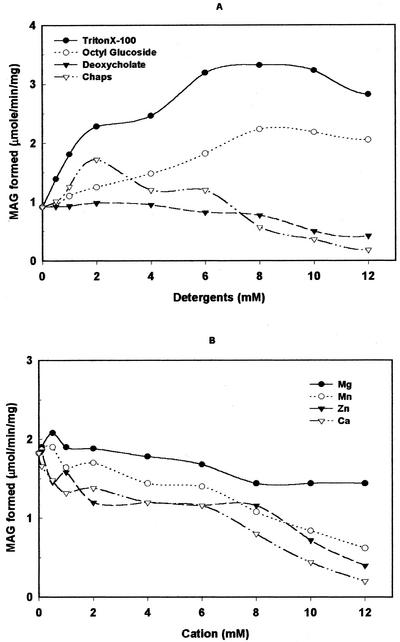
The influence of various detergents on the LPA phosphatase activity was studied (Fig. 6A). In presence of 1 mm Triton X-100, there was a 2-fold increase in the enzyme activity. At 10 mm concentration of Triton X-100 or octylglucoside the enzyme showed a 2- to 3-fold increase in activity. At the same concentration, deoxycholate and 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonic acid (CHAPS) showed a 50% inhibition of the enzyme activity. At a 12 mm concentration of CHAPS, the enzyme lost its activity completely. The effects of cations on the enzyme activity were determined in the presence of 0.1% (w/v) Triton X-100. At 8 mm concentration, Ca2+, Mn2+, and Zn2+ inhibited 40% to 50% of the LPA phosphatase activity, whereas no significant effect was observed with Mg2+ (Fig. 6B).
Figure 6.
Effect of detergents and cations on LPA phosphatase activity. The substrate was made in the indicated detergent to study the effect on LPA phosphatase activity. The purified enzyme activity was measured for 10 min using [3H]LPA and 3 μg of purified protein in the presence of various concentrations of detergents Triton X-100, CHAPS, octylglucoside, and deoxycholate (A) and cations (B). The effects of cations were monitored in the presence of 0.1% (w/v) Triton X-100. Each point is the average of two separate experiments, each performed in triplicate.
Effect of Fatty Acids, Phospholipids, and Sphingoid Bases on LPA Phosphatase Activity
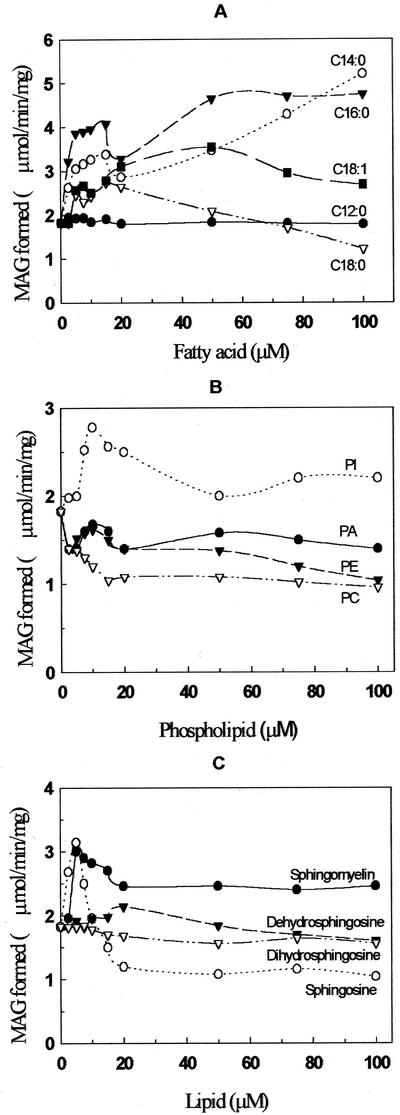
The effects of various lipids on the enzyme activity were determined in the presence of 0.1% (w/v) Triton X-100 (Fig. 7A). Myristic, palmitic, stearic, and oleic acids stimulated the LPA phosphatase activity, whereas dodecanoic acid had no effect. Among the fatty acids examined, palmitic acid showed a 2-fold stimulation at 5 μm, whereas myristic acid showed a 50% increase; and at the same concentration, oleic and stearic acids showed only a 25% increase in enzyme activity. However, the stimulation was not concentration dependent. At higher concentrations of stearic acid (100 μm), the activity was reduced to almost one-half of the original value. Phosphatidylinositol had a 40% stimulating effect at 10 μm on the purified LPA phosphatase activity. Phosphatidylcholine, phosphatidylethanolamine, and PA had no significant effect on the enzyme activity (Fig. 7B). Sphingomylein and lower concentrations (2.5–5 μm) of sphingosine activated the LPA phosphatase activity, whereas concentrations above 10 μm of sphingosine showed an inhibitory effect on the enzyme activity (Fig. 7C). The derivatives of sphingosine, dehydrosphingosine, and dihydrosphingosine showed no significant effect on the enzyme activity.
Figure 7.
Effect of fatty acids, phospholipids, and sphingoid bases on LPA phosphatase activity. The purified LPA phosphatase activity was measured using 2 to 3 μg of protein under the standard assay conditions with [3H]LPA in 0.1% (w/v) Triton X-100 as substrate and in the presence of various concentrations of fatty acids (A), phospholipids (B), and sphingoid bases (C). Each point is the average of two determinations and each performed in triplicate. PI, Phosphatidylinositol; PE, phosphatidylethanolamine; and PC, phosphatidylcholine.
DISCUSSION
The biosynthesis of DAG is known to occur by the sequential acylation of G3P followed by dephosphorylation of PA in the microsomal membranes (Kennedy, 1961; Browse and Somerville, 1991). In the developing peanut cotyledons, both DAG and TAG biosynthetic activities were higher in membrane fraction as compared with the corresponding soluble fraction (Fig. 1; Table II). In addition, we report here, for the first time to our knowledge, the presence of LPA phosphatase in the soluble fraction of developing oilseeds. The results reported here provide evidence for the origin of MAG from LPA by dephosphorylation, and the synthesis is NaF insensitive. The formation of LPA by the acylation of G3P in the soluble fraction is well established in oil accumulating systems such as cocoa seed (Fritz et al., 1986). The synthesized LPA from the soluble G3P acyltransferase is dephosphorylated to MAG by the LPA phosphatase.
This study also provides evidence for the soluble nature of LPA phosphatase in plants. First, the activity is associated with 150,000g supernatant. Second, the LPA phosphatase is purified by successive column chromatographic separations without detergent from the soluble fraction. It was reported earlier that both in animal (Bell and Coleman, 1980) and in plant (Murphy and Mukherjee, 1987) systems, some of the lipid biosynthetic enzymes are loosely bound to the membranes and are released during isolation procedures. It is likely that the enzyme identified in this study is released from the membranes. A few soluble enzymes such as G3P acyltransferase from plants (Frentzen et al., 1983; Nishida et al., 1987), LPA phosphatase from animal systems (Xie and Low, 1994; Hiroyama and Takenawa, 1998), DAG synthesizing system from oilseed (Murphy, 1988), PA phosphatase of Brewer's yeast (Saccharomyces cerevisiae; Hosaka and Yamashita, 1984), and a soluble TAG biosynthetic complex from oleaginous yeast (Gangar et al., 2001) have been reported.
The kinetic experiments revealed that the overall catalytic efficiency for oleoyl-LPA was 2-fold higher than that of saturated fatty acid containing LPA suggesting that the oleoyl-LPA was the preferred substrate (Table IV). The characterization of the purified bovine LPA phosphatase showed a complete inhibition of activity with 2.5 mm NaF (Hiroyama and Takenawa, 1998). On the contrary, peanut enzyme was not inhibited significantly even at 10 mm NaF.
The generation of MAG by the LPA phosphatase may serve as a precursor for TAG biosynthesis in plants. The synthesized MAG is acylated by a soluble MAG acyltransferase (Tumaney et al., 2001) to form DAG. The DAG is further acylated to form TAG either by the membranes or by the soluble DAG acyltransferase (A. W. Tumaney and R. Rajasekharan, unpublished data) from oilseeds. This study suggests that the additional TAG biosynthetic machinery could exist in the soluble fraction of oilseeds. The identified LPA phosphatase may cause the reduction of lipids through the removal of precursor lipid, LPA, which is the key intermediate for the biosynthesis of glycerolipids. However, the involvement of LPA phosphatase in TAG biosynthesis remains to be elucidated. The LPA phosphatase has significant implication in understanding the regulation of lipid biosynthesis and signal transduction.
MATERIALS AND METHODS
Materials
[9,10-3H(N)]Trioleoylglycerol (10 Ci mmol−1), [γ-32P]ATP (3,000 Ci mmol−1), [1-oleoyl-9,10-3H]LPA (50 Ci mmol−1), [2-palmitoyl-9,10-3H]phosphatidylcholine (92.3 Ci mmol−1), [glycerol-U-14C]PA (100 mCi mmol−1), and [1-14C]palmitoyl-CoA (54 mCi mmol−1), were obtained from PerkinElmer Biosystems (Foster City, CA). Superdex 75 (26/60) prep grade and Superdex 75 (10/30) FPLC columns, octyl-Sepharose 4 fast flow matrix, heparin-agarose, and molecular mass standards were purchased from Amersham Pharmacia Biotech (Uppsala). Protein concentrations were determined by the bicinchoninic acid method (Smith et al., 1985) using bovine serum albumin as the standard and the reagents were from Pierce (Rockford, IL). Liquid scintillation cocktail (UltimaGold) was from Packard Biosciences (Meriden, CT). Thin layer chromatography plates were from Merck (Rahway, NJ). DAG kinase was obtained from Calbiochem (San Diego). All other reagents were obtained from Sigma (St. Louis). [32P]LPA and PA were synthesized using DAG kinase as described (Nachiappan and Rajasekharan, 1994). [32P]G3P was synthesized enzymatically using glycerokinase. Phospholipids were quantified by liberating the organic phosphate with perchloric acid and the liberated phosphate was determined colorimetrically (Bartlett, 1959). Field-grown developing peanut (Arachis hypogaea) cotyledons were harvested at 20 to 25 d after flowering and were either used fresh or stored at −80°C until further use. Castor (Ricinus communis) seedpods were harvested from field grown castor plants at 35 to 44 d after flowering. Endosperm was collected and used either fresh or stored at −80°C until use.
Preparation of the Soluble and the Membrane Fractions
Frozen immature seeds (100 g) were ground in a prechilled mortar and pestle with 10 g of acid-washed sand and 250 mL of extraction buffer containing 50 mm Tris-HCl (pH 8.0), 1 mm EDTA, 10 mm KCl, 1 mm MgCl2, 1 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, 1 μg mL−1 leupeptin, and 0.25 m Suc. The extract was passed through two layers of cheesecloth and centrifuged at 18,000g for 15 min. The 18,000g supernatant was further centrifuged at 150,000g for 2.5 h. The pellet obtained was resuspended in a small volume of buffer containing 20 mm Tris-HCl (pH 7.0) and 1 mm β-mercaptoethanol. Soluble fraction (150,000g supernatant) was used as the enzyme source. All the operations were performed at 4°C.
Incorporation of [3H]LPA and [14C]PA into MAGs, DAGs, and TAGs
In the labeling experiments, 25 to 125 mg of freshly prepared soluble and membrane fractions were used in 0.1 mL of buffer containing 50 mm Tris-HCl (pH 7.0), 10 mm KCl, 1 mm MgCl2, 1 mm β-mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, 1 μg mL−1 leupeptin, and 0.25 μCi of labeled substrate (50 μm). Incubation was carried out at 30°C for various time points. The reaction was stopped by the addition of 50 μL of 6 n HCl followed by 400 μL of chloroform:methanol (1:2, v/v). After lipid extraction, the lower chloroform-soluble materials were separated by thin-layer chromatography on 250 μm silica gel G plates using chloroform:methanol:water (98:2:0.5, v/v) as the solvent system (Tumaney et al., 2001). Labeled MAG, DAG and TAG bands corresponding to the standards were scraped and counted in liquid scintillation counter. Boiled enzyme and zero time were used as control. Average control values were subtracted from the actual assay value and the specific activity was calculated after the correction. Each set of experiments was repeated four times.
Enzyme Assays
Lipid was suspended in 1% (w/v) Triton X-100 and sonicated for 5 min in the sonic bath. The substrate stock solution was diluted 10-fold in the assay. LPA phosphatase assay mixture consisted of 50 mm Tris-HCl (pH 7.0), 50 μm [3H]LPA (220,000 dpm) and 20 to 45 μg enzyme in a total volume of 100 μL. The incubation was carried out at 30°C for indicated time points and stopped by the addition of 50 μL of 6 n HCl followed by 400 μL of chloroform:methanol (1:2, v/v). After lipid extraction, the lower chloroform-soluble materials were separated by thin-layer chromatography on 250-μm silica gel G plates using chloroform:methanol:water (98:2:0.5, v/v) as the solvent system. The lipids were visualized with iodine vapor and spots of MAG were scraped off for determination of radioactivity by liquid scintillation counting. LPA phosphatase was also assayed using [32P]LPA (50 μm; 180,000–270,000 cpm). The assay components and conditions were same as described above, and the amount of [32P]Pi released was measured in the upper phase after lipid extraction. Boiled soluble and membrane fractions were used as a control, and control values were subtracted from the values measured in the presence of active enzyme sources.
Purification of LPA Phosphatase
All the operations were carried out at 4°C except FPLC purification step that was carried out at ambient temperature.
Octyl-Sepharose Chromatography
Solid ammonium sulfate was added to bring the developing peanut cotyledon soluble fraction to 1 m followed by centrifugation. The clear supernatant was loaded onto an octyl-Sepharose column (4.4 × 15 cm) that had been pre-equilibrated with 1 m ammonium sulfate in buffer A (50 mm Tris-HCl [pH 8.0], 1 mm EDTA, 100 mm KCl, 1 mm MgCl2, 1 mm β-mercaptoethanol, and 0.1 mm phenylmethylsulfonyl fluoride) with a flow rate of 1 mL min−1. The column was washed with the same buffer to remove unbound proteins until the effluent had very low A280. The enzyme was eluted with 400 mL of a linear reversed gradient from 1 to 0 m ammonium sulfate in buffer A to elute MAG acyltransferase (Tumaney et al., 2001). The column was further eluted with 200 mL of buffer A without salt and fractions of 5 mL were collected and assayed for LPA phosphatase activity.
Blue Sepharose Chromatography
Active fractions from the octyl-Sepharose were combined and applied onto a Blue Sepharose column (4.4 × 12 cm). The LPA phosphatase did not bind to the column.
Size-Exclusion Chromatography
The unbound fractions from the Blue Sepharose were concentrated and filtered. The filtrate was applied onto a preparative Superdex 75 FPLC column (HR 26/60) fitted with the BioLogic low-pressure chromatography system (Bio-Rad Hercules, CA). The column was eluted with the same buffer at a flow rate of 5 mL min−1. Molecular mass markers used for calibrating column were as follows: blue dextran 2000 (2,000 kD), bovine serum albumin (67 kD), ovalbumin (43 kD), carbonic anhydrase (29 kD), and lysozyme (14 kD).
Heparin-Agarose Chromatography
A 2.5-mL column matrix was preequilibrated with buffer A at room temperature, and the active fractions from the size exclusion column were mixed with matrix for 1 h at 4°C. The mixture was then poured into a column, washed with buffer A, and eluted with buffer A containing 0.25, 0.5, and 1 m NaCl. The LPA phosphatase activity was eluted with 0.5 m NaCl.
Molecular Mass Determination by Gel Filtration
The purified preparation was loaded onto a FPLC HR 10/30 Superdex 75 gel filtration column equilibrated on buffer A containing 0.25 m KCl. The column was calibrated with blue dextran 2000, albumin, ovalbumin, chymotrypsinogen A, and ribonuclease A. The column was eluted with 33 mL of the same buffer, and 0.5-mL fractions were collected. LPA phosphatase activity was assayed, and active fractions 16 to 18 were pooled and dialyzed against buffer A.
Reconstitution of LPA Phosphatase Activity
The purified preparation was resolved on a 12% (w/v) SDS polyacrylamide gel (10 × 10 cm) in the presence of 0.1% (w/v) SDS without boiling the sample, and the gel was resolved under constant current (100 V) at 4°C. After the run, the resolving gel (7 cm) was progressively cut into 0.5-cm slices, the protein was eluted from each segment by finely crushing the gel pieces in 10 mm Tris-HCl (pH 7.0), 1 mm EDTA, 100 mm KCl, 1 mm MgCl2, 1 mm β-mercaptoethanol, and 5% (w/v) Suc, and incubated the same overnight at 4°C. The eluted protein was subjected to dialysis against the same buffer (without Suc) overnight at 4°C. LPA phosphatase containing fraction was detected by estimating the capacity of the fraction to hydrolyze [3H]LPA to form MAG.
ACKNOWLEDGMENTS
We thank Dr. P.N. Rangarajan (Indian Institute of Science) for his helpful discussion and support. We are grateful to Dr. D.L. Savithramma (University of Agricultural Sciences, Bangalore, India) for providing us the immature peanut and castor seeds.
Footnotes
This work was supported by the seed grant from the Indian Institute of Science, Bangalore, India.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010654.
LITERATURE CITED
- Athenstaedt K, Paltauf F, Weys S, Daum G. Redundant systems of phosphatidic acid biosynthesis via acylation of glycerol-3-phosphate or dihydroxyacetone phosphate in the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181:1458–1463. doi: 10.1128/jb.181.5.1458-1463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett GR. Phosphorus assay, in column chromatography. J Biol Chem. 1959;234:466–468. [PubMed] [Google Scholar]
- Bell RM, Coleman RA. Enzymes of glycerolipid synthesis in eukaryotes. Annu Rev Biochem. 1980;49:459–487. doi: 10.1146/annurev.bi.49.070180.002331. [DOI] [PubMed] [Google Scholar]
- Browse J, Somerville C. Glycerolipid synthesis: biochemistry and regulation. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:467–506. [Google Scholar]
- Gait F, Fourcade O, Le Balle G, Gueguen G, Gaige B, Gassama-Digne A, Fauvel J, Salles JP, Mauco G, Simon M-F et al. Lysophosphatidic acid as a phospholipid mediator: pathways of synthesis. FEBS Lett. 1997;410:54–58. doi: 10.1016/s0014-5793(97)00411-0. [DOI] [PubMed] [Google Scholar]
- Frentzen M, Heinz E, McKeon TA, Stumpf PK. Specificities and selectivities of glycerol-3-phosphate acyltransferase and monoacylglycerol-3-phosphate acyltransferase from pea and spinach chloroplasts. Eur J Biochem. 1983;129:629–636. doi: 10.1111/j.1432-1033.1983.tb07096.x. [DOI] [PubMed] [Google Scholar]
- Fritz PJ, Kauffman JM, Robertson CA, Wilson MR. Cocoa butter biosynthesis: purification and characterization of a soluble sn-glycerol-3-phosphate acyltransferase from cocoa seeds. J Biol Chem. 1986;261:194–199. [PubMed] [Google Scholar]
- Gangar A, Karande AA, Rajasekharan R. Isolation and localization of a cytosolic 10 S triacylglycerol biosynthetic multienzyme complex from oleaginous yeast. J Biol Chem. 2001;276:10290–10298. doi: 10.1074/jbc.M009550200. [DOI] [PubMed] [Google Scholar]
- Gurr MI. Biosynthesis of triacylglycerols. In: Stumpf PK, Conn EE, editors. The Biochemistry of Plants. Vol. 4. New York: Academic Press; 1980. pp. 205–248. [Google Scholar]
- Hiroyama M, Takenawa T. Purification and characterization of a lysophosphatidic acid-specific phosphatase. Biochem J. 1998;336:483–489. doi: 10.1042/bj3360483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroyama M, Takenawa T. Isolation of a cDNA encoding human lysophosphatidic acid phosphatase that is involved in the regulation of mitochondrial lipid biosynthesis. J Biol Chem. 1999;274:29172–29180. doi: 10.1074/jbc.274.41.29172. [DOI] [PubMed] [Google Scholar]
- Hosaka K, Yamashita S. Regulatory role of phosphatidate phosphatase in triacylglycerol synthesis of Saccharomyces cerevisiae. Biochim Biophys Acta. 1984;796:110–117. [PubMed] [Google Scholar]
- Kanoh H, Iwata T, Ono T, Suzuki T. Immunological characterization of sn-1,2-diacylglycerol and sn-2-monoacylglycerol kinase from pig brain. J Biol Chem. 1986;261:5597–5602. [PubMed] [Google Scholar]
- Kennedy EP. Biosynthesis of complex lipids. Fed Proc Fed Am Soc Exp Biol. 1961;20:934–940. [PubMed] [Google Scholar]
- Murphy DE. A highly active soluble diacylglycerol synthesizing system from developing rapeseed, Brassica napus L. Lipids. 1988;23:157–163. doi: 10.1007/BF02535452. [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Mukherjee KD. Acyltransferases in subcellular fractions of developing seeds of rape (Brassica napus L.) Lipids. 1987;22:293–298. [Google Scholar]
- Nachiappan V, Rajasekharan R. Enzymatic synthesis of [32P]acyl-sn-glycerol 3-phosphate using diacylglycerol kinase. Anal Biochem. 1994;222:283–285. doi: 10.1006/abio.1994.1487. [DOI] [PubMed] [Google Scholar]
- Nishida I, Frentzen M, Ishizaki O, Murata N. Purification of isomeric forms of acyl-[acyl-carrier-protein]:glycerol-3-phosphate acyltransferase from greening squash cotyledons. Plant Cell Physiol. 1987;28:1071–1079. [Google Scholar]
- Pearce ML, Slabas AR. Phosphatidate phosphatase from avocado (Persea americana): purification, substrate specificity and possible metabolic implications for the Kennedy pathway and cell signalling in plants. Plant J. 1998;14:555–564. [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olsen BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Thompson FJ, Clark MA. Purification of a phosphatidic-acid-hydrolyzing phospholipase A2 from rat brain. Biochem J. 1995;306:305–309. doi: 10.1042/bj3060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumaney AW, Shekar S, Rajasekharan R. Identification purification and characterization of monoacylglycerol acyltransferase from developing peanut cotyledons. J Biol Chem. 2001;276:10847–10852. doi: 10.1074/jbc.m100005200. [DOI] [PubMed] [Google Scholar]
- van Crovan EJ, Groenink A, Jalink K, Eichholtz T, Moolenaar WH. Lysophosphatidate-induced cell proliferation: identification and dissection of signaling pathway mediated by G proteins. Cell. 1989;59:45–54. doi: 10.1016/0092-8674(89)90868-4. [DOI] [PubMed] [Google Scholar]
- Webber KO, Hajra AK. Dihydroxyacetone phosphate acyltransferase. Methods Enzymol. 1992;209:92–98. doi: 10.1016/0076-6879(92)09012-r. [DOI] [PubMed] [Google Scholar]
- Xie M, Low MG. Identification and characterization of an ecto-(lyso) phosphatidic acid phosphatase in PAM212 keratinocytes. Arch Biochem Biophys. 1994;312:254–259. doi: 10.1006/abbi.1994.1307. [DOI] [PubMed] [Google Scholar]