Abstract
Actin-related proteins (ARPs) share less than 60% amino acid sequence homology with conventional actins and have roles in diverse cytoskeletal processes in the cytoplasm and nucleus. The genome of Arabidopsis was explored for possible ARP gene family members. Eight potential ARP gene sequences were found dispersed on three of the five Arabidopsis chromosomes. AtARP2 and AtARP3 are protein orthologs of their similarly named counterparts in other kingdoms. AtARP4, AtARP5, and AtARP6 are orthologs of two classes of nuclear ARPs previously characterized in animals and fungi, BAF53s and ARP6s. AtARP7 and AtARP8 appear to be novel proteins that are not closely related to any known animal or fungal ARPs, and may be plant specific. The complex Arabidopsis ARP gene structures each contain from five to 20 exons. Expressed transcripts were identified and characterized for AtARP2 through AtARP8, but not for AtARP9, and transcripts representing two splice variants were found for AtARP8. The seven expressed genes are predicted to encode proteins ranging from 146 to 471 amino acids in length. Relative to conventional actin and the other ARPs, AtARP2 and AtARP3 transcripts are expressed at very low levels in all organs. AtARP5, AtARP6, and AtARP8 each have distinct transcript expression patterns in seedlings, roots, leaves, flowers, and siliques. Using isovariant-specific monoclonal antibodies, AtARP4 and AtARP7 proteins were shown to be most highly expressed in flowers. The likely involvement of plant ARPs in actin nucleation, branching of actin filaments, chromatin restructuring, and transcription are briefly discussed.
During the last decade, a series of actin-related proteins (ARPs) have been found in eukaryotes (Schafer and Schroer, 1999). ARP amino acid sequence identities vary 20% to 60% from conventional actins. ARPs usually share the actin fold for the nucleotide binding pocket with conventional actins, but they are generally tens to hundreds of amino acids longer. By comparison, conventional actins in all eukaryotes share at least 80% amino acid sequence identity and vary little in length from the 377 amino acid residues found among most plant actins (the only exceptions are found in the evolutionarily most distant protists). ARP functions range from specialized effects on conventional G- and F-actin structures to structural activities that are apparently independent from actin (Kreis and Vale, 1999; Schafer and Schroer, 1999). For example, Arp2 and Arp3 in animals are part of a protein complex that forms branch points on actin filaments, bind profilin, and nucleate and polymerize F-actin. These branched actin polymers generate force and structure at the leading edge of mobile animal cells, but are so far unreported during the tip growth of pollen tubes and root hairs in plants where such force generation might be expected. Yeast Arp4, Arp5, Arp6 (Act3), Arp7, Arp8, and Arp9 proteins and many homologs in animals are found in the cell nuclei (Harata et al., 2000). Genetic analysis of yeast Arp4 suggests it is involved in epigenetic alteration of gene transcription that operates through chromatin remodeling. The Arp6s form another sequence clade with known orthologs in animals and fungi. Fruitfly (Drosophila melanogaster) Arp6 (Frankel and Mooseker, 1996; Kato et al., 2001) is associated with heterochromatin and may also play a role in chromatin structure. With a few exceptions (Kreis and Vale, 1999; Schafer and Schroer, 1999), the functions of other newly discovered and more distantly related ARP sequences are essentially unknown.
The plant actins and in particular Arabidopsis actin gene family has been examined in great detail (Meagher, 1995; Meagher et al., 1999a, 1999b). The vegetative and reproductive classes of actins have not shared a common ancestor for 350 million years. The reproductive actins are expressed at high levels in pollen, ovules, and developing embryos. The vegetative actins are strongly expressed in all plant organs including root, leaves, stems, sepals, and petals. The small numbers of amino acid differences among the Arabidopsis actin isovariants play a significant role in protein function (Meagher et al., 1999a). For example, ectopic expression of a reproductive actin in vegetative organs has dramatic consequences on the cytoskeletal structures and plant development (Kandasamy et al., 2001), whereas overexpression of a vegetative actin is of little consequence.
There is almost nothing known about plant ARP sequences as compared with the plant actins. A detailed study was made of the expression of an Arabidopsis ARP2 gene ortholog AtARP2 (Klahre and Chua, 1999) examining both transcripts and promoter-driven reporter expression. The AtARP2 gene was expressed in only a small subset of vascular tissue types and pollen, and unlike any conventional actin, AtARP2 was expressed at very low levels. Thus, it seemed possible that AtARP2 and other subclasses of plant ARPs might each be contained in gene families with different members showing complementary expression patterns with activity in more cell types than shown by AtARP2 alone. In this study, we have surveyed the ARP genes and their expression in Arabidopsis.
Relative to most other plant species, Arabidopsis has many features that make it an exceptional laboratory plant for molecular cell biologists, including simple genetics, small size, rapid life cycle, and a small genome. The sequence of the 125-megabase genome has been completed, and it appears to contain approximately 25,500 genes (Arabidopsis Genome Initiative, 2000). This genome size suggests that all plants have a minimum complexity approaching 85% of that of the human genome with its 29,000 genes. The Arabidopsis genome is used as a model for the larger and more complex genomes of crop plants (Dennis and Surridge, 2000) and is believed to accurately represent 90% of the complement of most plant protein coding gene families.
In the following study Arabidopsis actin and ARP query sequences were used to identify potential Arabidopsis ARPs in the newly completed genomic database. The gene and transcript structures of seven expressed ARPs were characterized in detail. These ARP genes have far more complex structures than plant actin genes with their coding sequence split up into as many as 20 exons. The ARP transcripts were differentially expressed in a variety of plant organs, but the patterns of ARP expression had little relation to those observed for actins, actin binding proteins, or tubulins. The expression of two of the diverse ARP proteins, AtARP4 and AtARP7, was examined with specific monoclonal antibodies. The Arabidopsis ARP genes were all highly divergent and with one exception, there appears to be only one gene member in each ARP subclass.
RESULTS
Identifying Functional Arabidopsis ARP Homologs
The Arabidopsis actin AtACT2 protein sequence of 377 amino acids was used to query possible translation products from the Arabidopsis genome for all actins and actin-related sequences (Arabidopsis Genome Initiative, 2000). Besides the eight functional actin genes and two actin pseudogenes known from previous studies (McDowell et al., 1996), eight more highly divergent, actin-related gene sequences were also identified and they are listed in Table I. All of the Arabidopsis ARPs are highly divergent from conventional actin but share some sequence identity in the motifs that make up the conserved nucleotide binding pocket. The identity of the ARP amino acid sequences compared with Arabidopsis AtACT2 (Table I) ranges from 45% (e.g. AtARP2 and AtARP4) down to 27% (e.g. AtARP9). Arabidopsis AtARP2, AtARP3, AtARP4, AtARP5, AtARP6, and AtARP7 are likely homologs to characterized animal and yeast ARP genes (Schafer and Schroer, 1999), and these relationships will be discussed further. The most highly divergent Arabidopsis sequences, AtARP8 and AtARP9, show 29% and 27% amino acid identity, respectively, to AtACT2 in the regions of alignment. AtARP8 and AtARP9 could not, based on amino acid sequence identity or similarity, be identified as encoding true orthologs of any known ARP from other non-plant sources. The eight Arabidopsis ARP sequences are found dispersed on chromosomes 1, 3, and 5. Using a series of ARP coding sequences from animals and yeast to independently query the Arabidopsis genome, no other ARP genes were detected (not shown). No genes encoding orthologs to ARP1 or ARP11 were found in Arabidopsis. It appears that these ARPs and other conserved components of the dynactin complex found in animals, protist, and fungi, are not present in higher plants (Lawrence et al., 2001).
Table I.
Actin-related sequences in Arabidopsis
| Actin-Related Protein Gene No. | Chromosome No. | Gene Accession No. | Protein Accession No. | No. of Amino Acids | No. of Exons | Identity/Similarity to AtACT, % | EST Accession No. | EST Tissuea |
|---|---|---|---|---|---|---|---|---|
| AtARP2 Landsburg | 3 | AF095912 | AAC69601 | 389 | 15 | 44.4 /58.5 | None | Low levels all tissue |
| AtARP2 Columbia | 3 | AB026649 | BAB01081 | 389 | 15 | 45.7 /59.3 | Z34035Z34870 | Rl. BudsFl. Buds |
| AtARP3 | 1 | AC007357B19833 | ADD31071 | 427 | 9 | 37.8 /49.0 | None | |
| AtARP4 | 1 | AC013354 | AAF25998 | 442 | 20 | 45.5 /58.4 | R64821 | Variety |
| AA720074 | Variety | |||||||
| AV552990 | Roots | |||||||
| AV527857 | Above 2 to 3 weeks | |||||||
| AV553391 | Roots | |||||||
| AV535502 | Fl. buds | |||||||
| AA042724 | Seedling hypo | |||||||
| H35984 | Variety | |||||||
| AI992577 | ? | |||||||
| AV541926 | Roots | |||||||
| AV531042 | Fl. buds | |||||||
| AV566067 | Siliques | |||||||
| AV555243 | Siliques | |||||||
| AV521175 | Above 2 to 3 weeks | |||||||
| AtARP5 | 1 | AC016662 | AAG52523 | 146 | 5 | 43.1 /51.5 | None | |
| AtARP6 | 3 | AC009992 | AAF03459 | 422 | 6 | 34.5 /44.6 | BE524122 | Dev. seeds |
| AtARP7 | 3 | AL162295 | CAB82687 | 363 | 5 | 39.1 /45.9 | T21352 | Variety |
| AI993893 | ? | |||||||
| N37803 | Variety | |||||||
| T43586 | Variety | |||||||
| AA720292 | Variety | |||||||
| AV549368 | Roots | |||||||
| AV557759 | Siliques | |||||||
| AV557882 | Siliques | |||||||
| AA395654 | Rosettes | |||||||
| AI997687 | Variety | |||||||
| AtARP8 | 5 | AB011476 | BAB09300 | 471/242 | 12/7 | 29.4 /40.24 | BE52594 | Dev. Seed |
| N96055 | Variety | |||||||
| Z29158 | Seedling | |||||||
| AV559233 | Siliques | |||||||
| Z30864 | Seedling, 5 d | |||||||
| AtARP9 | 5 | AB025638 | BAA97429 | 596 | Not determined | 26.7 /39.8 | No EST | |
| No PCR | ||||||||
| Products w/cDNA libraries | ||||||||
| Fl. or leaf |
The tissue origin of the cDNA library from which the EST was derived as listed in GenBank.
Most of the Arabidopsis ARP sequences detected were expressed at the RNA level. The sequence database contains expressed transcripts for five of the eight ARP sequences including AtARP2, AtARP4, AtARP6, AtARP7, and AtARP8 (Table I, column 7). Based on genomic sequence, PCR primers were designed to amplify the full-length protein-coding region of each predicted transcript (Table II). Transcripts for seven of the eight potential ARP genes were found in flower cDNA libraries and sequenced; the exception was AtARP9. AtARP9 transcripts were not detected in leaf or flower cDNA libraries or preparations of seedling, root, leaf, flower, pollen, or silique RNA after reverse transcriptase (RT)-mediated PCR. AtARP9 transcripts also were not found in the database. However transcript sequences from the seven expressed genes enable the prediction of at least eight ARP protein products, because two splice variants of AtARP8 cDNA were found.
Table II.
ARP Gene Sequence Primers for PCRa
| ARP2-NcS | tagtgaaccATGGACAACAAAAACGTCGTCGTTTGC |
| ARP2-BgN | gtctagagatctTTAAGCTTGGCTCATTTTATTC |
| ARP2-333S | CACATGGTATACCTCGGAGGTGCT |
| ARP2-3′N1 | GAATTAAATCATCCTGACGGTAGTA |
| ARP2-3′N2 | ACCAATTGTACTCAAACTTTGA |
| ARP3-NcS | tagtgacccATGGATCCGACTTCTCGACCCGCTATT |
| ARP3-BgN | gtctagagatctTCAATACATTCCCTTGAACACCGGA |
| ARP3-376S | GTCGTGAGCCATCCTGTCCAGAGGT |
| ARP3-3′N1 | CTAGCTTACACTGTGTGTACTCA |
| ARP4-NcS | tagtgagccATGGGATACGGCGGAGATGAAGTGTCAGCT |
| ARP4-BgN | gtctagagatctTTAAGGGCATTTTCTCTGAATGTA |
| ARP4-415S | TCACTAGGCTCATTTCAACAGATG |
| ARP4-3′N1 | AGGTTCAGGCGGAATACTAATAATCT |
| ARP5-NcS | tagtgagccATGGGATACGGTGGAGACGAAGTGTCA |
| ARP5-BmN | gtctagggatccTCAGCATCGATAAAAGGTAGAGAAAC |
| ARP5B-138S | TCTCTACCTTTTATCGATGCTGA |
| ARP5-3′N1 | ATCCTAAACCCTCTGACTGTCTCCA |
| ARP5-3′N2 | AAGTGTTTAATATTTGCCTAAATA |
| ARP6-NcS | tagtgatccATGGGATCAAACATCGTTGTTCTAGACAAC |
| ARP6-BmN | gtctagggatccTCAATGAAAGAATCGTCTACGACA |
| ARP6-362S | GAGCTTCGACCACTTGTCCCAGAT |
| ARP6-N1 | GCATTACAATATACGACAAATAATGTG |
| ARP7-NcS | tagtgaaccATGGAAGCACTAGTTGTTGATGCTGGC |
| ARP7-BmN | gtctagggatccTCAGAAACATTTCCTGTGAACCA |
| ARP7-281S | GTACTTTGTGGTGGTACAACCTCCATG |
| ARP7-3′N1 | TACATAATCTGGTCCACATCTTCA |
| ARP8-NcS | tagtgaaccATGGGAATCCTGAAGAAAGTATGGGGATCG |
| ARP8-BmN | gtctagggatccTCACCACATGAGTCTTGACTTG |
| ARP8-361S | CACTGTGATGCAGCAGGACTTACA |
| ARP8-N1 | TGTACCTTATAACAGCTGTAACCTGCC |
| ARP8-N2 | ATCACCAAGAGTTTCAACAATCTCTCG |
| ARP9-NcS | tagtgatccATGGCAAAACCAAAATCCAATTCTCAT |
| ARP9-BmN | gtctagggatccCTAGGGGTTGATGAAGTACATTG |
Primer sequences are in capitals and lower case characters indicate added cloning sites and sequence clamps.
Determining ARP Gene and Protein Structures
Comparisons of the cloned AtARP2, AtARP3, AtARP4, AtARP5, AtARP6, AtARP7, and AtARP8 transcripts with genome sequences (Table I) were used to determine gene structure. The intron-exon structures of seven of the eight potential Arabidopsis ARP genes are shown in Figure 1. The structures determined for AtARP2, AtARP3, and AtARP5 agreed with the database predictions and were comprised of 15, 9, and 7 exons, respectively. The structure of the previously characterized AtARP2 gene from the Landsberg ecotype of Arabidopsis has been confirmed independently (Klahre and Chua, 1999) and differs by 10% in nucleotides and five nonsynonymous changes that cause amino acid differences from the sequence we found in the Columbia ecotype. This difference suggests a rapid rate of ARP sequence divergence, because nucleotide sequences in these two strains usually differ by less than 3% (Lister and Dean, 1993; McKinney and Meagher, 1998). The closely linked profilin AtPRF1 and AtPRF4 genes show similar exceptional and rapid rates of sequence divergence between these two ecotypes (McKinney and Meagher, 1998). The predicted Arabidopsis AtARP2 and AtARP3 protein sequences of 389 and 427 amino acids, respectively, are in close agreement with their orthologs in other kingdoms. Arabidopsis AtARP5, however, is predicted to encode one of the smallest ARPs ever reported, only 146 amino acids long.
Figure 1.
Physical structure of the expressed Arabidopsis actin-related genes. The structure of the ARP genes is presented based on comparison of genomic sequences and transcript sequences. Exons are boxed and the size of the protein product is indicated in parentheses. White and shaded boxes represent transcribed but untranslated regions (5′- and 3′-UTRs) and translated regions, respectively. Exons are numbered. Intervening and untranscribed flanking sequences are shown as lines. Start and stop codons are indicated. Percent nucleotide homology for regions of AtARP5 aligning with AtARP4 is shown above the AtARP5 sequence. The splice variant of AtARP8 is shown with a dotted line connecting the donor and acceptor sites found in the shorter transcript.
The structures of the AtARP4, AtARP6, AtARP7, and AtARP8 genes were determined unambiguously from expressed sequence tags (ESTs) and/or RT-PCR-amplified and sequenced transcripts (Fig. 1; Table I). However, their structures were different from those proposed for these genomic sequences in the Arabidopsis database. They are comprised of 20, 6, 5, and 12 exons and encode proteins of 442, 422, 363, and 471 amino acids, respectively. Transcripts resulting from an alternate shorter splice variant of AtARP8 were also found in flower, but not leaf cDNA libraries. In the shorter AtARP8 transcript, the exon 6 donor site is spliced to the exon 12 acceptor site, skipping exons 7 through 11, or 729 nucleotides of primary transcript. The shortened AtARP8 protein is only 242 amino acids long.
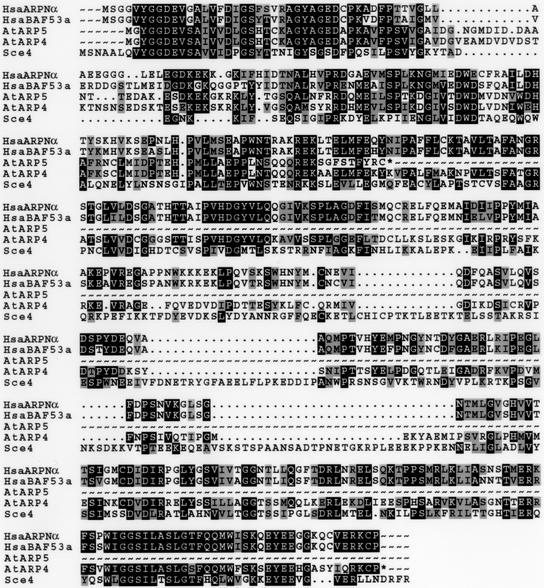
The relationship of the Arabidopsis ARP sequences to each other and to known ARPs from animals and fungi was explored using protein sequence comparison algorithms, such as the neighbor-joining tree shown in Figure 2. Arabidopsis AtARP2 is a clear ortholog of the ARP2 proteins from many organisms, sharing about 62% amino acid sequence identity with human ARP2. AtARP3 is similarly a clear ortholog of ARP3 proteins from many sources, sharing about 60% amino acid sequence identity with human ARP3. Arabidopsis AtARP4 and AtARP5 proteins are clear orthologs of the BAF53 proteins and yeast ARP4. AtARP4 shares 48% identity with human BAF53, as shown in Figure 3. Arabidopsis AtARP4 and AtARP5 share 76% sequence identity with each other in the regions where these two very different-sized proteins align.
Figure 2.
ARP protein sequence phylogram. The seven expressed Arabidopsis ARP protein sequences (Table I) were aligned to known ARP sequences, and their distance relationships are resolved using the neighbor-joining tree-building method. The ARPs examined formed several distinct and ancient clades that are easily resolved from conventional actins. The groups of non-plant actin and ARP sequences are listed with their species of origin and accession numbers. Conventional actins: AtACT2 (Arabidopsis, U41998); Cre (Chlamydomonas reinhardtii, T09103); and Dme53D (fruitfly, S47986). ARP1s: Dme87D (fruitfly, CAA55240) and NcrRo4 (Neurospora crassa, A54802). Arp2s: Aca2 (Acanthamoeba castellanii, U29609); Ddi2 (slime mold [Dictyostelium discoideum], AF095929); Dne14 (fruitfly, S47987); Gga (Gallus gallus, X73971); Hsa2 (human [Homo sapiens], AAB64187); and Sce2 (Saccharomyces cerevisiae, X61502). Arp3s: Aca3 (A. castellanii, U29610); Bta3 (Bos taurus, D12816); Ddi3 (slime mold, Z46418); Dme66B (fruitfly, X71789); Fru3 (Fugu rubripes, AF034581); Hsa3 (human, AF0060830); Ncr3 (N. crassa, U79737); Sce3 (S. cerevisiae, Z49565); and Spo3 (fission yeast[Schizosaccharomyces pombe], P32390). Arp4: HsaARPNα (BAF53α; human, AB015906); HsaBAF53A (human, AF041474); MusBAF53A (Mus musculus, AAC94992); and Sce4 (S. cerevisiae, CAA53066). Arp5s: Sce5 (S. cerevisiae, CAA95933). Arp6s: CelArp6 (Caenorhabditis elegans, Q09443); Dme13E (fruitfly, P45890); GgaArp6X (G. gallus, AB038230); HsaArp6X (human, AB038229); Osa6 (rice [Oryza sativa], BAA89581); Sce6 (S. cerevisiae, CAA97645); and Spo6 (fission yeast, CAA19116). Other Arps: TbrArp (Trypanosoma brucei, AJ132925); MusArp11 (M. musculus, Q9QZB7); Osa7 (rice, AAK09223); Sce7 (S. cerevisiae, Q12406); Sce8 (S. cerevisiae, YOR141C); Sce9 (S. cerevisiae, Q12406); and Sce10 (S. cerevisiae, S52672). Bootstrap values are shown at the base of important branch points.
Figure 3.
Sequence comparisons of Arabidopsis AtARP4 and AtARP5 with BAF53 homologs and yeast ARP4 (Sce4). Amino acid residues outlined in black are identical and those in gray are functionally similar in all sequences. Accession numbers and species of origin for these sequences are give in Figure 2.
AtARP6 is a likely ortholog to a group of less well characterized ARPs including the ARP6s from yeast, fission yeast, and C. elegans and ARP13E from fruitfly (Fig. 2). AtARP6 is 35% identical to the fruitfly ARP13E amino acid sequence. Although the amino acid identity of AtARP4 and AtARP6 with their animal counterparts is not significantly greater than their identity to conventional actin AtACT2, they have greater sequence similarity to their animal counterparts and their Clustal alignments each differ from AtACT2 by many more sequence gaps than do alignments with their nearest ARP relatives (not shown). Therefore, they group with their nearest ARP relatives. AtARP7 could not be grouped reliably with any known class of ARPs (Fig. 2) using several tree-building methods, but did group with an ARP sequence from rice that we named Osa7 (Fig. 2). Other tree-building methods such as maximum parsimony produce similar or consistent topological relationships among most of the ARP sequences compared, with the exception of AtARP8, which was not consistently more related to the yeast ARP8 (Sce8, Fig. 2). The highly divergent AtARP9 sequence could not be aligned efficiently with the other ARP or actin sequences without seriously disrupting closer relationships in the sequence alignments used in these comparisons and, hence, was not included in the phylogram shown in Figure 2.
Differential Expression of Arabidopsis ARP Transcripts and Proteins
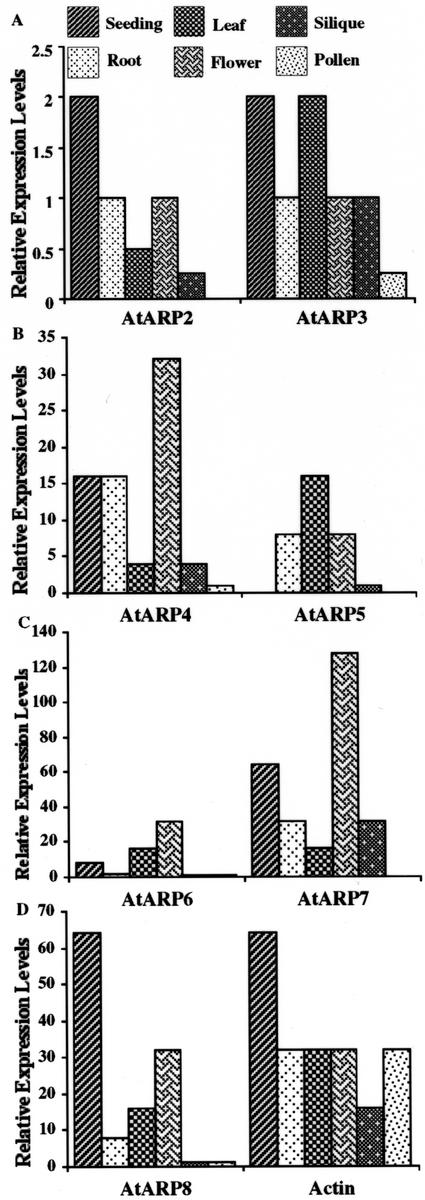
All ARP transcripts characterized from a variety of sources, including the one previously characterized ARP2 transcript from Arabidopsis (Klahre and Chua, 1999), are expressed at very low levels relative to conventional actins. Therefore, organ-specific transcript expression patterns were compared using quantitative RT-PCR of a 300-nucleotide region from the 3′ end of each gene (see “Materials and Methods”). cDNA samples were prepared and quantified from seedlings, roots, leaves, flowers, pollen, and siliques. The levels of transcripts were measured by performing PCR assays on a 2-fold dilution series of these cDNAs with 5 ng of input cDNA in the first sample. The dilution end point for these assays was used to give a quantitative comparison of RNA expression among various organs and pollen (An et al., 1996). AtARP2 and AtARP3 transcripts were expressed at very low levels in all RNA samples examined as shown in Figure 4. This is in agreement with the transcript data of Klahre and Chua (1999) for AtARP2. The highest levels were found in seedlings, roots, leaves, flowers, and siliques. AtARP2 transcripts were not detected in pollen, in contrast to the promoter-reporter fusion data of Klahre and Chua (1999) that suggest reasonable expression of AtARP2 in pollen. Transcripts for the other five Arabidopsis ARPs appear to be at higher levels, based on the greater ease of detection by RT-PCR. For AtARP4, AtARP6, and AtARP7, the highest transcript levels were found in flower, lower levels were found in the other organs, and levels were undetectable in pollen. The transcript levels of conventional actin are shown for comparison in Figure 4D. The frequencies of ARP ESTs in the database are in reasonable agreement with the levels of RT-PCR products we detected, with two exceptions. No ESTs encoding AtARP5 and only one EST encoding AtARP6 was found, contrasting with their moderate expression levels shown by RT-PCR.
Figure 4.
Relative ARP transcript expression levels. Relative ARP transcript expression levels were determined by RT-PCR (see “Materials and Methods”) on a dilution series of cDNA samples prepared from total seedling, root, leaf, flower, silique, and pollen RNA. The dilution half-point at which a product was still obtained is reported for each sample. For example, the dilution half-point for AtARP4 cDNA amplification from seedling and root were 32 and 16, respectively. The lowest dilution contained 5 ng of cDNA. Total cDNA for conventional actin was amplified using degenerate primers as a control (bottom right).
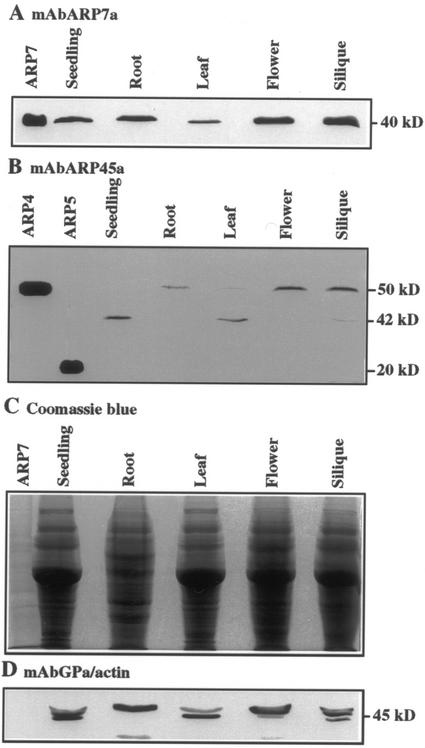
ARP-specific monoclonal antisera were prepared to examine protein expression for three of the Arabidopsis ARPs, as shown in Figure 5. Using the approach described by Li et al. (2001), synthetic peptide immunogens were designed from the N-terminal amino acid residues of AtARP4 and AtARP7 (Fig. 5A). A monoclonal antibody mAbARP7a was obtained that bound to the correct-sized AtARP7 recombinant protein of 40 kD but not to the most closely related AtARP4, AtARP5, or to diverse conventional actins, AtACT2 and AtACT11 (Fig. 5D). Similarly, monoclonal antibody mAbARP45a was obtained that bound to the correct-sized AtARP4 recombinant protein of 50 kD (Fig. 5C). AtARP4 and AtARP5 share identical amino-terminal sequences, thus, mAb45a also reacts with 20-kD AtARP5 protein on the same blot. MAbARP45a does not react with the closely related AtARP7 or to diverse conventional actins, AtACT2 and AtACT11 (Fig. 5C). Figure 5B shows reaction of a general actin-specific monoclonal antibody with the two conventional actins on an identical blot.
Figure 5.
Monoclonal antibodies mAbARP45a and mAbARP7a are specific for their ARP target proteins. A, The N-terminal 26 amino acid sequences of ARPs AtARP4, AtARP5, and AtARP7 and conventional Arabidopsis actins AtACT2 and AtACT11 are compared. The synthetic peptide immunogens for AtARP4 and AtARP7 were comprised of 25 ARP amino acids and omitted the N-terminal methionein (M) residue. B through D, Escherichia coli extracts containing recombinant proteins for actins AtACT2 and AtACT11 and ARPs AtARP4, AtARP5, and AtARP7 were loaded on a 12% (w/v) acrylamide gel, resolved by SDS-PAGE, and transferred to membrane for western blots. Membranes were reacted with monoclonal antisera to all actins mAbGPa (B), mAbARP45a to AtARP5 and AtARP5 (C), and mAbARP7a reacting with AtARP7 (D). The sizes of the ARPs 50, 20, and 40 kD, respectively, and actin (45 kD) are indicated on the relevant blots.
These two-ARP specific monoclonal antibodies were used to characterize relative protein expression levels in various plant organs using western blots presented in Figure 6. Reaction with mAbARP7a showed that AtARP7 was most highly expressed in flowers and siliques, less strongly expressed in seedlings, roots, and leaves (Fig. 6A). Equivalent 45-μg samples of protein were loaded in each lane as shown in the Coomassie Blue-stained panel (Fig. 6B). Equal transfer and sample integrity was confirmed using a control blot probed with a general actin-specific monoclonal antibody, mAbGPa, known to react with all eight Arabidopsis actins (Fig. 6C) and a few common actin degradation products (Kandasamy and Meagher, 1999). These protein expression data were in reasonable agreement with the RNA expression data except that AtARP7 protein appears higher in silique than expected from RNA levels.
Figure 6.
AtARP4 and AtARP7 protein expression. Western blot analysis was performed on total proteins extracted from seedlings, roots, leaves, flowers, and siliques. Monoclonal antibodies specific to AtARP7 (A) and AtARP4 (B) were used to assay the levels of each protein. Replicas of the gel blotted in A were stained with Coomassie Blue (C) or blotted and probed with a general actin antibody mAbGPa (D) to show equal protein loading. The respective sizes of ARP and actin proteins are marked to the right.
Western blots designed to examine AtARP4 and AtARP5 protein expression showed more complex patterns than expected when reacted with mAbARP45a (Fig. 6D). No 20-kD plant protein, the size of recombinant AtARP5 and predicted from sequence, was detected in any organ examined. Protein of the expected 50-kD size for AtARP4 was easily detected at moderate levels in flower and silique and weakly in roots. But the predominant protein detected in seedling and leaf was only 42 kD. In longer exposures of independent blots probed with mAbARP45a bands of both 50 and 42 kD were detected in seedlings. The peptide immunogen used to illicit mAbARP45a is identical for the AtARP4 and AtARP5 proteins, and no other closely related amino acid sequence is found in the Arabidopsis database. In addition, mAbARP45 does not react with any of the other recombinant ARPs (AtARP2, 3, 6, 7, or 8, not shown) or actins. The control blot (Fig. 6C) does not show significantly more degradation of actin in seedling and leaf samples than the other organs, so significant protein degradation is unlikely to have occurred during sample preparation. However, the most likely interpretation is still that the 42-kD band detected is a degradation product of AtARP4. Assuming the 50- and 42-kD bands both represent AtARP4 expression, the protein was moderately expressed in flowers, siliques, and seedlings, and weakly expressed in leaves and roots. Again these results are in agreement with the transcript data shown in Figure 2, except that protein expression in siliques is unexpectedly high relative to RNA levels.
DISCUSSION
Relationships among the Arabidopsis ARP Sequences
Conventional plant actin is contained in a complex gene family. The eight expressed actins in Arabidopsis have complementary expression patterns divided first into vegetative and reproductive classes and further subdivided into ancient subclasses (Meagher et al., 1999b, 2000). In contrast, in Arabidopsis only one or two plant orthologs were found for each of four ancient classes of ARPs (ARP2, ARP3, ARP4 [BAF53s], and ARP6) as shown in Figure 2. This protein sequence tree suggests that the plant sequences are more closely related to ancient classes of ARPs from other kingdoms than they are to other plant ARPs or to conventional actins. Three Arabidopsis ARP protein sequences (AtARP7, AtARP8, and AtARP9) did not group reliably with any known ancient class of ARPs. AtARP7 grouped tightly with Osa7 from rice. The great distance between Arabidopsis and this monocot suggests that plant-specific classes of ARPs may exist.
AtARP4 and AtARP5 clearly belong to the same ARP or BAF53 subclass; however, they have different gene structures and encode proteins of very different lengths (Fig. 1; Table I). AtARP5 is one of the smallest expressed ARPs ever reported. Exon 5 from AtARP5 is a fusion of exons 5, 17, and 19 from AtARP4, whereas exon 6 from AtARP5 is homologous with exon 20 from AtARP4. The counterparts to AtARP4's exons 6 to 16 and exon 18 are apparently deleted from AtARP5. Although more related to each other than to any other ARPs, it appears that these two genes have not had a common ancestor for millions of years. A 381-nucleotide protein-encoding region from the two genes can be aligned unambiguously. AtARP4 and AtARP5 are 9.6% diverged in nonsynonymous nucleotides (Ka = 0.096) that change codons (Fig. 3), but 63% in synonymous positions (Ks = 0.63). Because the unselected mutation rate in plants that would apply to synonymous positions is about 1% to 2% per million years (Meagher et al., 1989; Wolfe et al., 1989), these two genes are tens of millions of years from sharing a common ancestor. Thus, for their common amino acid sequences to be well conserved and their synonymous nucleotide positions to be nearly saturated with changes, they must have been under selective constraint conserving both protein sequences for most of the time since their divergence. However, these data do not reveal when the deletion and fusion of exons occurred in AtARP5. If the reduction in size and rearrangement of AtARP5were also ancient, then the small novel ARP5 isovariant could be functionally quite important and distinct from AtARP4.
Transcripts were found for seven of the eight Arabidopsis ARP sequences identified with the exception being AtARP9 gene, which does not appear to be expressed based on sensitive RT-PCR assays for transcripts. This suggests that the other seven ARP genes may be functional. The expression patterns observed for the various Arabidopsis AtARPs are not concordant with any of the plant actins (Meagher et al., 1999b, 2000), which are either similarly high in all vegetative organs and low in pollen, or the converse. This suggests there has not been the same concordance between the evolution of actin and ARP gene expression patterns as there appears to be for plant profilins and actin depolymerizing factors (Meagher et al., 1999a, 1999b).
Conventional plant and animal actin genes are normally comprised of four or five exons. Only one Arabidopsis ARP (AtARP7) had such a simple structure (five exons), whereas the others were much more complex with as many as 20 exons in AtARP4 (Fig. 1, Table I), the BAF53 ortholog. Some animal ARPs also have complex gene structures such as the human BAF53 ortholog Arp4α with 14 exons (Kato et al., 2001).
Functions of Plant ARPs
Arp2 and Arp3 in animals are part of the ARP2/3-protein complex that forms branch points on actin filaments, cap actin filaments, bind profilin, and nucleate and polymerize actin. Considering the high degree of homology between Arabidopsis AtARP2 and AtARP3 with orthologs from distant eukaryotic kingdoms (Fig. 2), it seems likely that these plant proteins will share in many common functions. In animal cells the greater activity of the ARP2/3-protein complex at the leading edge of cells and the resulting massive branching of actin filaments are thought to generate force and structure in the direction of cell movement or extension. However, nothing like the degree of actin filament branching seen at the leading edge of motile animal cells has ever been observed in any plant cell type including rapidly elongating pollen tubes, trichomes, or root hairs (Parthasarathy, 1985; Parthasarathy et al., 1985; Kandasamy et al., 1999; Kandasamy and Meagher, 1999). This difference in the plant cytoskeleton is not likely to be due to the infamous instability of plant actin filaments, because these various studies include a wide variety of fixation protocols including rapid freeze substitution. These data suggests that activity of AtARP2 and AtARP3 in branching as a mechanism of increasing force generation may be less important in plant cells. Considering the very low level of AtARP2 and AtARP3 transcripts detected, one consistent interpretation would be that their protein products are needed in all cells for the nucleation of actin filaments.
A number of ARP proteins have been localized to the nucleus (Harata et al., 2000), including (a) the BAF53-related proteins characterized in vertebrates; (b) yeast ARP4 (Sce4); and (c) the Arp6-related proteins from yeast, fission yeast, C. elegans, and fruitfly (Fig. 2). Arabidopsis AtARP4 and AtARP5 appear to be sequence orthologs of the BAF53s and yeast ARP4 (Sce4, Fig. 2). The vertebrate BAF complex of which the BAF53 ARP is a component is involved in chromatin remodeling. For example, during lymphocyte activation after antigen recognition, signal transduction is accompanied by association of the BAF53 complex with chromatin and drastic changes in chromatin structure (Zhao et al., 1998). The yeast ARP4 gene is involved in epigenetic alteration of gene transcription, consistent with a role in chromatin remodeling. However, this yeast protein is a subunit of the NuA4 histone acetyltransferase complex, which through nucleosome binding, controls overall gene expression levels (Galarneau et al., 2000). This suggests two potential and nonexclusive roles for the Arabidopsis sequence homologs.
AtARP6 appears to be a sequence ortholog of the fungal and lower animal Arp6s (Fig. 2). The fruitfly ARP6-related protein ARP13E is associated with heterochromatin and may also play a role in chromatin structure (Frankel and Mooseker, 1996; Kato et al., 2001). In yeast, green fluorescent protein fusions of ARP6 are found primarily in the nucleus, but the hydrophobicity of its nuclear export signals suggests some of this protein may move back out to the cytoplasm (Harata et al., 2000). Thus, it may be anticipated that the plant proteins AtARP4, AtARP5, and AtARP6 will be found in the nucleus, playing direct roles in chromatin restructuring and indirect roles in transcription. Considering the location of AtARP8 between the ARP6s and BAF53s in the protein sequence tree (Fig. 2), AtARP8 is likely to be a nuclear ARP protein.
CONCLUSIONS
Arabidopsis contains a small, complex ARP gene family of seven genes falling into four known ARP subclasses and two unknown subclasses (AtARP7 and AtARP8). The ancestry of the four recognizable gene subclasses all predate the divergence of plants from common eukaryotic ancestry, and even the most closely related Arabidopsis ARPs AtARP4 and AtARP5 appear to be separated by tens of millions of years from a common ancestry with each other. The ARP transcripts have distinct organ-specific expression patterns that are quite different from the actins. Based on homology with animal and fungal proteins, two of the plant ARP proteins, AtARP2 and AtARP3, should be found predominantly in the cytoplasm, whereas AtARP4, 5, 6, and AtARP8 should be found in the nucleus.
MATERIALS AND METHODS
Strains and Growth Conditions
The Columbia ecotype of Arabidopsis used in all expression studies was grown with 12 h each of light and darkness each day at 24°C on soil or agar.
Transcript Identification, Quantification, and Cloning
The full-length protein coding region of each transcript from start to stop codon was PCR amplified from a flower cDNA library (Invitrogen, Carlsbad, CA) using the primers described in Table II. Each coding region was cloned into the NcoI/BamHI replacement region of a pET-15b vector (Novagen, Madison, WI). Protein was expressed from this vector in E. coli as per the manufacturer's instructions.
Transcript levels were assayed by RT-PCR. RNA was prepared as described in Huang et al. (1996). cDNA was prepared by reverse transcription of total RNA from Arabidopsis seedlings, roots, leaves, flowers, pollen, and siliques using a First Strand cDNA Synthesis kit (Roche, Indianapolis). cDNA product was quantified in each sample using a sensitive fluorescent microtiter plate assay (PicoGreen dsDNA Quantification kit, Molecular Probes, Eugene, OR) and a Biolumin 960 microtiter plate reader (Molecular Dynamics, Sunnyvale, CA). Gene-specific primers were designed to amplify the C-terminal end of each coding region, including the last intron and a portion of the 3′-untranslated region (Table II). PCR amplification across an intron allows a distinction to be made between PCR products from transcripts and those from contaminating genomic DNA. RT-PCR was quantified as described in An et al. (1996), except that PCR was performed on a 2-fold cDNA dilution series starting with 5 ng of cDNA in the first diluted sample. The dilution half-point at which a RT-PCR product was still obtained was reported as the relative transcript expression level.
Antibodies and Western Blots
Monoclonal antibodies specific to the common N-terminal sequence of AtARP4 and AtARP5 and specific to the unique N-terminal sequence of AtARP7 were obtained using the rapid method described in Li et al. (2001). Two peptides comprised of the first 25 residues of AtARP4 or AtARP5 and AtARP7 proteins were synthesized as 4-fold redundant multiple antigenic peptides (MAP) joined at their C-terminal residues by a Lys core (Tam, 1988). The linear peptide sequences were YGGDEVSAIV VDLGSHTCKA GYAGE (N-MAP-ARP4) and EALVVDAGSK FLKAGAAIPD QSPAM (N-MAP-ARP7), respectively. The peptides were mixed at 1 mg/mL each and a total of 100 μg of protein was injected into each mouse. Sera from mice and monoclonal antibodies produced by hybridomas were screened first using the ARP MAP peptides and then using the E. coli expressed ARP proteins as antigens as described earlier (Li et al., 2001). Western blots were prepared according to Kandasamy et al. (1999) with a buffer containing 5 mg/mL Complete Mini EDTA-Free protease inhibitor (Roche, Mannheim, Germany). Monoclonal antisera were used at 0.5 to 1 μg/mL in blocking mixture.
Sequence Comparisons and Phylograms
Small numbers of closely related sequences were aligned in PileUp as an extension of the Genetics Computer Group (Madison, WI) package and displayed under Boxshade. For phylograms comparing more distant proteins, the various ARP sequences were aligned using ClustalX (Thompson et al., 1994), which is also an extension of the Genetics Computer Group package. In ClustalX the gap extension and gap opening penalties were set at 0.20 and 10.00, respectively. Neighbor-joining trees and maximum parsimony trees (not shown) were constructed using PAUP (Phylogenetic Analysis Using Parsimony, version 4.0, Sinaur Associates, Sunderland, MA).
ACKNOWLEDGMENTS
We thank Kelly Dawe and Gay Gragson for their comments on the manuscript, Greg Derda and Bonnie McCaig for their help with computational and tree-building methods, the Molecular Genetics Facility at the University of Georgia for synthesizing and purifying the synthetic peptide immunogens, and the Monoclonal Facility at the University of Georgia for their help in making monoclonal antibodies.
Footnotes
This work was funded by the National Institutes of Health (grant no. GM36397–15).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010906.
LITERATURE CITED
- An Y-Q, Huang S, McDowell JM, McKinney EC, Meagher RB. Conserved expression of the Arabidopsis ACT1 and ACT3actin subclass in organ primordia and mature pollen. Plant Cell. 1996;8:15–30. doi: 10.1105/tpc.8.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Dennis C, Surridge C. Arabidopsis thalianagenome: introduction. Nature. 2000;408:791. doi: 10.1038/35048677. [DOI] [PubMed] [Google Scholar]
- Frankel S, Mooseker MS. The actin-related proteins. Curr Opin Cell Biol. 1996;8:30–37. doi: 10.1016/s0955-0674(96)80045-7. [DOI] [PubMed] [Google Scholar]
- Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- Harata M, Oma Y, Tabuchi T, Zhang Y, Stillman DJ, Mizuno S. Multiple actin-related proteins of Saccharomyces cerevisiaeare present in the nucleus. J Biochem. 2000;128:665–671. doi: 10.1093/oxfordjournals.jbchem.a022799. [DOI] [PubMed] [Google Scholar]
- Huang S, An Y-Q, McDowell JM, McKinney EC, Meagher RB. The Arabidopsis ACT4/ACT12actin gene subclass is strongly expressed in post-mitotic pollen. Plant J. 1996;10:189–202. doi: 10.1046/j.1365-313x.1996.10020189.x. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney E, Meagher RB. The late pollen specific actins in angiosperms. Plant J. 1999;18:681–691. doi: 10.1046/j.1365-313x.1999.00487.x. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB (2002) Functional non-equivalency of actin isovariants in Arabidopsis. Mol Biol Cell (in press) [DOI] [PMC free article] [PubMed]
- Kandasamy MK, Meagher RB. Actin-organelle interactions: association with chloroplast in Arabidopsisleaf mesophyll cells. Cell Motil Cytoskeleton. 1999;44:110–118. doi: 10.1002/(SICI)1097-0169(199910)44:2<110::AID-CM3>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Kato M, Sasaki M, Mizuno S, Harata M. Novel actin-related proteins in vertebrates: similarities of structure and expression pattern to Arp6 localized on Drosophilaheterochromatin. Gene. 2001;268:133–140. doi: 10.1016/s0378-1119(01)00420-6. [DOI] [PubMed] [Google Scholar]
- Klahre U, Chua NH. The Arabidopsisactin-related protein 2 (AtARP2) promoter directs expression in xylem precursor cells and pollen. Plant Mol Biol. 1999;41:65–73. doi: 10.1023/a:1006247600932. [DOI] [PubMed] [Google Scholar]
- Kreis T, Vale R. Guidebook to the Cytoskeletal and Motor Proteins. New York: Oxford University Press; 1999. Arp2/3 complex; pp. 45–52. [Google Scholar]
- Lawrence CJ, Morris NR, Meagher RB, Dawe RK. Dyneins have run their course in plant lineage. Traffic. 2001;2:362–363. doi: 10.1034/j.1600-0854.2001.25020508.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Kandasamy MK, Meagher RB. Rapid isolation of monoclonal antibodies: monitoring enzymes in the phytochelatin synthesis pathway. Plant Physiol. 2001;127:711–719. [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Huang S, McKinney EC, An Y-Q, Meagher RB. Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics. 1996;142:587–602. doi: 10.1093/genetics/142.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EC, Meagher RB. Members of the Arabidopsisactin gene family are widely dispersed in the genome. Genetics. 1998;149:663–675. doi: 10.1093/genetics/149.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB. The impact of historical contingency on gene phylogeny: plant actin diversity. In: Hecht M, MacIntyre R, Clegg M, editors. Evolutionary Biology. New York: Plenum Press; 1995. pp. 195–215. [Google Scholar]
- Meagher RB, Berry-Lowe S, Rice K. Molecular evolution of the small subunit of ribulose bisphosphate carboxylase: nucleotide substitution and gene conversion. Genetics. 1989;123:845–863. doi: 10.1093/genetics/123.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB, McKinney EC, Kandasamy MK. Isovariant dynamics expand and buffer the responses of complex systems: the diverse plant actin gene family. Plant Cell. 1999a;11:995–1006. doi: 10.1105/tpc.11.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB, McKinney EC, Kandasamy MK. The significance of diversity in the plant actin gene family: studies in Arabidopsis. In: Staiger CJ, Baluska F, Volkman D, Barlow P, editors. Actin: A Dynamic Framework for Multiple Plant Cell Functions. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 3–27. [Google Scholar]
- Meagher RB, McKinney EC, Vitale AV. The evolution of new structures: clues from plant cytoskeletal genes. Trends Genet. 1999b;15:278–284. doi: 10.1016/s0168-9525(99)01759-x. [DOI] [PubMed] [Google Scholar]
- Parthasarathy MV. F-actin architecture in coleoptile epidermal cells. Eur J Cell Biol. 1985;39:1–12. [Google Scholar]
- Parthasarathy MV, Perdue TD, Witzmun A, Alvernaz J. Actin network as a normal component of the cytoskeleton in many vascular plant cells. Am J Bot. 1985;72:1318–1323. [Google Scholar]
- Schafer DA, Schroer TA. Actin-related proteins. Annu Rev Cell Dev Biol. 1999;15:341–363. doi: 10.1146/annurev.cellbio.15.1.341. [DOI] [PubMed] [Google Scholar]
- Tam JP. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc Natl Acad Sci USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM, Li W-H. Rates of synonymous substitution in plant nuclear genes. J Mol Evol. 1989;29:208–211. [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]