Abstract
Arabidopsis contains 34 genes that are predicted to encode calcium-dependent protein kinases (CDPKs). CDPK enzymatic activity previously has been detected in many locations in plant cells, including the cytosol, the cytoskeleton, and the membrane fraction. However, little is known about the subcellular locations of individual CDPKs or the mechanisms involved in targeting them to those locations. We investigated the subcellular location of one Arabidopsis CDPK, AtCPK2, in detail. Membrane-associated AtCPK2 did not partition with the plasma membrane in a two-phase system. Sucrose gradient fractionation of microsomes demonstrated that AtCPK2 was associated with the endoplasmic reticulum (ER). AtCPK2 does not contain transmembrane domains or known ER-targeting signals, but does have predicted amino-terminal acylation sites. AtCPK2 was myristoylated in a cell-free extract and myristoylation was prevented by converting the glycine at the proposed site of myristate attachment to alanine (G2A). In plants, the G2A mutation decreased AtCPK2 membrane association by approximately 50%. A recombinant protein, consisting of the first 10 amino acids of AtCPK2 fused to the amino-terminus of β-glucuronidase, was also targeted to the ER, indicating that the amino terminus of AtCPK2 can specify ER localization of a soluble protein. These results indicate that AtCPK2 is localized to the ER, that myristoylation is likely to be involved in the membrane association of AtCPK2, and that the amino terminal region of AtCPK2 is sufficient for correct membrane targeting.
The predominant calcium-stimulated protein kinase activity in plant extracts is attributed to calcium-dependent protein kinases (CDPK), a group of enzymes identified only in plants and some protists. Calcium-stimulated kinase activity has been detected in both the soluble and microsomal fractions of plant cells. In many cases, CDPK-like activity was associated with the plasma membrane; for example, in oat (Avena sativa; Schaller et al., 1992), red beet (Beta vulgaris; Baizabal-Aguirre and de la Vara, 1997), zucchini (Cucurbita pepo; Verhey et al., 1993), and tobacco (Nicotiana tabacum; Iwata et al., 1998). In other studies, calcium-stimulated kinase activity was reported to be associated with plant microsomes, although the specific membrane was not determined (Battey, 1990; Klimczak and Hind, 1990; Abo-El-Saad and Wu, 1995; MacIntosh et al., 1996; Martin and Busconi, 2000). Thus, it is possible that CDPKs are associated with other cellular membranes in addition to the plasma membrane.
Whereas the roles of individual CDPKs have not yet been elucidated, it has been hypothesized that each CDPK isoform is functionally specialized. Sev eral lines of evidence support this hypothesis. First, three soybean (Glycine max) CDPKs have different susceptibilities to protein kinase inhibitors (Lee et al., 1998). Second, these soybean CDPKs differ in their calcium-binding properties (Lee et al., 1998). Third, Arabidopsis CDPKs have overlapping but distinct expression patterns (E. Hrabak, unpublished data). In addition, it is possible that CDPKs could be targeted to different subcellular locations, thereby enabling them to interact with different substrates.
All CDPK proteins contain three domains with well-characterized functions: the Ser/Thr kinase catalytic, autoregulatory, and calcium-binding domains (Harmon et al., 2000). The fourth amino-terminal variable domain is the most divergent region of these proteins, ranging in length from 20 to 200 amino acids and usually exhibiting little sequence similarity between different CDPK isoforms. The function of the variable domain is largely unknown but the majority of CDPK proteins contain a potential myristoylation site at the beginning of the variable domain (Harmon et al., 2000; Hrabak, 2000).
Myristate, a C14:0 fatty acid, can be covalently attached to the amino-terminal Gly residue of a protein when the Gly is found in the context of a short myristoylation consensus sequence (Towler et al., 1988). Many myristoylated proteins are membrane associated but they also can be soluble or alternate between membrane and cytosol (Johnson et al., 1994; Bhatnagar and Gordon, 1997). In addition to a role in mediating protein-lipid interactions, myristoylation can be important for protein-protein interactions or protein stability (Yonemoto et al., 1993; Kennedy et al., 1996; Herberg et al., 1997; Taniguchi, 1999).
Myristoylation, catalyzed by N-myristoyltransferase (NMT), has been intensively studied in fungal and animal cell systems (for review, see Johnson et al., 1994; Bhatnagar and Gordon, 1997). In contrast, until recently there were few examples of protein myristoylation in plants (Thompson and Okuyama, 2000). An Arabidopsis NMT gene has been cloned and shown to myristoylate amino-terminal peptides derived from a CDPK and from the Fen kinase (Qi et al., 2000). Ellard-Ivey et al. (1999) demonstrated in vitro myristoylation of a CDPK from zucchini and confirmed the requirement for an amino-terminal Gly residue. Rice (Oryza sativa) CDPK OsCPK2 was shown to be myristoylated in a heterologous maize (Zea mays) protoplast system and this acyl modification was critical for membrane binding (Martin and Busconi, 2000). The importance of myristoylation for correct protein function in plants was suggested by mutation of the putative myristoylation site in the tomato (Lycopersicon esculentum) Fen gene, which abolished its ability to confer sensitivity to the insecticide fenthion (Rommens et al., 1995). In addition, the myristoylation site of the Arabidopsis SOS3 protein was required for its role in salt tolerance (Ishitani et al., 2000).
Many proteins involved in signal transduction in eukaryotes are myristoylated, including the alpha subunits of heterotrimeric G proteins, members of the Src family of Tyr protein kinases, and the protein phosphatase calcineurin (Casey, 1995; Resh, 1996; Taniguchi, 1999; Thompson and Okuyama, 2000). Acyl groups, and the properties conferred by these hydrophobic modifications, are critical for the proper functioning of these proteins in signaling pathways. In plants, many signaling pathways are known to involve CDPKs, including the response to drought stress, the regulation of carbon and nitrogen metabolism, and the control of seed germination (for review, see Harmon et al., 2000; Hrabak, 2000). Because many CDPKs have predicted acylation sites, the subcellular localization of these enzymes and the role of myristoylation in membrane binding and protein function is of interest. We have focused on Arabidopsis CDPK isoform 2 (AtCPK2) which does not contain any significant transmembrane domains, signal sequences, or targeting signals when analyzed by the PSORT program for predicting protein localization (Nakai and Kanehisa, 1992). However, AtCPK2 does have a predicted amino-terminal myristoylation consensus sequence (Towler et al., 1988).
In this paper, we demonstrate that the AtCPK2 protein in plant cells is associated with the ER membrane. To our knowledge, this is the first example of a CDPK localized to the endoplasmic reticulum (ER). AtCPK2 can be myristoylated in vitro and mutation of the myristoylation site prevents addition of the fatty acid. Mutation of the myristoylation site also decreases membrane association of AtCPK2 in plants. The first 10 amino acids of AtCPK2 are shown to be sufficient to direct a soluble protein to the ER membrane, indicating that this region can be used for protein targeting.
RESULTS
AtCPK2 Genomic Clones
A clone containing the AtCPK2 genomic region was isolated from a genomic library by low-stringency hybridization and a 5.8-kb region of this clone was sequenced (see GenBank accession no. AF286222 for the complete sequence). The predicted open reading frame contained all of the characteristic features of a CDPK (Hrabak, 2000), including a calmodulin-like domain with four predicted calcium-binding EF hands. AtCPK2 also contains the largest amino-terminal variable domain (187 amino acids) of any CDPK characterized to date (Hrabak, 2000). The genomic sequence, rather than the cDNA, was chosen for these experiments because we have evidence that regions downstream of the promoter are important for full expression of AtCPK2 (E. Hrabak, unpublished data).
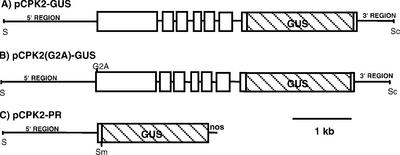
For some experiments where it was important to accurately quantitate levels of AtCPK2 protein or distinguish between the endogenous AtCPK2 and a modified transgenic protein, constructs were made in which the AtCPK2 gene was tagged with the β-glucuronidase (GUS) reporter gene. pCPK2-GUS contains an AtCPK2 genomic DNA fragment of 4.9 kb into which the GUS coding sequence was inserted in-frame at an introduced BamHI site (Fig. 1A). This plasmid contains 1.6 kb of DNA upstream of the translation start codon and 0.6 kb downstream of the translation stop codon. Although the precise transcription start site for the AtCPK2 gene is not known, this construct is predicted to contain the entire AtCPK2 promoter and 5′-untranslated region because there is less than 1.1 kb of intergenic DNA between the AtCPK2 translational start codon and the stop codon of the preceding gene, based on the completed Arabidopsis genome sequence. pCPK2-GUS should also contain sufficient DNA downstream of the stop codon to encompass the typical plant polyadenylation signals (Li and Hunt, 1997) because the 3′ region of this clone is 385 bp larger than the 3′ region of the largest cDNA clone identified. In the resulting 138-kD CPK2-GUS fusion protein, the GUS protein is fused to the AtCPK2 protein at a position ten amino acids from the carboxy terminus of AtCPK2.
Figure 1.
AtCPK2 constructs used for plant transformation. Lines represent introns or non-coding regions. White boxes are AtCPK2 coding regions. Striped boxes are the coding region for GUS. Most vector sequences are not shown. S, SalI; Sm, SmaI; Sc, SacI.
Membrane Association of AtCPK2 in Plants
To eliminate the possibility of detecting multiple CDPK isoforms simultaneously and to accurately quantitate levels of a single isoform, transgenic plants expressing the CPK2-GUS fusion protein were used in initial experiments to assess membrane localization. Two-week-old transgenic plants were homogenized, debris was removed by low-speed centrifugation, and the membranes were pelleted by ultracentrifugation. GUS enzyme activity was assayed in both the soluble and membrane fractions using a sensitive fluorimetric assay. Controls for these experiments included wild-type plants and transgenic plants expressing the GUS protein alone. No GUS activity was detected in extracts from wild-type plants. In plants expressing the GUS protein alone, 2% of the GUS activity was found in the membrane fraction (data not shown), which probably represents protein trapped in vesicles during homogenization or nonspecifically bound to membranes. In extracts from CPK2-GUS transgenic plants, 40% of the GUS activity was detected in the membrane fraction (Table I), providing evidence that some of the AtCPK2 protein in these plants is membrane associated.
Table I.
Membrane association of GUS-tagged AtCPK2 proteins in transgenic plants
| Protein Expressed in Transgenic Plants | No. of Independent Experiments | No. of Independent Transgenic Lines | GUS-Specific Activity in Membrane Fractiona | Membrane-Associated GUS Activityb |
|---|---|---|---|---|
| % | ||||
| CPK2-GUS | 7 | 3 | 67.1–154.2 | 40 ± 2.3 |
| CPK2(G2A)-GUS | 7 | 3 | 37.5–76.6 | 18 ± 1.7 |
| CPK2-PR | 4 | 2 | 5.2–8.3 | 46 ± 2 |
After low-speed centrifugation, plant extracts were ultracentrifuged to pellet microsomes. The supernatant after ultracentrifugation contains primarily soluble proteins, whereas the pellet contains primarily membrane-bound proteins. Both the supernatant and the pellet were assayed fluorimetrically for GUS enzymatic activity.
Specific activity units are nmol methylumbelliferone produced min−1 mg protein−1.
Values were calculated from the total enzyme activity data and are presented as mean ± sd.
Treatments to Dissociate AtCPK2 from Membranes
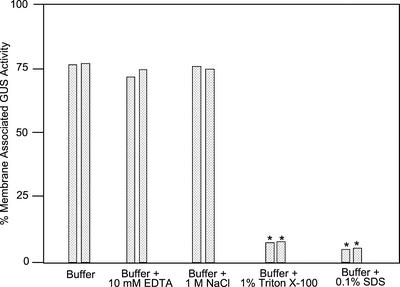
To investigate the interaction of AtCPK2 with membranes, isolated membranes from plants containing GUS-tagged AtCPK2 constructs were incubated in the presence of buffer alone or buffer containing a chelating agent (EDTA), high ionic strength (NaCl), a nonionic detergent (Triton X-100), or an ionic detergent (SDS). After 30 min, the samples were recentrifuged and the amount of GUS activity in the pellet and supernatant was determined with the GUS fluorimetric assay (Fig. 2). In transgenic plants expressing CPK2-GUS protein, approximately 75% of the membrane-associated GUS activity remained in the pellet after treatment of the membranes with buffer containing EDTA or NaCl, whereas the remainder of the CPK2-GUS protein was now found in the supernatant. These results were not significantly different from membranes treated with buffer alone (Fig. 2), indicating that AtCPK2 protein may exist in an equilibrium between soluble and membrane-bound states. Almost all of the GUS activity was released from the membranes of these transgenic plants by detergent treatment. Thus, treatments that disrupt ionic or electrostatic interactions were not effective at dissociating AtCPK2 from membranes, whereas treatments that disrupted most types of hydrophobic interactions efficiently solubilized AtCPK2. Because Triton X-100 was able to release AtCPK2 from membranes, AtCPK2 is unlikely to be associated with detergent-resistant membranes or lipid rafts (Moffett et al., 2000).
Figure 2.
AtCPK2 associated with the membrane fraction after various treatments. Microsomal membranes were isolated from transgenic plants expressing CPK2-GUS. Membrane pellets were homogenized in resuspension buffer alone or resuspension buffer containing EDTA, NaCl, Triton X-100, or SDS and incubated at 4°C for 30 min before repelleting. The resulting supernatant and pellet were assayed for GUS activity to assess the effect of the treatment on AtCPK2 membrane binding. Data shown are the percentage of GUS activity remaining in the pellet. Results from two independent experiments are shown. Asterisks indicate values that were significantly different from the buffer control (P ≤ 0.05)
AtCPK2 Is Associated with the ER Membrane
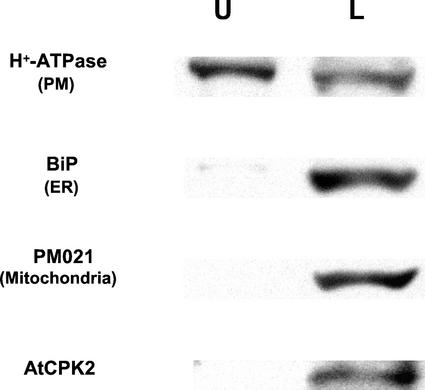
Because calcium-stimulated protein kinase activity previously has been detected in association with the plasma membrane fraction in plants (Schaller et al., 1992; Verhey et al., 1993; Baizabal-Aguirre and de la Vara, 1997; Iwata et al., 1998), we used aqueous two-phase partitioning to analyze membranes from wild-type plants (Fig. 3). This technique enriches for plasma membranes in the upper phase and other cellular membranes in the lower phase. Both phases were analyzed by immunoblotting to locate AtCPK2 protein, as well as markers for the ER, mitochondrial, vacuolar, and plasma membranes. Golgi membranes were assayed enzymatically for latent UDPase, whereas chloroplast membranes were detected by measuring chlorophyll concentration. The plasma membrane marker, as expected, was enriched in the upper phase, whereas ER and mitochondrial markers, as well as AtCPK2 protein, were located in the lower phase (Fig. 3). All other membrane markers were also found predominantly in the lower phase (data not shown). These results indicate that AtCPK2 is not associated with the plasma membrane.
Figure 3.
Two-phase separation of membranes from wild-type Columbia plants showing AtCPK2 accumulation in the lower phase. Equal proportions of the upper and lower phases were separated by SDS-PAGE and assayed by immunoblotting with antibodies specific for AtCPK2, H+-ATPase (plasma membrane marker), BiP (ER marker), and PM021 (mitochondrial membrane marker). U, Upper phase; L, lower phase.
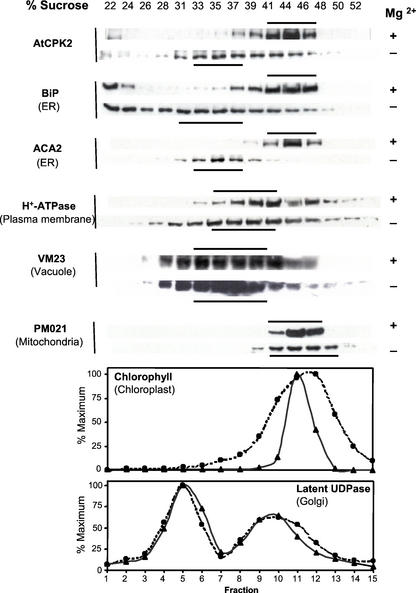
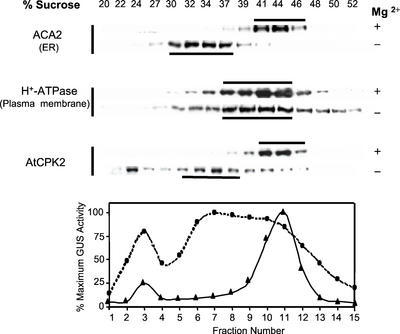
To localize AtCPK2 to a specific cellular membrane, Suc gradients were used to separate microsomes based on their buoyant density. Microsomes and Suc gradients were prepared in buffers containing EDTA alone (−Mg2+) or EDTA plus excess MgCl2 (+Mg2+). In the presence of Mg2+, ribosomes remain associated with the ER membranes that migrate at 40% to 46% (w/w) Suc. Chelation of Mg2+ by EDTA dissociates the ribosomes, shifting the ER membranes to a lower Suc concentration (Lord, 1987). Gradient fractions were analyzed as described for the phase partitioning experiments.
In the presence of Mg2+, AtCPK2 was detected in fractions containing 41% to 46% (w/w) Suc, similar to the ER, chloroplast, and mitochondrial markers. This sedimentation pattern could be easily distinguished from vacuolar, Golgi, and plasma membranes (Fig. 4), but did not permit the unambiguous localization of AtCPK2. In the absence of Mg2+, the ER markers BiP (a major ER-resident binding protein) and ACA2, as well as AtCPK2, shifted to 33% to 37% (w/w) Suc, consistent with a change in buoyant density of the ER after dissociation of ribosomes (Fig. 4). Although we consistently observed that the absence of Mg2+ broadened the sedimentation profiles for some of the membrane marker proteins, there was not a shift of the peak fractions to the extent observed for AtCPK2 or the ER membrane markers. Data from the Suc gradients are consistent with localization of membrane-associated AtCPK2 to the ER membrane.
Figure 4.
Suc gradient fractionation of membranes from wild-type Columbia plants showing colocalization of AtCPK2 with ER markers. Fractions from parallel gradients, with and without Mg2+, were separated by SDS-PAGE and assayed by immunoblotting with antibodies specific for AtCPK2, BiP and ACA2 (ER markers), H+-ATPase (plasma membrane marker), Vm23 (vacuolar membrane marker), or PM021 (mitochondrial membrane marker). Horizontal bars indicate the peak fractions. Graphs show chlorophyll absorbance (chloroplast marker) and enzyme assay data for latent UDPase (Golgi marker). ▴, +Mg2+ gradients; ●, −Mg2+ gradients. The fraction with the highest activity was assigned a value of 100%.
All of the membrane marker proteins detected in this study are integral membrane proteins with the exception of BiP, which is associated with the lumenal face of the ER but does not contain any membrane-spanning domains. In our experiments, some BiP protein is usually detected near the top of the gradients (22%–24% [w/w] Suc). This most likely represents BiP that dissociated during resuspension of the pelleted membranes before loading onto the gradients. It is probably not a result of proteolysis because BiP shows no apparent change in Mr in different gradient fractions. Some AtCPK2 is usually detected in these low-density gradient fractions also, consistent with our previous results that some AtCPK2 protein is solubilized during resuspension in buffer (Fig. 2). The other membrane protein markers, which represent integral membrane proteins, are not found in these low-density gradient fractions.
AtCPK2 Protein Tagged with GUS Localizes to the ER
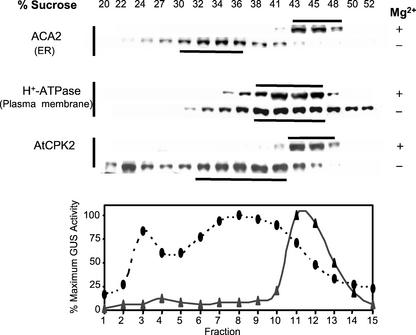
To determine whether a GUS tag would affect the localization of AtCPK2, Suc gradients were performed on membranes from transgenic plants expressing CPK2-GUS protein (Fig. 5). As expected, the ER marker protein ACA2 was detected at 41% to 46% (w/w) Suc in the presence of Mg2+ and shifted to lower buoyant density when the Mg2+ was chelated by EDTA. The location of the plasma membrane marker protein was similar in the presence and absence of Mg2+, as were all other membrane markers tested (data not shown). Sedimentation of the membranes containing the wild-type AtCPK2 protein, detected by immunoblotting, was most similar to that of the ER membrane markers. Distribution of GUS enzyme activity, representing the CPK2-GUS fusion protein, closely resembled the distribution of ACA2 and wild-type AtCPK2. These results indicate that the 600-amino acid GUS tag did not interfere with localization of the AtCPK2 protein to the ER membrane (Fig. 5). Because the CPK2-GUS fusion protein is localized in a manner comparable with the wild-type AtCPK2 protein, we conclude that transgenic plants expressing CPK2-GUS can be used interchangeably with wild-type plants to monitor the location of AtCPK2.
Figure 5.
CPK2-GUS, a full-length AtCPK2 protein tagged with GUS, is localized to the ER in transgenic Arabidopsis plants. Fractions from parallel Suc gradients, with and without Mg2+, were separated by SDS-PAGE and assayed by immunoblotting with antibodies specific for various membrane markers. AtCPK2-GUS fusion protein was assayed fluorimetrically. Horizontal bars indicate the peak fractions. ▴, +Mg2+ gradients; ●, −Mg2+ gradients. The fraction with the highest activity was assigned a value of 100% which corresponds to 188 nmol min−1 mL−1 for the +Mg2+ gradients and 76 nmol min−1 mL−1 for the −Mg2+ gradients. Total GUS activity loaded onto the gradient was 654 nmol min−1 for the +Mg2+ gradients and 734 nmol min−1 for the −Mg2+ gradients.
AtCPK2 Is Myristoylated in Vitro
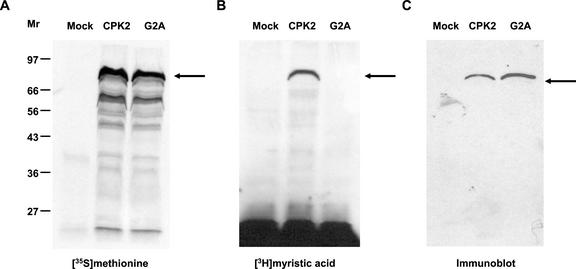
The AtCPK2 protein has no predicted transmembrane domains that would account for its observed membrane localization, but it does contain a predicted amino-terminal myristoylation sequence (MGNACVGPN). To determine if the AtCPK2 protein could be a substrate for plant NMT, plasmid pCPK2-ORF, which contains the AtCPK2 coding sequence downstream of the viral T7 promoter, was used in an in vitro myristoylation experiment. A coupled transcription-translation system from wheat germ was used to transcribe and translate the AtCPK2 cDNA sequence after addition of T7 RNA polymerase. Wheat germ extract has been shown to contain NMT activity (Heuckeroth et al., 1988; Ellard-Ivey et al., 1999). The reactions were performed in the presence of either [35S]Met for detection of total protein synthesis or [3H]myristate to detect myristoylated proteins. A prominent protein of approximately 80 kD was synthesized in the [35S]Met-containing reaction (Fig. 6A, lane 2). The identity of this protein was confirmed by immunoblot analysis with AtCPK2-specific antibody (Fig. 6C, lane 2). A protein of similar size was labeled in the [3H]myristate-containing reaction (Fig. 6B, lane 2). These results indicate that the AtCPK2 protein can be myristoylated in vitro.
Figure 6.
In vitro transcription and translation reactions demonstrate that AtCPK2 can be myristoylated, whereas a G2A mutation prevents myristoylation. Mock reactions contained no plasmid template, CPK2 reactions contained pCPK2-ORF, and G2A reactions contained pCPK2(G2A). A, Proteins synthesized in the presence of [35S]Met. B, Proteins synthesized in the presence of [3H]myristate. C, Proteins detected by immunoblotting with antiserum against the first 90 amino acids of AtCPK2 fused to GST.
No proteins recognized by AtCPK2 antibodies were detected by immunoblotting of mock reactions that contained no plasmid template (Fig. 6C, lane 1). Likewise, no radiolabeled proteins were synthesized when the plasmid template was omitted from the reaction mix (Fig. 6, A and B, lane 1). The smaller proteins routinely observed in [35S]Met-labeled reactions (Fig. 6A, lane 2) might be AtCPK2 degradation products or translation products from alternative start sites within the CPK2 mRNA. Because they are not myristoylated and are not detected by immunoblotting, these proteins probably represent amino-terminally truncated products produced from pCPK2-ORF.
A G2A Mutation Prevents Myristoylation of AtCPK2 in Vitro
To confirm that the amino-terminal Gly of AtCPK2 was the site of myristoylation, site-directed mutagenesis was used to convert the second position Gly of pCPK2-ORF to an Ala (G2A) to create the plasmid pCPK2-G2A. A coupled transcription-translation reaction was performed in the presence of either [35S]Met or [3H]myristate using pCPK2-G2A as the template. The G2A mutation did not affect the synthesis of the AtCPK2 protein (Fig. 6A, lane 3 and Fig. 6C, lane 3), but prevented the addition of [3H]myristate (Fig. 6B, lane 3), consistent with the Gly at position 2 of the native AtCPK2 protein being the site of myristoylation.
Effect of CPK2(G2A) Mutation in Plants
To investigate the effect of a G2A mutation in Arabidopsis, transgenic plants were created using the pCPK2(G2A)-GUS construct (Fig. 1B). pCPK2(G2A)- GUS is identical to pCPK2-GUS except for a single nucleotide change that converted the second codon from Gly to Ala. The amount of GUS activity in the membrane fraction from plants expressing CPK2(G2A)-GUS was reduced to 18%, compared with 40% in plants expressing CPK2-GUS (Table I). Suc gradients were used to determine the location of CPK2(G2A)-GUS protein in these plants. The distribution of membrane-bound GUS activity paralleled the distribution of the ER membrane markers (data not shown). Thus, although mutation of the myristoylation site decreased the proportion of AtCPK2 in the membrane fraction, the G2A mutation did not affect ER localization.
The First 10 Amino Acids of AtCPK2 Are Sufficient for ER Localization
To determine whether the amino terminus of AtCPK2 contains the ER-targeting information, we tested whether this region was able to direct a soluble protein to the ER membrane. Arabidopsis plants were stably transformed with DNA from plasmid pCPK2-PR containing 1.6 kb of AtCPK2 genomic DNA upstream of the translational start site followed by the first 30 bp of the AtCPK2 coding sequence in a translational fusion with the GUS gene and nos terminator (Fig. 1C). These plants expressed the GUS protein preceded by the first 10 amino acids of AtCPK2. This 10-amino acid region was chosen because it contains the myristoylation consensus sequence as defined by Towler et al. (1988). The proportion of GUS activity found in the membrane fraction from plants expressing the CPK2-PR protein was 46%, which is similar to the results from plants expressing CPK2-GUS, the full-length CPK2 protein tagged with GUS (Table I). The specific activity of the GUS enzyme detected in extracts from plants expressing CPK2-PR was consistently lower than in extracts from plants containing the full-length CPK2-GUS constructs (Table I). The lower activity may be because of missing regulatory sequences downstream of the promoter that are required for higher levels of expression or to differences between Arabidopsis ecotypes because CPK2-PR transgenic plants are in the RLD genetic background, whereas the CPK2-GUS plants are in the Columbia ecotype. Regardless of the reason for the lower expression levels in CPK2-PR plants, the results demonstrate that the first 10 amino acids of AtCPK2 were sufficient to allow direct membrane targeting of the normally soluble GUS protein.
Suc gradients were used to determine the location of CPK2-PR protein in microsomes from transgenic CPK2-PR-expressing plants. As observed previously for the wild-type AtCPK2 protein (Fig. 4) and for plants expressing CPK2-GUS (Fig. 5), the distribution of membrane-bound GUS activity was most similar to the location of the ER membrane marker (Fig. 7). Thus, a 10-amino acid region from the amino terminus of AtCPK2 was sufficient to direct localization of the GUS protein to the ER in a manner indistinguishable from the intact AtCPK2 protein.
Figure 7.
CPK2-PR, consisting of the GUS protein preceded by the first 10 amino acids of AtCPK2, is localized to the ER in transgenic Arabidopsis plants. Fractions from parallel Suc gradients, with and without Mg2+, were separated by SDS-PAGE and assayed by immunoblotting with antibodies specific for various membrane markers. AtCPK2-GUS fusion protein was assayed fluorimetrically. Horizontal bars indicate the peak fractions. ▴, +Mg2+ gradients; ●, −Mg2+gradients. The fraction with the highest activity was assigned a value of 100% that corresponds to 41.2 nmol min−1 mL−1 for the +Mg2+ gradients and 21.8 nmol min−1 mL−1 for the −Mg2+ gradients. Total GUS activity loaded onto the gradient was 146 nmol min−1 for the +Mg2+ gradients and 199 nmol min−1 for the −Mg2+ gradients.
DISCUSSION
CDPKs are known to be involved in many cellular processes such as pollen tube growth (Moutinho et al., 1998), mobilization of starch during seed germination (Ritchie and Gilroy, 1998), regulation of actin tension (Grabski et al., 1998), plant defense (Romeis et al., 2000), and responses to water stress (Shinozaki and Yamaguchi-Shinozaki, 1997). However, the details of how specific CDPKs function in plant cells are not well understood. It has been proposed that individual CDPKs may function in specific cell types, respond to different calcium concentrations, or be targeted to specific subcellular locations (Harmon et al., 2000; Hrabak, 2000).
Targeting of a protein kinase to a membrane can serve to increase the local concentration of the enzyme manyfold and to enhance the phosphorylation of substrate proteins found at that location, while limiting interaction with proteins in other parts of the cell. CDPK substrates include both soluble proteins, such as nitrate reductase (Douglas et al., 1998) and Suc phosphate synthase (Huber et al., 1996), and integral membrane proteins, such as the ER-localized calcium pump ACA2 (Hwang et al., 2000), a vacuolar chloride channel (Pei et al., 1996), and a proton pump and a potassium channel in the plasma membrane (Schaller and Sussman, 1988; Li et al., 1998; Lino et al., 1998). In addition, calcium-stimulated protein kinase activity has been detected in many subcellular locations, including the cytosol (Battey, 1990; Klimczak and Hind, 1990; Putnam-Evans et al., 1990; DasGupta, 1994; MacIntosh et al., 1996; Frylinck and Dubery, 1998), the nucleus (Li et al., 1991), the cytoskeleton (Putnam-Evans et al., 1989), and the plasma membrane (Schaller et al., 1992; Verhey et al., 1993; Baizabal-Aguirre and de la Vara, 1997; Iwata et al., 1998).
We investigated the membrane localization of AtCPK2 from Arabidopsis. Initial evidence that AtCPK2 was membrane associated was obtained using transgenic plants expressing AtCPK2 tagged with the GUS reporter protein. The sensitive fluorometric GUS assay enabled specific and quantitative detection of AtCPK2 in the presence of other members of the CDPK family. When plant extracts were separated into membrane and soluble fractions, about 40% of the GUS enzyme activity was detected in the insoluble fraction. A similar distribution has been reported for myristoylated Src kinase (Resh, 1989). Some AtCPK2 was removed from the membrane fraction by treatment with buffer but removal was not increased in the presence of EDTA or NaCl, indicating that ionic or electrostatic interactions are not essential for binding. Comparable results have been reported for the Src protein (Resh, 1989) and a CDPK from tobacco (Romeis et al., 2000), which suggests that these proteins may exist in an equilibrium between membrane-bound and soluble forms. The GUS enzyme can also be assayed histochemically. Microscopic examination of these plants revealed that the colored product of the histochemical assay accumulated slowly, an indication that AtCPK2 expression levels are low. Expression was limited to a few cell types during vegetative growth such as developing trichomes and cells in the elongation zone of the root (E. Hrabak, unpublished data). The restricted expression of AtCPK2 explains why the native AtCPK2 protein could only be detected on western blots using the most sensitive chemiluminescent reagents currently available. The fluorimetric GUS assay was similar in sensitivity to chemiluminescence and provided a separate means to confirm localization studies.
To determine the specific membrane(s) to which AtCPK2 was bound, the membrane fraction of wild-type and transgenic plants was further analyzed by aqueous two-phase partitioning and Suc density gradients. The phase partitioning experiments indicated that AtCPK2 was not associated with the plasma membrane, but was enriched in the intracellular membranes. Suc gradient fractionation in buffer containing Mg2+ was able to narrow the location of AtCPK2 to several membrane types with similar buoyant densities. Incorporation of EDTA into the buffers used to prepare plant microsomes and Suc gradients allowed us to distinguish ER membranes from other membranes based on a characteristic buoyant density shift and demonstrated that the distribution of AtCPK2 most closely paralleled the distribution of the ER membrane markers.
Whereas many acylated proteins are associated with the plasma membrane in yeast or animal cells, few myristoylated proteins have been localized to the ER. Because the ER lumen is a site of calcium sequestration (Malho et al., 1998; Trewavas, 1999), localized calcium release could activate CDPKs located on or near the ER. Whereas the specific substrate(s) of AtCPK2 have not yet been identified, the ER-localized calcium pump ACA2 has been shown to be an in vitro substrate for a closely related CDPK, AtCPK1 (Hong et al., 1999; Hwang et al., 2000). It is tempting to speculate that AtCPK2 might be one of the kinases that phosphorylates ACA2 in plants, although it is not yet known if these two proteins are expressed in the same cell types.
Although most CDPKs contain one slightly hydrophobic region in the kinase domain, there is no evidence that CDPKs are integral membrane proteins. One mechanism that can allow membrane binding of peripheral membrane proteins is acquisition of a hydrophobic domain, like prenyl, myristate, or palmitate groups or glycosylphosphatidylinositol anchors (Casey, 1995). Whereas CDPKs do not contain prenylation or glycosylphosphatidylinositol anchor motifs, the majority of CDPKs, including AtCPK2, contain potential amino-terminal myristoylation and palmitoylation sites (Hrabak, 2000).
An NMT gene has recently been cloned and characterized from Arabidopsis (Qi et al., 2000) and wheat germ extract contains NMT activity (Heuckeroth et al., 1988), providing evidence that plants contain the enzyme needed to perform the myristoylation reaction. We demonstrated that AtCPK2 could be myristoylated in vitro using a cell-free wheat germ extract to transcribe and translate the AtCPK2 protein in the presence of radiolabeled myristic acid. Whereas these results are not definitive proof that AtCPK2 is myristoylated in vivo, they indicate that AtCPK2 is a substrate for plant NMT. This approach has been used previously to demonstrate that CDPKs from zucchini (Ellard-Ivey et al., 1999) and from rice (Martin and Busconi, 2000) were able to be myristoylated. An AtCPK2 protein containing a G2A mutation could not be myristoylated, indicating that the amino-terminal Gly residue was the site of myristate attachment.
Based on the in vitro myristoylation assays, we predict that the CPK2(G2A)-GUS protein would not be myristoylated in plants. This prediction is supported by experiments with transgenic plants expressing CPK2(G2A)-GUS in which membrane-bound GUS activity was reduced from 40% to 18% by the G2A mutation. This value is above the 2% background level for membrane binding of the GUS protein alone, indicating that a significant portion of AtCPK2 is still membrane associated in the G2A mutant. There are at least two potential explanations for these results. First, all Arabidopsis CDPKs that contain a myristoylation consensus sequence also contain at least one nearby Cys residue that may serve as a palmitoylation site (Hrabak, 2000). Palmitate is more hydrophobic than myristate and binds more tightly to lipid bilayers (Shahinian and Silvius, 1995; Bhatnagar and Gordon, 1997) and therefore most palmitoylated proteins are membrane bound (Resh, 1996). Modification by both myristate and palmitate has been demonstrated for many acylated proteins, including heterotrimeric G protein alpha subunits and Src family proteins (Milligan et al., 1995; Resh, 1996), and recently a rice CDPK, OsCPK2, was shown to be both myristoylated and palmitoylated in a maize protoplast expression system (Martin and Busconi, 2000). Because preventing myristoylation usually decreases or inhibits palmitoylation (Galbiati et al., 1994; Hallak et al., 1994; Wilson and Bourne, 1995; Morales et al., 1998), the decreased membrane binding of the G2A mutant may reflect incomplete palmitoylation. The second potential explanation is that ER targeting of AtCPK2 might be mediated by interaction with specific receptor(s) in or associated with the ER membrane that recognize the AtCPK2 amino terminus. In this case, the hydrophobicity of the myristoyl group might function to facilitate initial membrane binding leading to association with the receptor. This scenario is attractive because it would help to explain how AtCPK2 is targeted specifically to the ER, rather than to many of the other membrane types in the cell as might be expected if the binding was strictly a result of hydrophobic interactions. Thus, the lower membrane binding observed for the G2A mutant could be because of a decreased tendency of the non-myristoylated protein to be close to membranes.
Because the myristoylation site of AtCPK2 is located at the amino terminus of the protein, we investigated whether the first 10 amino acids of AtCPK2 were sufficient to direct the soluble GUS protein to the ER membrane. Several other proteins whose amino termini are sufficient for specific membrane binding have been described including poliovirus VP4 (Martin-Belmonte et al., 2000), CAP23/NAP22 (Takasaki et al., 1999), p59fyn (Gauen et al., 1992), and p60src (Resh and Ling, 1990). In addition, acylated amino termini can target green fluorescent protein to specific subcellular locations (McCabe and Berthiaume, 1999). In our experiments, the percentage of total GUS activity in extracts of transgenic plants that was associated with the membrane fraction was similar in plants expressing either the full-length AtCPK2 protein tagged with GUS (CPK2-GUS) or the GUS protein preceded by the first 10 amino acids of AtCPK2 (CPK2-PR). These results demonstrated that the first 10 amino acids of AtCPK2 retained the membrane targeting ability of the intact protein and indicated that this region may be useful for targeting other proteins to the ER.
In this report, we have demonstrated that Arabidopsis CDPK isoform AtCPK2 is associated with the ER membrane in plants. AtCPK2 is myristoylated in vitro and a G2A mutation prevented myristoylation and decreased membrane binding in plants. These findings support the hypothesis that myristoylation contributes to membrane association of AtCPK2, but is not the only factor involved. The region of AtCPK2 necessary for ER targeting is located within the first 10 amino acids of the protein. An understanding of the role of acylation in the proper functioning of this kinase awaits more information about the function of AtCPK2 in plants. However, the identification of a CDPK associated with the ER membrane presents many directions for future research. It will be of interest to determine whether AtCPK2 binds to or phosphorylates substrates on the ER, to understand the function of the membrane-bound and soluble forms of AtCPK2, and to delineate the specific residue(s) at the amino terminus of AtCPK2 that are required for ER targeting.
MATERIALS AND METHODS
DNA cloning was done in Escherichia coli strain DH5α (Life Technologies, Rockville, MD). Standard molecular cloning techniques were used throughout, according to Sambrook et al. (1989). GUS activity was determined with a fluorimetric assay as previously described (Gallagher, 1992). Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
AtCPK2 cDNA Clones
To identify an AtCPK2 cDNA clone, a size-fractionated Arabidopsis ecotype Columbia cDNA library (Schindler et al., 1992) was screened by hybridization with an AtCPK2 genomic DNA fragment as the probe. Three 1.6-kb clones (pE1-5, pE17-3, and pE17-4) were isolated but all of them were missing approximately 400 bp at the beginning of the open reading frame, as predicted from the genomic sequence. A 1.4-kb region of cDNA beginning at the translation initiation codon was amplified by PCR using the cDNA library as template, an upstream primer 5′GGATCCATGGGTAATGCT containing an introduced BamHI site, and the downstream primer 5′GGTTAGTCTTCG. The amplified 5′ region and the truncated cDNA clone pE1-5 overlapped in a region containing a unique BglII restriction site, which was used to ligate both segments of the AtCPK2 coding sequence to create the plasmid pCPK2-ORF containing the entire AtCPK2 coding sequence. The GenBank accession number for the AtCPK2 cDNA sequence is U31833.
An AtCPK2 cDNA clone with a G2A mutation was made by PCR with pCPK2-ORF as the template, a forward primer with a single base change (bold font) to convert the Gly at codon 2 to an Ala (5′ AGTGGATCCATGGCTAATGCTTGCGT), and a reverse primer in the pBluescript vector. The PCR product was recloned into pBluescript to give pCPK2-G2A and sequenced to confirm the G2A mutation.
AtCPK2 Genomic Clones
An AtCPK2 genomic clone was isolated by hybridization of a cosmid library of Arabidopsis ecotype Columbia genomic DNA in the vector pOCA18 (Olszewski et al., 1988) under low stringency conditions with a radiolabeled DNA probe corresponding to a portion of the closely related AtCPK1 gene (Harper et al., 1993). An 11-kb XbaI fragment containing the AtCPK2 genomic region was subcloned from one of the hybridizing cosmids to pBluescript (Stratagene, La Jolla, CA) and designated pgAK19. A 5.6-kb region of pgAK19 was sequenced on both strands using either Sequenase 2.0 (U.S. Biochemicals, Cleveland) for manual sequencing or Dye-deoxy Terminators (Perkin-Elmer Applied Biosystems, Foster City, CA) for automated sequencing.
To facilitate detection of the AtCPK2 protein in wildtype plant extract, the AtCPK2 genomic sequence (including 1.6 kb upstream of the translational start site) was tagged near the end of the coding sequence with the 1.8-kb GUS (uidA) gene. A 2-kb EcoRI fragment from the 3′ end of pgAK19 was subcloned to pALTER-1 (Promega Corp., Madison, WI). Site-directed mutagenesis was performed according to the manufacturer's instructions using the oligonucleotide 5′ GGGAGGACCTCTGAAGATGGATCCAGAGAACAGCATTAGCATTTCTC in which bold type indicates nucleotide changes from the original sequence. This procedure introduced a BamHI restriction site 30 bp upstream of the AtCPK2 stop codon. The 2-kb EcoRI fragment was sequenced to confirm that no unintended errors had been introduced by the mutagenesis procedure. To eliminate the unsequenced upstream and downstream regions from pgAK19, a multistep process was used to construct a clone that contained most of the 5.6-kb sequenced region. pCPK2-PR (described below) was digested with MluI and XbaI and ligated to a 6-kb MluI-XbaI fragment from pgAK19 to yield pPGS. pPGS contains the 5.6-kb sequenced region as well as 1.3 kb of downstream DNA. The 2-kb EcoRI fragment in pPGS was replaced with the EcoRI fragment containing the introduced BamHI site to yield pPGS-M and the fragment was confirmed to be in the correct orientation by restriction enzyme digestion. pPGS-M was digested with SalI and KpnI and the 4.9-kb fragment containing 1.6 kb of AtCPK2 sequence upstream of the start codon, the entire coding sequence including introns, and 0.6 kb of AtCPK2 sequence downstream from the stop codon was cloned into pUC18 that had been digested with SalI and KpnI. A GUS cassette (DeWitt et al., 1996) with BamHI ends was ligated into the introduced BamHI site, which fused the GUS sequence in frame with the AtCPK2 reading frame, to yield pGMG-GUS. Orientation and correct fusion of the cassettes were confirmed by sequencing through the fusion junctions. Finally, the tagged AtCPK2 genomic construct was subcloned into the vector pBIN19 as a SalI-SacI fragment to yield pCPK2-GUS.
To create an AtCPK2 genomic clone with Ala (GCT) instead of Gly (GGT) as the second codon (G2A mutation), site-directed mutagenesis was performed with the QuikChange kit (Stratagene) according to manufacturer's instructions using pGMG-GUS as the template DNA. The mutated AtCPK2 gene was subcloned into the vector pBIN19 as described above and named pCPK2(G2A)-GUS.
To fuse the promoter and first 10 codons of AtCPK2 to the GUS reporter gene, PCR was used to amplify a 1.6-kb region upstream of the AtCPK2 genomic sequence. A SalI site was added at the 5′ end of the upstream primer 5′ ACTGTCGACTTATATGTCTTCATATCTCT and a SmaI site was added to the downstream primer at a position immediately after the 10th full codon of the coding sequence 5′ CCACCCGGGAAATGTGGTGTCCAACGCA. Products from the PCR reaction were cloned into pBluescript to yield pPRM and sequenced to confirm that no PCR errors had occurred. The SalI-SmaI fragment was then cloned upstream of the GUS reporter gene in pBI101.2 (CLONTECH, Palo Alto, CA) to produce pCPK2-PR. The correct reading frame across the translational fusion junction was confirmed by DNA sequencing.
Production of Transgenic Plants and Plant Culture
Arabidopsis ecotype RLD roots were transformed as described previously (Valvekens et al., 1988), followed by plant regeneration. Arabidopsis ecotype Columbia plants were transformed using a vacuum infiltration procedure (Bent and Clough, 1998). Transformed plants were confirmed to contain the correct transgene using a rapid PCR method (Klimyuk et al., 1993).
Plants for membrane isolation were grown from surface-sterilized seeds in liquid Murashige and Skoog basal medium (Sigma, St. Louis), pH 5.7, containing Gamborg's B-5 vitamins and 1% (w/v) Suc at 100 rpm, 22°C, and an 18-h-light/dark cycle.
AtCPK2-Specific Antibodies
AtCPK2 rabbit polyclonal antibody was made against a purified fusion protein consisting of the first 90 amino acids of the 185 amino acid AtCPK2 variable domain fused to glutathione S-transferase in vector pGEX-KT (Hakes and Dixon, 1992). Recombinant protein expressed in E. coli was purified on a glutathione-agarose matrix (Pharmacia, Piscataway, NJ) and used for immunization of New Zealand white female rabbits. The antibody did not cross-react with AtCPK1 protein, the CDPK isoform most closely related to AtCPK2, or with AtCPK4 or AtCPK5 protein.
Membrane Isolation
Cellular membranes were prepared as previously described with minor modifications (Schaller and DeWitt, 1995). All procedures were conducted at 4°C. Two-week-old, liquid-grown Arabidopsis plants were homogenized in a mortar and pestle in 1 to 2 mL of homogenization buffer (50 mm Tris-HCl, pH 8.2; 20% [v/v] glycerol; 1 mm phenylmethylsulfonyl fluoride; 1 mm dithiothreitol; 10 μg mL−1 leupeptin; 1 μg mL−1 pepstatin; and 10 μg mL−1 aprotinin) per gram of tissue. Homogenates were filtered through Miracloth and centrifuged at 5,000g for 5 min. The supernatant was centrifuged at 125,000g for 30 min to pellet microsomes. The remaining supernatant contains primarily soluble proteins.
Membrane-Binding Assays
To investigate the membrane binding affinity of AtCPK2, microsomal membranes were resuspended at 0.5 mg mL−1 in resuspension buffer (25 mm Tris-HCl, pH 7.5; 10% [w/v] Suc; and protease inhibitors as described above) alone or in resuspension buffer containing one of the following: 10 mm EDTA, 1 m NaCl, 1% (v/v) Triton X-100, or 0.1% (w/v) SDS. After incubation at 4°C for 30 min, samples were re-centrifuged at 125,000g at 4°C for 30 min to pellet membrane vesicles. The supernatants were saved and the pellets were resuspended in resuspension buffer. Results were analyzed via one-way ANOVA in Systat 9.0. Treatment means were compared with the control via Dunnett's test.
Two-Phase Separation
An aqueous two-phase system (Larsson, 1983) was used to separate plasma membrane from intracellular membranes. Total membranes, prepared as described above, were resuspended in 200 μL of SPK buffer (0.33 m Suc, 5 mm KPO4, and 3 mm KCl, pH 7.8) and added to a 4-g phase system prepared in the same buffer. The final composition of the phase system was 6.3% (w/w) dextran (Mr = 413,000) and 6.3% (w/w) polyethylene glycol (Mr = 3,350). After thorough mixing by inverting the tube 20 to 30 times, the phases were separated by centrifugation at 1,400g for 5 min. The upper phase (enriched for plasma membrane) was removed to a clean tube and repartitioned twice with lower phase. Likewise, the lower phase, containing primarily intracellular membranes, was repartitioned twice with upper phase. Fresh upper and lower phase were obtained from a bulk-phase system of identical composition prepared separately. The final upper and lower phases were diluted with buffer containing 10 mm Tris-HCl (pH 7.0), 1 mm EGTA, and 1 mm EDTA and centrifuged at 125,000g for 30 min. Pellets were resuspended in equal volumes of SPK buffer. After separation by SDS-PAGE, proteins were transferred to polyvinylidene fluoride (PVDF) membranes and analyzed by immunoblotting as described below.
Suc Gradient Fractionation
Membrane pellets were resuspended in 1 mL of resuspension buffer per 10 g wet weight of starting material using a ground glass homogenizer. Resuspended membranes were layered onto linear Suc gradients (20% to 50% [w/w]) prepared in centrifugation buffer (10 mm Tris-HCl, pH 7.6, and protease inhibitors). For density gradients performed in the presence of Mg2+, 2 mm EDTA and 5 mm MgCl2 were added to homogenization, resuspension, and centrifugation buffers. For density gradients performed in the absence of Mg2+, 5 mm EDTA was added to homogenization and resuspension buffers, whereas 2 mm EDTA was used in centrifugation buffer. After centrifugation in a swinging bucket rotor at 125,000g for 16 h at 4°C, 1-mL fractions were collected.
The Suc concentration of each fraction was measured with a refractometer (Fisher Scientific, Pittsburgh). Protein concentrations were determined according to the method of Lowry as described previously (Schaller and DeWitt, 1995). Chlorophyll, a marker for chloroplast thylakoid membranes, was measured spectrophotometrically (Schaller and DeWitt, 1995). Latent UDPase, a marker for Golgi membranes, was assayed as described by Schaller and DeWitt (1995) using the Malachite Green method to detect released phosphate. All other membrane markers were detected by immunoblot analysis. After separation of proteins on 10% (w/v) SDS-polyacrylamide gels (Sambrook et al., 1989), proteins were electrophoretically transferred to PVDF membrane (Millipore, Bedford, MA) and the membranes were blocked overnight at 4°C in TBS (20 mm Tris-HCl, pH 7.6, and 137 mm NaCl) containing 2% (w/v) nonfat dry milk. The PVDF membranes were then treated sequentially with primary antibodies in TBST (TBS containing 0.05% [v/v] Tween 20) containing 2% (w/v) nonfat dry milk for 1 h, TBST for 5 min (3 times), secondary antibodies in TBST containing 2% (w/v) nonfat dry milk for 45 min, TBST for 5 min (three times), and TBS for 5 min. Immunodecorated proteins were detected with SuperSignal chemiluminescent substrate (Pierce, Rockford, IL) on x-ray film. Quantitation of immunodetected proteins was done using the NIH Image program. Membranes were stripped between detections following manufacturer's instructions. Antisera were used at the following dilutions: plasma membrane H+-ATPase (DeWitt and Sussman, 1995), 1:10,000 (v/v); ER lumenal binding protein BiP (gift of Maarten Chrispeels, University of California, San Diego), 1:1,000 (v/v); ER integral membrane protein ACA2 (Arabidopsis calcium ATPase; Harper et al., 1998), 1:2,000 (v/v); mitochondrial membrane protein β-ATPase D (Luethy et al., 1993), 1:100 (v/v); vacuolar membrane protein VM23 (Maeshima, 1992), 1:3,000 (v/v); and AtCPK2 variable domain antibody (described above), 1:10,000 (v/v).
In Vitro Myristoylation Assays
A cell-free wheat germ extract system was used to transcribe and translate the AtCPK2 gene in the presence of either radiolabeled methionine to assess total protein synthesis or radiolabeled myristate to detect myristoylated proteins. One microgram of plasmid, linearized with EcoRI, was used as the template for transcription by T7 RNA polymerase in the TNT Coupled Transcription-Translation Wheat Germ Extract System (Promega Corp.). Control reactions contained no plasmid. Reactions were prepared according to the manufacturer's instructions in the presence of either 50 μCi of [9,10-3H]myristic acid (54 Ci mmol−1; Amersham, Piscataway, NJ) or 10 μCi of l-[35S]Met (1,000 Ci mmol−1; Amersham). Immediately before beginning the 1.5-h reaction, the [3H]myristic acid was dried under nitrogen and resuspended by vortexing in DEPC-treated water at a concentration of 10 μCi μL−1. Reaction products were separated on 10% (w/v) SDS-polyacrylamide gels (Sambrook et al., 1989), stained with Coomassie Blue, and then treated with Entensify autoradiography enhancer (New England Nuclear, Boston) before detection on x-ray film.
ACKNOWLEDGMENTS
We thank Eric Schaller, Dennis Mathews, Xiang Qu, and Subhash Minocha for critical reading of the manuscript, Chris Neefus for statistical analyses, Neil Olszewski for the genomic library, Joe Kieber and Joe Ecker for the cDNA library, Maarten Chrispeels, Tom Elthon, and Masayoshi Maeshima for antibodies, Jeff Harper for antibodies and for cloning pgAK19, and Michael Sussman for financial support from the U.S. Department of Agriculture during initial stages of this work.
Footnotes
This work was supported by the U.S. Department of Agriculture-National Research Initiative (grant no. 9801263 to E.M.H.). This is paper no. 2,073 of the New Hampshire Agriculture Experiment Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010770.
LITERATURE CITED
- Abo-El-Saad M, Wu R. A rice membrane calcium-dependent protein kinase is induced by gibberellin. Plant Physiol. 1995;108:787–793. doi: 10.1104/pp.108.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baizabal-Aguirre VM, de la Vara LEG. Purification and characterization of a calcium-regulated protein kinase from beet root (Beta vulgaris) plasma membranes. Physiol Plant. 1997;99:135–143. [Google Scholar]
- Battey NH. Calcium-activated protein kinase from soluble and membrane fractions of maize coleoptiles. Biochem Biophys Res Commun. 1990;170:17–22. doi: 10.1016/0006-291x(90)91234-j. [DOI] [PubMed] [Google Scholar]
- Bent AF, Clough SJ. Agrobacterium germ-line transformation: transformation of Arabidopsis without tissue culture. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual, Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 1–14. [Google Scholar]
- Bhatnagar RS, Gordon JI. Understanding covalent modifications of proteins by lipids: where cell biology and biophysics mingle. Trends Cell Biol. 1997;7:14–20. doi: 10.1016/S0962-8924(97)10044-7. [DOI] [PubMed] [Google Scholar]
- Casey PJ. Protein lipidation in cell signaling. Science. 1995;268:221–225. doi: 10.1126/science.7716512. [DOI] [PubMed] [Google Scholar]
- DasGupta M. Characterization of a calcium-dependent protein kinase from Arachis hypogea (groundnut) seeds. Plant Physiol. 1994;104:961–969. doi: 10.1104/pp.104.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, Hong B, Sussman MR, Harper JF. Targeting of two Arabidopsis H+-ATPase isoforms to the plasma membrane. Plant Physiol. 1996;112:833–844. doi: 10.1104/pp.112.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, Sussman MR. Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell. 1995;7:2053–2067. doi: 10.1105/tpc.7.12.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P, Moorhead G, Hong Y, Morice N, MacKintosh C. Purification of a nitrate reductase kinase from Spinacia oleracea leaves, and its identification as a calmodulin-domain protein kinase. Planta. 1998;206:435–442. doi: 10.1007/s004250050419. [DOI] [PubMed] [Google Scholar]
- Ellard-Ivey M, Hopkins RB, White T, Lomax TL. Cloning, expression and N-terminal myristoylation of CpCPK1, a calcium-dependent protein kinase from zucchini (Cucurbita pepo L.) Plant Mol Biol. 1999;39:199–208. doi: 10.1023/a:1006125918023. [DOI] [PubMed] [Google Scholar]
- Frylinck L, Dubery IA. Protein kinase activities in ripening mango, Mangifera indica L., fruit tissue: III. Purification and characterization of a calcium-regulated protein kinase. Biochim Biophys Acta. 1998;1387:342–354. doi: 10.1016/s0167-4838(98)00149-6. [DOI] [PubMed] [Google Scholar]
- Galbiati F, Guzzi F, Magee AI, Milligan G, Parenti M. N-terminal fatty acylation of the α-subunit of the G-protein Gi1: Only the myristoylated protein is a substrate for palmitoylation. Biochem J. 1994;303:697–700. doi: 10.1042/bj3030697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. Quantitation of GUS activity by fluorometry. In: Gallagher SR, editor. GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression. New York: Academic Press; 1992. pp. 47–59. [Google Scholar]
- Gauen LKT, Kong A-NT, Samelson LE, Shaw AS. p59fyn tyrosine kinase associates with multiple T-cell receptor subunits through its unique amino-terminal domain. Mol Cell Biol. 1992;12:5438–5446. doi: 10.1128/mcb.12.12.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabski S, Arnoys E, Busch B, Schindler M. Regulation of actin tension in plant cells by kinases and phosphatases. Plant Physiol. 1998;116:279–290. [Google Scholar]
- Hakes DJ, Dixon JE. New vectors for high level expression of recombinant proteins in bacteria. Anal Biochem. 1992;202:293–298. doi: 10.1016/0003-2697(92)90108-j. [DOI] [PubMed] [Google Scholar]
- Hallak H, Brass LF, Manning DR. Failure to myristoylate the α subunit of Gz is correlated with inhibition of palmitoylation and membrane attachment, but has no affect on phosphorylation of protein kinase C. J Biol Chem. 1994;269:4571–4576. [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF. CDPKs: a kinase for every Ca2+ signal? Trends Plant Sci. 2000;5:154–159. doi: 10.1016/s1360-1385(00)01577-6. [DOI] [PubMed] [Google Scholar]
- Harper JF, Binder BM, Sussman MR. Calcium and lipid regulation of an Arabidopsis protein kinase expressed in Escherichia coli. Biochemistry. 1993;32:3282–3290. doi: 10.1021/bi00064a010. [DOI] [PubMed] [Google Scholar]
- Harper JF, Hong B, Hwang I, Guo HQ, Stoddard R, Huang JF, Palmgren MG, Sze H. A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J Biol Chem. 1998;273:1099–1106. doi: 10.1074/jbc.273.2.1099. [DOI] [PubMed] [Google Scholar]
- Herberg FW, Zimmermann B, McGlone M, Taylor SS. Importance of the A-helix of the catalytic subunit of cAMP-dependent protein kinase for stability and for orienting subdomains at the cleft interface. Protein Sci. 1997;6:569–579. doi: 10.1002/pro.5560060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuckeroth RO, Towler DA, Adams SP, Glaser L, Gordon JI. 11-(Ethylthio) undecanoic acid: a myristic acid analogue of altered hydrophobicity which is functional for peptide N-myristoylation with wheat germ and yeast acyltransferase. J Biol Chem. 1988;263:2127–2133. [PubMed] [Google Scholar]
- Hong B, Ichida A, Yuwen Wang, Gens JS, Pickard BG, Harper JF. Identification of a calmodulin-regulated Ca2+-ATPase in the endoplasmic reticulum. Plant Physiol. 1999;119:1165–1175. doi: 10.1104/pp.119.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM. Calcium-dependent protein kinases and their relatives. Adv Bot Res. 2000;32:185–223. [Google Scholar]
- Huber SC, Huber JL, Liao P-C, Gage DA, Robert W, McMichael J, Chourey PS, Hannah LC, Koch K. Phosphorylation of serine-15 of maize leaf sucrose synthase: occurence in vivo and possible regulatory significance. Plant Physiol. 1996;112:793–802. doi: 10.1104/pp.112.2.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sze H, Harper JF. A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc Natl Acad Sci USA. 2000;97:6224–6229. doi: 10.1073/pnas.97.11.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim C-S, Shi W, Zhu J-K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 2000;12:1667–1677. doi: 10.1105/tpc.12.9.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Kuriyama M, Nakakita M, Kojima H, Ohto M, Nakamura K. Characterization of a calcium-dependent protein kinase of tobacco leaves that is associated with the plasma membrane and is inducible by sucrose. Plant Cell Physiol. 1998;39:1176–1183. doi: 10.1093/oxfordjournals.pcp.a029318. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Bhatnagar RS, Knoll LJ, Gordon JI. Genetic and biochemical studies of protein N-myristoylation. Annu Rev Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- Kennedy MT, Brockman H, Rusnak F. Contributions of myristoylation to calcineurin structure/function. J Biol Chem. 1996;271:26517–26521. doi: 10.1074/jbc.271.43.26517. [DOI] [PubMed] [Google Scholar]
- Klimczak LJ, Hind G. Biochemical similarities between soluble and membrane-bound calcium-dependent protein kinases of barley. Plant Physiol. 1990;92:919–923. doi: 10.1104/pp.92.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JDG. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 1993;3:493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Larsson C. Partition in aqueous polymer two-phase systems: a rapid method for separation of membrane particles according to their surface properties. In: Hall JL, Moore AL, editors. Isolation of Membranes and Organelles from Plant Cells. London: Academic Press; 1983. pp. 277–309. [Google Scholar]
- Lee J-Y, Yoo B-C, Harmon AC. Kinetic and calcium-binding properties of three calcium-dependent protein kinase isoenzymes from soybean. Biochem. 1998;37:6801–6809. doi: 10.1021/bi980062q. [DOI] [PubMed] [Google Scholar]
- Li H, Dauwalder M, Roux SJ. Partial purification and characterization of a Ca2+-dependent protein kinase from pea nuclei. Plant Physiol. 1991;96:720–727. doi: 10.1104/pp.96.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee Y-RJ, Assmann SM. Guard cells possess a calcium-dependent protein kinase that phosphorylates the KAT1 potassium channel. Plant Physiol. 1998;116:785–795. doi: 10.1104/pp.116.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Hunt AG. The polyadenylation of RNA in plants. Plant Physiol. 1997;115:321–325. doi: 10.1104/pp.115.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino B, Baizabal-Aguirre VM, de la Vara LEG. The plasma-membrane H+-ATPase from beet root is inhibited by a calcium-dependent phosphorylation. Planta. 1998;204:352–359. doi: 10.1007/s004250050266. [DOI] [PubMed] [Google Scholar]
- Lord JM. Isolation of endoplasmic reticulum: general principles, enzymatic markers, and endoplasmic reticulum-bound polysomes. Methods Enzymol. 1987;148:542–558. [Google Scholar]
- Luethy MH, Horak A, Elthon TE. Monoclonal antibodies to the α- and β-subunits of the plant mitochondrial F1-ATPase. Plant Physiol. 1993;101:931–937. doi: 10.1104/pp.101.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC, Ulloa RM, Raices M, Tellez-Inon MT. Changes in calcium-dependent protein kinase activity during in vitro tuberization in potato. Plant Physiol. 1996;112:1541–1550. doi: 10.1104/pp.112.4.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M. Characterization of major integral protein of vacuolar membrane. Plant Physiol. 1992;98:1248–1254. doi: 10.1104/pp.98.4.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malho R, Moutinho A, van der Luit A, Trewavas AJ. Spatial characteristics of calcium signalling: the calcium wave as a basic unit in plant cell calcium signalling. Philos Trans R Soc Lond B. 1998;353:1463–1473. [Google Scholar]
- Martin ML, Busconi L. Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 2000;24:429–435. doi: 10.1046/j.1365-313x.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F, Lopez-Guerrero JA, Carrasco L, Alonso MA. The amino-terminal nine amino acid sequence of poliovirus capsid VP4 protein is sufficient to confer N-myristoylation and targeing to detergent-insoluble membranes. Biochemistry. 2000;39:1083–1090. doi: 10.1021/bi992132e. [DOI] [PubMed] [Google Scholar]
- McCabe JB, Berthiaume LG. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol Biol Cell. 1999;10:3771–3786. doi: 10.1091/mbc.10.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan G, Parenti M, Magee AI. The dynamic role of palmitoylation in signal transduction. Trends Biol Sci. 1995;20:181–186. doi: 10.1016/s0968-0004(00)89004-0. [DOI] [PubMed] [Google Scholar]
- Moffett S, Brown DA, Linder ME. Lipid-dependent targeting of G proteins into rafts. J Biol Chem. 2000;275:2191–2198. doi: 10.1074/jbc.275.3.2191. [DOI] [PubMed] [Google Scholar]
- Morales J, Fishburn CS, Wilson PT, Bourne HR. Plasma membrane localization of Gαz requires two signals. Mol Biol Cell. 1998;9:1–14. doi: 10.1091/mbc.9.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho A, Trewavas AJ, Malho R. Relocation of a Ca2+-dependent protein kinase activity during pollen tube reorientation. Plant Cell. 1998;10:1499–1509. doi: 10.1105/tpc.10.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski NE, Martin FB, Ausubel FM. Specialized binary vector for plant transformation: expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 1988;16:10765–10782. doi: 10.1093/nar/16.22.10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Ward JM, Harper JF, Schroeder JI. A novel chloride channel in Vicia faba guard cell vacuoles activated by the serine/threonine kinase, CDPK. EMBO J. 1996;15:6564–6574. [PMC free article] [PubMed] [Google Scholar]
- Putnam-Evans C, Harmon AC, Palevitz BA, Fechheimer M, Cormier MJ. Calcium dependent protein kinase is localized with F-actin in plant cells. Cell Motil Cytoskelet. 1989;12:12–22. [Google Scholar]
- Putnam-Evans CL, Harmon AC, Cormier MJ. Purification and characterization of a novel calcium-dependent protein kinase from soybean. Biochemistry. 1990;29:2488–2495. doi: 10.1021/bi00462a008. [DOI] [PubMed] [Google Scholar]
- Qi Q, Rajala RVS, Anderson W, Jiang C, Rozwadowski K, Selvaraj G, Sharma R, Datla R. Molecular cloning, genomic organization, and biochemical characterization of myristoyl-CoA:protein N-myristoyltransferase from Arabidopsis thaliana. J Biol Chem. 2000;275:9673–9683. doi: 10.1074/jbc.275.13.9673. [DOI] [PubMed] [Google Scholar]
- Resh MD. Specific and saturable binding of pp60v-src to plasma membranes: evidence for a myristyl-src receptor. Cell. 1989;58:281–286. doi: 10.1016/0092-8674(89)90842-8. [DOI] [PubMed] [Google Scholar]
- Resh MD. Regulation of cellular signalling by fatty acid acylation and prenylation of signal transduction proteins. Cell Signal. 1996;8:403–412. doi: 10.1016/s0898-6568(96)00088-5. [DOI] [PubMed] [Google Scholar]
- Resh MD, Ling H-p. Identification of a 32K plasma membrane protein that binds to the myristylated amino-terminal sequence of p60v-src. Nature. 1990;346:84–86. doi: 10.1038/346084a0. [DOI] [PubMed] [Google Scholar]
- Ritchie S, Gilroy S. Calcium-dependent protein phosphorylation may mediate the gibberellic acid response in barley aleurone. Plant Physiol. 1998;116:765–776. doi: 10.1104/pp.116.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Jones JDG. Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell. 2000;12:803–815. doi: 10.1105/tpc.12.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommens CMT, Salmeron JM, Baulcombe DC, Staskawicz BJ. Use of a gene expression system based on potato virus X to rapidly identify and characterize a tomato Pto homolog that controls fenthion sensitivity. Plant Cell. 1995;7:249–257. doi: 10.1105/tpc.7.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaller GE, DeWitt ND. Analysis of the H+-ATPase and other proteins of the Arabidopsis plasma membrane. Methods Cell Biol. 1995;50:129–148. [PubMed] [Google Scholar]
- Schaller GE, Harmon AC, Sussman MR. Characterization of a calcium- and lipid-dependent protein kinase associated with the plasma membrane of oat. Biochemistry. 1992;31:1721–1727. doi: 10.1021/bi00121a020. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Sussman MR. Phosphorylation of the plasma-membrane H+-ATPase of oat roots by a calcium-stimulated protein kinase. Planta. 1988;173:509–518. doi: 10.1007/BF00958964. [DOI] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahinian S, Silvius JR. Doubly lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry. 1995;34:3813–3822. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki A, Hayashi N, Matsubara M, Yamauchi E, Taniguchi H. Identification of the calmodulin-binding domain of neuron-specific protein kinase C substrate protein CAP-22/NAP-22. J Biol Chem. 1999;274:11848–11853. doi: 10.1074/jbc.274.17.11848. [DOI] [PubMed] [Google Scholar]
- Taniguchi H. Protein myristoylation in protein-lipid and protein-protein interactions. Biophys Chem. 1999;82:129–137. doi: 10.1016/s0301-4622(99)00112-x. [DOI] [PubMed] [Google Scholar]
- Thompson GA, Okuyama H. Lipid-linked proteins of plants. Prog Lipid Res. 2000;39:19–39. doi: 10.1016/s0163-7827(99)00014-4. [DOI] [PubMed] [Google Scholar]
- Towler DA, Gordon JI, Adams SP, Glaser L. The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem. 1988;57:69–99. doi: 10.1146/annurev.bi.57.070188.000441. [DOI] [PubMed] [Google Scholar]
- Trewavas A. Le calcium, c'est la vie: calcium makes waves. Plant Physiol. 1999;120:1–6. doi: 10.1104/pp.120.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Montagu MV, Lijsebettens MV. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey SD, Gaiser JC, Lomax TL. Protein kinases in zucchini: characterization of calcium-requiring plasma membrane kinases. Plant Physiol. 1993;103:413–419. doi: 10.1104/pp.103.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PT, Bourne HR. Fatty acylation of αz. Effects of palmitoylation and myristoylation on αz signaling. J Biol Chem. 1995;270:9667–9675. doi: 10.1074/jbc.270.16.9667. [DOI] [PubMed] [Google Scholar]
- Yonemoto W, McGlone M, Taylor SS. N-Myristylation of the catalytic subunit of cAMP-dependent protein kinase conveys structural stability. J Biol Chem. 1993;268:2348–2352. [PubMed] [Google Scholar]