Abstract
The 20S proteasome (multicatalytic proteinase) was purified from maize (Zea mays L. cv DEA 1992) roots through a five-step procedure. After biochemical characterization, it was shown to be similar to most eukaryotic proteasomes. We investigated the involvement of the 20S proteasome in the response to carbon starvation in excised maize root tips. Using polyclonal antibodies, we showed that the amount of proteasome increased in 24-h-carbon-starved root tips compared with freshly excised tips, whereas the mRNA levels of α3 and β6 subunits of 20S proteasome decreased. Moreover, in carbon-starved tissues, chymotrypsin-like and caseinolytic activities of the 20S proteasome were found to increase, whereas trypsin-like activities decreased. The measurement of specific activities and kinetic parameters of 20S proteasome purified from 24-h-starved root tips suggested that it was subjected to posttranslational modifications. Using dinitrophenylhydrazine, a carbonyl-specific reagent, we observed an increase in carbonyl residues in 20S proteasome purified from starved root tips. This means that 20S proteasome was oxidized during starvation treatment. Moreover, an in vitro mild oxidative treatment of 20S proteasome from non-starved material resulted in the activation of chymotrypsin-like, peptidyl-glutamyl-peptide hydrolase and caseinolytic-specific activities and in the inhibition of trypsin-like specific activities, similar to that observed for proteasome from starved root tips. Our results provide the first evidence, to our knowledge, for an in vivo carbonylation of the 20S proteasome. They suggest that sugar deprivation induces an oxidative stress, and that oxidized 20S proteasome could be associated to the degradation of oxidatively damaged proteins in carbon starvation situations.
Living organisms are subjected to numerous biotic or abiotic stresses. Daily, at a cellular level, changing environmental growth conditions trigger the synthesis of new sets of proteins necessary for the acclimation response, and the degradation of regulatory proteins, damaged proteins, and proteins that have become useless. Thus, in plant cells subjected to carbon starvation, the activity of enzymes involved in sugar metabolism and respiration (Journet et al., 1986; Brouquisse et al., 1991; Irving and Hurst, 1993), nitrogen reduction and assimilation (Brouquisse et al., 1992; Peeters and Van Laere, 1992), regulation of cell division and growth (Chevalier et al., 1996), or protein synthesis (Webster and Henry, 1987; Tassi et al., 1992) decreases and, in most cases, the corresponding proteins are likely subjected to proteolysis. In contrast, the activity of enzymes related to the catabolism of proteins (Tassi et al., 1992; James et al., 1993, 1996; Chevalier et al., 1995; Moriyasu and Ohsumi, 1996), amino acids (Brouquisse et al., 1992), or lipids (Dieuaide et al., 1992; Ismail et al., 1997) increases. Genes encoding enzymes involved in protein and lipid catabolism have been shown to be induced by sugar depletion (Koch, 1996), and it is clear that the selective synthesis and degradation of individual proteins are important components of the coordinated response to sugar starvation (Koch, 1996).
In plant cells, protein breakdown is mediated by different proteolytic systems: (a) vacuolar proteolysis, (b) organellar proteolysis, and (c) selective nuclear and cytosolic proteasome-dependent proteolysis (for review, see Vierstra, 1996; Brouquisse et al., 2000). Nonselective vacuolar proteolysis has been reported to increase in plant tissues submitted to sugar starvation (Aubert et al., 1996; Moriyasu and Ohsumi, 1996), and proteolytic enzymes (endopeptidases and carboxypeptidases) are induced during carbon starvation (Tassi et al., 1992; James et al., 1993, 1996; Chevalier et al., 1995). Similarly, plastid aminopeptidases and endopeptidases are induced in sugar beet (Beta vulgaris) cotyledons during prolonged dark growth (El Amrani et al., 1998, and refs. therein). However, the concomitant synthesis and degradation of proteins present in the same compartment (Brouquisse et al., 1992), as well as the tight control of the cell division cycle (Chevalier et al., 1996; Genschik et al., 1998), suggest that, in plant cells, selective proteasome-dependent proteolysis should occur in the response to carbon starvation as already suggested in animal tissues and yeast (Saccharomyces cerevisiae). Thus, in rat (Rattus norvegicus) skeletal muscles submitted to starvation, increased proteolysis has been shown to be related to an increase in proteasome and ubiquitin mRNAs and ubiquitin-protein conjugates (Medina et al., 1995). In yeast, Hilt and Wolf (1992) reported that double mutants defective in two proteasome genes, PRE1 and PRE2, accumulate ubiquitinated proteins upon exposure to starvation, and that ubiquitin mutants are hypersensitive to starvation.
The structure and functions of 20S and 26S proteasomes have been extensively reviewed (Coux et al., 1996; Hershko and Ciechanover, 1998; Voges et al., 1999). Both 20S and 26S forms have been shown to coexist in eukaryotic cells (Yang et al., 1995). The 26S proteasome complex is involved in the rapid ATP-dependent degradation of many rate-limiting enzymes, transcription regulators, regulatory proteins, abnormal proteins, and more generally in the slower degradation of the bulk of proteins in animal cells (for review, see Coux et al., 1996; Voges et al., 1999). It was reported that, besides its function as the proteolytic core of the 26S complex, the 20S proteasome could be involved in the degradation of oxidatively modified proteins (Rivett, 1985; Giulivi et al., 1994; Grune et al., 1995). Oxygen radicals and other activated oxygen species generated as by-products of cellular metabolism or from environmental sources cause modifications to the amino acids of proteins (Dean et al., 1997). Oxidatively modified proteins can undergo chemical fragmentations or form aggregates because of covalent cross-linking reactions and increased surface hydrophobicity. The recognition of hydrophobic amino acid residues of oxidized proteins and their subsequent degradation by the 20S proteasome could be a selective mechanism to remove oxidatively damaged proteins from the cell (Grune et al., 1997). As a consequence of the potential role of the 20S proteasome in the degradation of oxidized proteins, the effects of oxidative treatments on 20S proteasome structure and activities have also been investigated in vitro (Strack et al., 1996; Conconi et al., 1998; Reinheckel et al., 1998). However, whereas the 20S proteasome from chicken (Gallus gallus) erythrocytes has been report to be activated by mild oxidative treatment with either hydrogen peroxide (H2O2) or FeSO4-EDTA-ascorbate (Strack et al., 1996), no change in activity was observed with the 20S proteasome from human erythrocyte (Reinheckel et al., 1998). Moreover, whether the proteasome is modified in vivo by oxidative conditions is not known.
In the context of our protein degradation studies of carbon-starved plant cells, we investigated the fate of the 20S proteasome, and we focused our attention on the occurrence of secondary oxidative effects on its activities. We first tested the hypothesis that the proteasome could be involved in the response of plant cells to carbon starvation. To attain this goal, we purified and characterized the proteasome from maize (Zea mays L. cv DEA 1992) roots, and investigated the changes in 20S proteasome amounts and mRNA levels, in parallel with its proteolytic activities and kinetic properties, in carbon-starved and carbon-fed root tips. Second, we studied the occurrence of potential oxidative modifications of the 20S proteasome in starvation conditions. We report the first biochemical evidence for an in vivo oxidation of the 20S proteasome that activates its proteolytic activity in carbon-starved plant tissues.
RESULTS
Purification of 20S Proteasome from Whole Maize Root
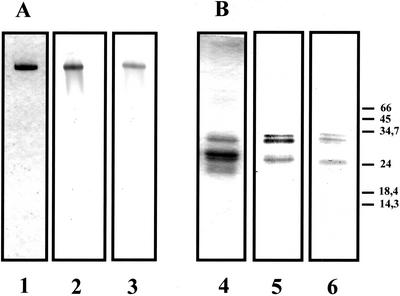
The purification of 20S proteasome (multicatalytic proteinase) was performed through a five-step purification procedure (Table I). The application of the crude extract to Sepharose DEAE Fast Flow (step 2) resulted in the retention of the whole chymotrypsin-like activity and the removal of polyphenols and microsomal fractions that were not retained. The elution of the bound proteins with an NaCl gradient resulted in a single activity peak, with a recovery close to 100%. Active fractions were subjected to gel filtration on Sephacryl S-300 (step 3), which separated two activity peaks. The first peak, which contains the 20S proteasome, eluted in the range of the 600- to 1,000-kD proteins, immediately after the void volume. The second activity peak corresponds to proteins close to 400 to 500 kD, and represents 50% to 60% of the loaded activity that is not because of proteasome—as indicated by a check with anti-20S proteasome antibodies. It should be mentioned that when the 20S proteasome is purified from maize root tips, instead of whole roots, the second activity peak represents less than 20% of the loaded activity (data not shown). The removal of some chymotrypsin-like activity resulted in a low apparent purification factor (Table I), but this step allowed the separation of the proteasome from the bulk of lower molecular mass contaminating proteins. Gradient used for the Mono-Q step was optimized to obtain the proteasome in an individualized sharp peak (around 250 mm NaCl), therefore removing 80% of the unrelated proteins (Table I). The active fractions were brought to 800 mm (NH4)2SO4 and applied to a Phenyl-Superose column. The proteasome did not bind to the column and this step allowed removal of minor contaminants. This last step provided a purified 20S proteasome, showing a single band of protein after native PAGE (Fig. 1A). Under denaturing conditions, 20S proteasome subunits were found to range between 20 and 35 kD (Fig. 1B), and at least 13 different subunits, with pI ranging between 5 and 7, were observed after two-dimensional electrophoresis (data not shown). Native molecular mass of the 20S proteasome was estimated to be 700 ± 30 kD after gel filtration on a Sephacryl S-300 HR column (data not shown).
Table I.
Purification of 20S proteasome from maize roots
| Step | Protein | Total Activity | Specific Activity | Yield | Purification Factor |
|---|---|---|---|---|---|
| mg | nmol h−1 | nmol mg−1 h−1 | % | ||
| Crude extract | 2,175 | 16,706 | 7.7 | 100 | 1 |
| DEAE-Sepharose | 681 | 16,154 | 24 | 97 | 3 |
| S-300 HR | 162 | 4,128 | 25 | 25 | 3.2 |
| MONO-Q | 12.6 | 1,073 | 85 | 6.4 | 11 |
| Phenyl-superose | 2.1 | 430 | 205 | 2.6 | 27 |
The activity was determined through the chymotrypsin-like activity of the 20S proteasome, with N-succinyl-L-L-V-Y-7-amido-4-methylcoumarin (Suc-L-L-V-Y-AMC) as substrate (final concentration 100 μm), as described in “Materials and Methods.”
Figure 1.
Native-PAGE (A) and SDS-PAGE (B) analysis of purified 20S proteasome preparation obtained from maize roots. The purified 20S proteasome was revealed by Coomassie Brillant Blue coloration after PAGE (6% [w/v] gel; lane 1) or SDS-PAGE (12.5% [w/v] gel; lane 4). Purified antibodies were used to immunodetect 20S proteasome in purified preparation (lanes 2 and 5) or in crude extracts of maize roots (lanes 3 and 6). Pure 20S proteasome (3 μg) was loaded in tracks 1, 2, 4, and 5 and 50 μg of total protein in tracks 3 and 6. The molecular masses of markers are indicated in kilodaltons.
Final purification factor and apparent yield of activity were 27% and 2.6%, respectively (Table I). However, these factors are underestimates because of the presence of the chymotrypsin-like activity removed at the gel filtration step. The five-step purification procedure typically yielded 2 mg of purified 20S proteasome from 60 g of maize roots (Table I). Rabbit polyclonal antibodies were raised against this 20S proteasome. Immunoprecipitation experiments showed that the activity of 20S proteasome was more than 95% inhibited by the addition of 15 μL of crude antiserum (data not shown). Typical western-blot patterns obtained in native and denaturing conditions are shown in Figure 1.
Biochemical Characterization
Substrate Specificity, Optimal pH and Temperature, and Autolysis Test
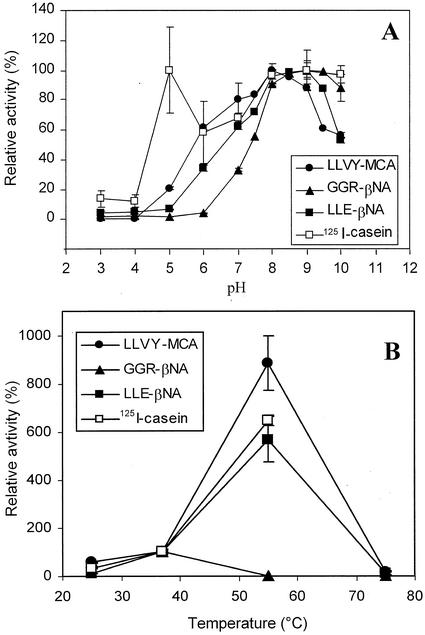
Purified maize root 20S proteasome was assayed against various endopeptidase and aminopeptidase substrates. As shown in Table II, it was found to possess classical chymotrypsin-like, trypsin-like, and peptidyl glutamyl peptide hydrolase (PGPH) activities against fluorescent synthetic substrates, and to degrade 125I-casein. No activity was found against various aminopeptidase substrates (Table II). Chymotrypsin-like, trypsin-like, PGPH, and caseinase activities were maximum at pH 8 to 9.5 against fluorescent substrates (Suc-L-L-V-Y-AMC, Cbz-G-G-R-AMC, and Cbz-L-L-E-βNA), and at pH 8 to 9 against 125I casein (Fig. 2A). The other maximum at pH 5 against casein was probably because of a change in the casein conformation at low pH, leading to a better susceptibility to proteolysis, rather than to a true acidic endopeptidase activity of the 20S proteasome itself.
Table II.
Hydrolysis of different substrates by purified 20S proteasome
| Substrate | Final Concentration | Specific Activity |
|---|---|---|
| μm | nmol mg−1 h−1 | |
| Suc-LLVY-AMC | 100 | 135 ± 3.5 |
| N-Carbobenzoxy (Cbz)-A-A-P-AMC | 100 | 54 ± 5 |
| Cbz-G-G-R-β-napthylamide (βNA) | 170 | 224 ± 12 |
| Cbz-Leu-Leu-Glu(LLE)-βNA | 170 | 59 ± 2 |
| L-Ala-βNA | 100 | n.d. |
| L-Leu-βNA | 100 | n.d. |
| L-Arg-βNA | 100 | n.d. |
| L-Phe-βNA | 100 | n.d. |
| L-Pro-βNA | 100 | n.d. |
| cpm μg−1 | μg casein mg−1 h−1 | |
| Casein-125I | 2.104 | 359 ± 25 |
Assays were performed as described in “Materials and Methods.” n.d., Not detected.
Figure 2.
Determination of 20S proteasome optimal pH (A) and temperature effects (B). Chymotrypsin-like, trypsin-like, PGPH, and caseinolytic activities of purified 20S proteasome (1 μg) were measured in 70 μL of pH 3 to 10 tri-buffer mixture (50 mm acetic acid, 50 mm MES, and 100 mm Tris) for the determination of the pH effects (A) or in Tris-HCl buffer, pH 7.5, and 20 mm NaCl at 25°C, 37°C, 55°C, and 75°C for the determination of the temperature effects (caseinolytic activities were not measured at 75°C because of casein precipitation; B). Activities measured at 37°C were normalized to 100%. Values are means of three independent experiments.
Chymotrypsin-like, PGPH, and caseinase activities were found to be significantly stimulated at 37°C and 55°C compared with 25°C (3- and 18-fold, respectively), whereas trypsin-like activity was stimulated 2-fold at 37°C but totally inhibited at 55°C (Fig. 2B). All the activities were inhibited at 75°C. Thus, proteasome activities were routinely measured at a pH of 8.1 and 37°C.
No autolytic degradation was observed when purified proteasome was incubated at 37°C; however, when incubated in the presence of a crude extract of maize roots, the amount of 20S proteasome decreased (data not shown). These data suggest that the 20S proteasome can be degraded by other proteases in the maize root tips extract, but not by autoproteolysis.
Effects of Inhibitors and Activators
The effects of several effectors and protease inhibitors were tested on the peptidic and caseinolytic activities of purified 20S proteasome (Table III). As already reported for proteasomes purified from other plant or animal sources (Rivett et al., 1994), maize root proteasome activities were inhibited to various extents by hemin, chymostatin, and PMSF, and activated by poly-Lys. SDS, at the concentration of 0.02% (w/v), was shown to activate the chymotrypsin-like, PGPH, and caseinolytic activities and to inhibit the trypsin-like activity. The peptide aldehyde inhibitor N-acetyl-leucyl-leucyl-norleucinal (MG132) was more effective against the trypsin-like activity (100% inhibition with 12.5 μm inhibitor) than against the chymotrypsin-like and caseinolytic activities, but was ineffective against the PGPH activity. Another peptide aldehyde, Z-Ile-Glu(OtBu)-Ala-Leucinal, known as proteasome inhibitor 1 (PI1), was more effective against the chymotrypsin-like activity (100% inhibition with 20 μm PI1) than the trypsin-like and caseinolytic activities, and was found to moderately stimulate the PGPH activity (Table III). Other effectors such as ATP-Mg, Cys-protease inhibitors E64 and iodoacetamide, or EDTA (except for trypsin-like activity) did not affect significantly the activities of this 20S proteasome.
Table III.
Effect of various inhibitors or activators on 20S proteasome activities
| Inhibitors | Concentrations | Activity (% of Control)
|
|||
|---|---|---|---|---|---|
| Chymotrypsin like (LLVY-AMC) | Trypsin like (GGR-βNA) | PGPH (LLE-βNA) | Caseinolytic 125I-casein | ||
| ATP-Mg | 3 mm | 98 | 98 | 90 | 100 |
| Poly-Lys | 0.1% (w/v) | 181 | 121 | 225 | 98 |
| Hemin | 50 μm | 49 | 69 | 60 | 7 |
| EDTA | 2 mm | 100 | 153 | 96 | 99 |
| Indole-3-acetic acid | 0.5 mm | 90 | 91 | 96 | 103 |
| E64 | 250 μm | 97 | 104 | 96 | – |
| Chymostatin | 25 μm | 45 | 83 | 86 | – |
| Phenylmethylsulfonyl fluoride (PMSF) | 5 mm | 80 | 44 | 82 | – |
| MG132 | 5 μm | 27 | 10 | 100 | – |
| 12.5 μm | 19 | 0 | 100 | – | |
| 25 μm | 13 | 0 | 100 | – | |
| 50 μm | 7 | 0 | – | 37 | |
| PI1 | 5 μm | 8 | 30 | – | 79 |
| 10 μm | 4 | 22 | 138 | 65 | |
| 20 μm | 0 | 14 | 124 | 62 | |
| 50 μm | 0 | 10 | 128 | 65 | |
| 100 μm | 0 | 5 | 123 | 67 | |
| 200 μm | 0 | 0 | 128 | 61 | |
| SDS | 0.02% (w/v) | 176 | 52 | 320 | 156 |
Except with SDS, 20S proteasome (5 μg) was pre-incubated with different inhibitors or activators for 15 min before the addition of substrates. With SDS, substrates and SDS were added simultaneously. Activities are expressed as a percentage of the control (with solvent alone). Values are the mean of three to five independent experiments. sds are below 10%. –, Not determined.
Physiological Characterization
Changes in 20S Proteasome in Sugar-Starved and Non-Starved Excised Root Tips
Maize root tips, either freshly excised (T0), or incubated for 24h in the presence or absence of Glc (T24 + Glc and T24-starved), were used for a study of enzymic activities, protein content, and mRNA levels of 20S proteasome.
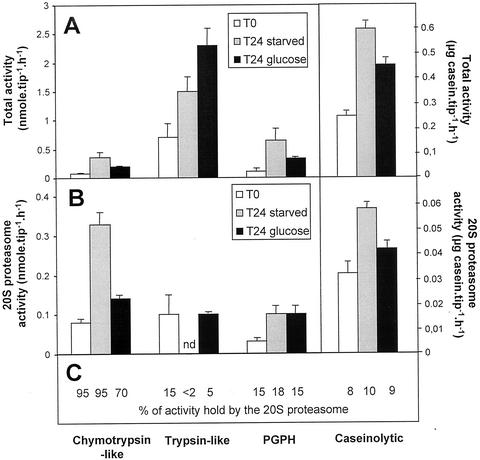
First, the chymotrypsin-like, trypsin-like, and PGPH activities (measured respectively against Suc- L-L-V-Y-AMC, Cbz-G-G-R-βNA, and Cbz-L-L-E-βNA), and the caseinolytic activities, have been measured in desalted crude extracts of T0, T24-starved, and T24 + Glc root tips, after incubation with either pre-immune or immune anti-20S proteasome IgG (see “Materials and Methods”). The activities measured in the pre-immune IgG-treated samples accounted for total proteolytic activities present in the extracts (Fig. 3A), whereas those measured in the supernatant from immune anti-20S IgG treated samples accounted for non-proteasomal activities. Thus, the proteolytic activities because of the 20S proteasome (Fig. 3B) were obtained from the difference between the activities measured in the pre-immune and the immune IgG-treated samples. PI1 and MG132 inhibitors were also tested for the inhibition of chymotrypsin- and trypsin-like activities in root tip extracts. Each, at a concentration of 30 μm in the extracts, triggered an inhibition of the activity equal to that observed with anti-20S proteasome antibodies (data not shown). This means that, at this concentration, PI1 and MG132 inhibit only the chymotrypsin- and trypsin-like activities of the 20S proteasome, respectively. Thus, they were used routinely to follow these two activities of the 20S proteasome in root tip extracts. For each activity type, the percentage of the total activity due to the 20S proteasome is reported in Figure 3C. After a 24-h incubation period, all the total activities measured in the extracts were increased both in starved and Glc-fed root tips (Fig. 3A). In starved root tips, chymotrypsin-like and PGPH activities were increased 4- and 5-fold, respectively, whereas trypsin-like activity was increased only 2-fold compared with T0 root tips. However, the three activities were similarly enhanced (around 3-fold) in T24 + Glc root tips. This shows that mechanical stress (excision and/or incubation) possibly stimulated proteolytic activities in the root tips. Figure 3B shows that the chymotrypsin-like, PGPH, and caseinolytic activities related to the 20S proteasome were enhanced (4.5-, 3.2-, and 1.8-fold, respectively) in T24-starved root tips, whereas its trypsin-like activity became non-detectable. In T24 + Glc root tips, 20S proteasome activities were slightly or not increased, except for PGPH activity, which increased by a factor of 3 (Fig. 3, A and B). It may be noted that in the root tips, the 20S proteasome accounted for most of the total chymotrypsin-like activity (70%–95%), but for a minor part of the trypsin-like, PGPH, and caseinolytic activities (<2%–15%, 15%–18%, and 8%–11%, respectively). These data signify that in excised root tips, sugar starvation leads to an increase of chymotrypsin-like activity and a decrease in trypsin-like activity of the 20S proteasome, whereas PGPH activity is similarly increased in both starved and non-starved root tips.
Figure 3.
Total and 20S proteasome activities in starved and non-starved maize root tips. A, Chymotrypsin-like, trypsin-like, PGPH, and caseinolytic total activities were measured in desalted crude extracts of non starved (T0), 24-h Glc-starved (T24 starved), and 24-h Glc-fed (T24 + Glc) root tips. B, The 20S proteasome activities were determined after immunoprecipitation of desalted crude extracts with anti-20S proteasome antibodies. For each activity, the percentage of total activity because of the 20S proteasome is reported in C. n.d., Non-detected, because of the detection limit of the immunoprecipitation method. Values are means of six independent experiments.
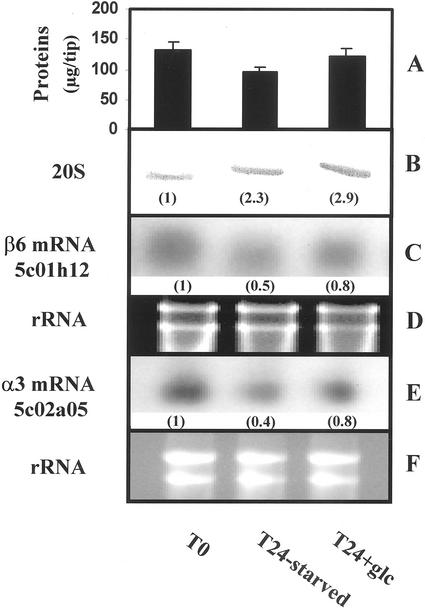
These changes in 20S proteasome activities might be because of either a change in proteasome subunits expression or posttranslational activation/inhibition of preexisting proteasome. We first investigated the modifications in proteasome content by western-blot analysis (Fig. 4B). Compared with freshly excised root tips, 20S proteasome amount increased, by a factor of 2 to 3, in both T24-starved and T24 + Glc root tips, whereas total protein content decreased in the same time (from 38 and 8 μg tip−1, respectively, Fig. 4A). This may be due either to increased proteasome synthesis, or to decreased degradation, or both. However, northern-blot analysis of two maize 20S proteasome subunit mRNA levels (encoding the β6- and α3-type subunits) showed a 50% and 60% decrease in transcript levels in starved root tips, and only a 20% decrease in Glc-fed root tips, compared with T0 levels (Fig. 4, C and E). These data show that, in carbon-starved maize root tips, a decrease in 20S proteasome subunits mRNA levels may occur concomitantly to an increase in proteasome amount. This result, together with the differential regulation of proteasome activities in starved root tips (Fig. 3), led us to purify proteasomes from starved and non-starved materials and to investigate their kinetic parameters.
Figure 4.
Western- and northern-blot analysis of the 20S proteasome in starved and non-starved maize root tips. A, Total protein content in T0, T24-starved, and T24 + Glc root tips. Values are means of three independent experiments. B, Protein extracts (1 root tip/track) were separated by PAGE, and the proteasome was immunodetected with anti-20S proteasome antibodies. Relative immunosignal intensities are indicated in brackets. C and E, Northern-blot analysis of two transcripts encoding for β6- and α3-subunits of the maize 20S proteasome (10 μg total RNA/track). D and F, rRNA were used as a loading control. Values are representative of four independent experiments.
Table IV reports specific activities and kinetic parameters of proteasomes purified from T0, T24-starved, and T24 + Glc maize root tips. The chymotrypsin-like specific activity of proteasome was found to be higher in T24-starved than in T0 and T24 + Glc root tips, whereas it was the opposite for the trypsin-like specific activity. The caseinase specific activity, which is thought to reflect the coordinated action of the three peptidic activities, was 20% higher in the T24-starved than in the control root tips. These modifications in specific activities may be partly explained by changes in affinity (Km) and turnover number (kcat) values of 20S proteasomes (Table IV). As a consequence, on the basis of the calculated kcat/Km ratios, it may be estimated that Suc- L-L-V-Y-AMC was a better substrate for the proteasomes from T24-starved than for T0 and T24 + Glc root tips, whereas Cbz-G-G-R-βNA was better for the proteasome in T0 root tips. No clear-cut changes were observed for PGPH specific activity because of opposite changes in Km and kcat. These data show that besides changes in RNA or protein turnover, kinetic properties of 20S proteasomes were modified during the incubation in the presence or absence of Glc.
Table IV.
Specific activities and kinetic parameters of 20S proteasomes purified from T0, T24-starved, and T24 + Glc maize root tips
| Biochemical Parameters | T0 | T24 Starved | T24 + Glc |
|---|---|---|---|
| Specific activity (nmole mg−1 h−1) | |||
| Suc-L-L-V-Y-AMC | 104 ± 3 | 192 ± 0 | 124 ± 3 |
| Cbz-G-G-R-βNA | 204 ± 8 | 159 ± 15 | 185 ± 7 |
| Cbz-L-L-E-βNA | 54 ± 5 | 58 ± 9 | 42 ± 6 |
| 125I-Casein (μgcasein mg−1 h−1) | 381 ± 15 | 471 ± 25 | 388 ± 20 |
| Km (μm) | |||
| Suc-L-L-V-Y-AMC | 160 | 105 | 113 |
| Cbz-G-G-R-βNA | 274 | 397 | 199 |
| Cbz-L-L-E-βNA | 367 | 213 | 254 |
| kcat (s−1) | |||
| Suc-L-L-V-Y-AMC | 0.086 | 0.095 | 0.050 |
| Cbz-G-G-R-βNA | 0.085 | 0.100 | 0.040 |
| Cbz-L-L-E-βNA | 0.040 | 0.025 | 0.024 |
| kcat/Km (m−1 s−1) | |||
| Suc-L-L-V-Y-AMC | 537 | 905 | 442 |
| Cbz-G-G-R-βNA | 310 | 252 | 201 |
| Cbz-L-L-E-βNA | 109 | 117 | 94 |
Kinetic parameters were determined with the Lineweaver-Burk plot and linear fitting of four plots obtained from four different measurements. Ranges of substrates concentrations are: 10 to 200 μm for Suc-L-L-V-Y-AMC, and 20 to 170 μm for Cbz-G-G-R-βNA and Cbz-L-L-E-βNA.
Characterization of Oxidized Proteasome in Sugar-Starved Root Tips
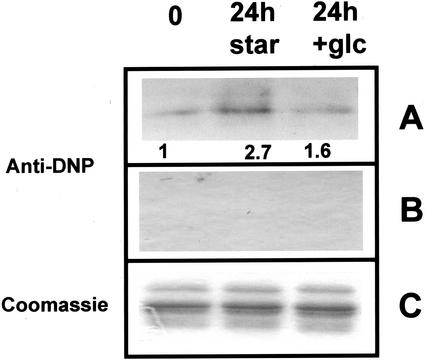
In animal cells, sugar deprivation has been shown both to induce an oxidative stress, via an increase in oxidized glutathione and intracellular pro-oxidant levels (Blackburn et al., 1999), and to enhance the sensitivity of the cells to oxidative stress (Zhang et al., 1996). Thus, we examined the possibility that the proteasome was subjected to oxidative damage. Oxidation of proteins is known to produce carbonyl modifications in certain amino acids (Chao et al., 1997; Dean, et al., 1997). To assess the level of oxidative damages generated during starvation, 20S proteasome preparations from T0, T24-starved, and T24 + Glc-fed tips were treated with 2,4-dinitrophenylhydrazine (DNPH), a carbonyl-specific reagent. As shown in Figure 5, for an equal deposit of the three types of purified proteasomes (Fig. 5C), carbonyl moieties were detected at a significantly higher level in T24-starved proteasome (2.7-fold higher compared with T0). Proteasome isolated from T24 + Glc root tips produced 1.6-fold stronger signal (Fig. 5A). Un-derivatized proteins showed no cross reactions with anti-DNP antibodies (control, Fig. 5B). It appears that oxidative modifications of proteasome may occur during starvation without major changes in the protein structure. To check that the changes in specific activities of proteasome (Table IV) could be related to oxidative alterations, purified proteasomes from T0 and T24-starved root tips were submitted in vitro to a mild oxidative treatment, through a metal-catalyzed oxidation (MCO) in the absence of H2O2 (see “Material and Methods”), and then analyzed for enzymic activities. Chymotrypsin-like and PGPH-specific activities of proteasomes fromT0 and T24 + Glc root tips were found to increase by factors of 2.1 and 1.25, respectively, after oxidative treatment, whereas trypsin-like specific activities decreased by 20% to 30% (Table V). Furthermore, the analysis of DNPH treated proteasomes with anti-DNP antibodies (Fig. 6) showed that the immunosignal of the T0 proteasome submitted to mild oxidative treatment (lane 3) is of the same order of intensity as the immunosignal linked to proteasome from T24-starved root tips (lane 5). Taken together, these data show that a mild oxidation in vitro of proteasome isolated from non-starved material mimicked the modifications in specific activities and carbonylation observed in vivo during the starvation.
Figure 5.
Immunodetection of carbonyl residues in 20S proteasomes from starved and non-starved maize root tips. Purified 20S (10 μg) proteasomes from T0, T24-starved, and T24 + Glc-fed root tips were incubated with (A) or without (B) DNPH and separated by 12.5% (w/v) SDS-PAGE. Carbonyl residues were then immunodected with anti-DNP antibodies as described in “Materials and Methods.” A, Relative immunosignal intensities are indicated. C, Coomassie Brillant Blue coloration of aliquot fractions of DNPH-treated proteasomes after SDS-PAGE (2 μg/track) was used as loading controls.
Table V.
Effects of MCO on the three peptidic activities of 20S proteasomes isolated from starved and non-starved maize root tips
| 20S Proteasome | Chymotrypsin Like | Trypsin Like | PGPH |
|---|---|---|---|
| nmol mg−1 h−1 | |||
| T0 | |||
| Nonoxidized | 104 ± 3 | 204 ± 8 | 54 ± 5 |
| Oxidized | 220 ± 7 | 140 ± 12 | 67 ± 1 |
| T24 starved | |||
| Nonoxidized | 192 ± 0 | 159 ± 15 | 58 ± 9 |
| Oxidized | 162 ± 1 | 124 ± 8 | 45 ± 1 |
| T24 + Glc | |||
| Nonoxidized | 124 ± 3 | 185 ± 7 | 42 ± 6 |
| Oxidized | 237 ± 4 | 154 ± 8 | 77 ± 12 |
Isolated 20S proteasomes from T0, T24-starved, and T24 + Glc root tips were oxidized for 3 h at 37°C in the presence of Fe2+/ascorbate. Control proteasomes (nonoxidized) were incubated in parallel without Fe2+/ascorbate. Peptidic activities of oxidized and nonoxidized proteasomes were then measured as described in “Materials and Methods.” Values are the mean of six measurements from three independent experiments.
Figure 6.
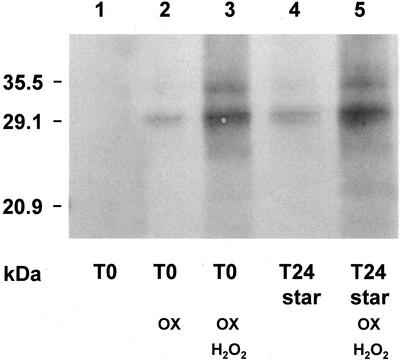
Immunodetection of carbonyl residues in 20S proteasomes from starved and non-starved maize root tips after oxidative treatment. Purified 20S proteasomes (2 μg) from T0 and T24-starved root tips were submitted to various oxidative treatments, incubated with DNPH, and separated by 12.5% (w/v) SDS-PAGE. Carbonyl residues were then immunodected with anti-DNP antibodies as described in “Materials and Methods.” Lanes 1 and 4, Nonoxidized 20S proteasomes from T0 and T24 starved root tips. Lane 2, 20S proteasome from T0 root tips treated for 2 h at 37°C with 50 mm ascorbate/200 μm FeSO4. Lanes 3 and 5, 20S proteasome from T0 and T24 starved root tips treated for 2 h at 37°C with 50 mm ascorbate/200 μm FeSO4/5 mm H2O2.
It is interesting that the three specific activities of the proteasome from T24-starved root tips decreased by 20% after the oxidative treatment (Table V). Such decrease probably resulted from the cumulative effects of starvation and of artificial oxidation because we observed a similar decrease in the proteolytic activity of 20S proteasome (not shown), and a dramatic increase in carbonyl moieties (Fig. 6), after a strong oxidative treatment in the presence of 5 mm H2O2.
DISCUSSION
Proteasome Purification and Characterization
The 20S proteasome from maize roots was purified through a five-step procedure (Table I) close to procedures classically used for animal (Rivett et al., 1994) or plant materials (Ozaki et al., 1992; Skoda and Malek, 1992; Fernandez Murray et al., 1997). After biochemical characterization, maize root proteasome was shown to be similar to most eucaryotic proteasomes with a molecular mass of 700 kD, and containing at least 13 subunits ranging between 20 and 35 kD. When assayed with fluorogenic peptide substrates, it exhibited the three main activities found in all 20S proteasomes: chymotrypsin like, trypsin like, and PGPH, and possessed an endopeptidase activity against casein (Table II). It exhibited optimal pH around 8 to 9, and was not subject to autolysis. Finally, it was sensitive to classical proteasome inhibitors such as PI1, MG132, or chymostatin, and activated by poly-Lys and SDS (Table III), as already reported for other proteasomes (Rivett et al., 1994).
Modifications of 20S Proteasome in Carbon-Starved Tissues
It has been already shown that, in maize roots, carbon starvation stops cell divisions and tissue growth (Chevalier et al., 1996; Brouquisse et al., 1998). In such a situation, proteins are remobilized to supply carbon skeletons to respiration and residual biosynthesis through nonselective autophagic processes involving vacuolar proteolysis (Aubert et al., 1996; James et al., 1996; Moriyasu and Ohsumi, 1996). However, during starvation like in other stress situations, the cessation of cell growth and the remobilization of proteins are accompanied by selective degradation and synthesis of specific proteins (Chevalier, et al., 1996; Brouquisse et al., 1998; and refs. therein). Such selective proteolysis suggests that the 20S proteasome, either as a component of the ubiquitin-dependent proteolysis or by itself, could be involved in the process of acclimation to starvation.
The involvement of proteasome in carbon starvation processes is suggested by the increase in 20S proteasome total activities and amounts in sugar-starved maize root tips (Figs. 3 and 4). These data are slightly different from previous observations showing that 20S proteasome amount levels off, instead of increasing, in roots from whole plants submitted to dark-induced starvation (Brouquisse et al., 1998). This difference may be attributed to different development stages of the roots and to milder starvation conditions in the whole plant. On the other hand, 20S proteasome activities and amounts have also been observed to increase in the nucleus of cancer cells submitted to Glc deprivation (Ogiso et al., 1999). However, contrary to a previous report showing that 20S proteasome-subunit mRNAs increased in starved rat skeletal muscles (Medina et al., 1995), α3- and β6-subunit mRNA levels clearly decreased in 24-h-starved maize root tips (Fig. 4). In Arabidopsis, six α-subunits and three β-subunits of the 20S proteasome are encoded by at least two genes (Fu et al., 1998). Among them, the α3-subunit is encoded by two genes, PAC1 and PAC2. Thus, in starved maize root tips, the decrease in PAC1-like mRNA level could be balanced by an increase in PAC2-like mRNA level to maintain or increase the amount of 20S proteasome. However, considering (a) the decrease in mRNA level of β6-subunit, which is encoded by only one gene in Arabidopsis (Fu et al., 1998), and (b) the fact that the presence of the 14 subunits is essential for the 20S proteasome structure (Coux et al., 1996; Hochstrasser, 1996), it appears unlikely that the α3-subunit can be expressed from a PAC2-like gene when the β6-subunit is not. Nevertheless, this point, as well as the change in mRNA level of the other 20S proteasome subunits during carbon starvation, will have to be checked when the sequences of the corresponding genes become available. To date, because in most animal, yeast, and plant materials studied so far the expression of the various proteasome subunits seems to occurs via a concerted mechanism (Genschik et al., 1994; Ichihara and Tanaka, 1995; Fu et al., 1998), it may be reasonably assumed that the expression of the other subunits of maize 20S proteasome were similarly decreased. This suggests that, in carbon-starved maize root tips, 20S proteasome expression was modified presumably through: (a) a decrease in proteasome gene transcription, and (b) an increase in proteasome stability and/or in mRNA translation efficiency. In addition to possible changes in translation efficiency, the modification of specific peptidic activities and kinetic parameters in T24-starved root tips (Table IV) suggest that 20S proteasome was subjected to posttranslational modifications during starvation treatment.
In eukaryotic cells, 20S proteasome has been reported to be submitted to various posttranslational modifications such as processing (Schmidtke et al., 1996; Groll et al., 1997), glycosylation (Schliephacke et al., 1991), phosphorylation (Umeda et al., 1997; Bose et al., 1999), or ADP-ribosylation (Ullrich et al., 1999). In the present work, we focused our attention on potential oxidative modifications of the proteasome. As indicated by the increase in carbonyl residues in T24-starved proteasome (Fig. 5), we report, to our knowledge, the first evidence for in vivo oxidation of the 20S proteasome. Sugar deprivation is known to induce metabolic oxidative stress via an increase in oxidized glutathione and intracellular prooxidant levels (Blackburn et al., 1999), and to enhance the sensitivity of the cells to oxidative stress (Zhang et al., 1996). Reactive oxygen species (peroxyl, alkoxyl, and hydroxyl radicals) react with proteins and generate oxidation products such as carbonyl compounds (Chao et al., 1997; Dean et al., 1997). Thus, obviously proteasome may be oxidized, like other intracellular proteins. Furthermore, the changes in specific activities of proteasome during starvation (Table IV) may be related to oxidative modifications because, after a mild oxidative treatment in vitro: (a) the specific activities of proteasomes purified from non-starved tissues (T0 and T24 + Glc) became similar to those of T24-starved proteasome: trypsin-like specific activities decreased, whereas chymotrypsin-like and PGPH ones increased more or less markedly (Table V); and (b) the level of carbonylation is similar between oxidized proteasome from T0 root tips and proteasome from T24 starved once (Fig. 6). Conflicting results have been reported about the chymotrypsin-like activity of 20S proteasome, which was found to be either activated (Strack et al., 1996) or not (Reinheckel et al., 1998) by in vitro oxidative treatment. It has been hypothesized that the activation by H2O2 found by Strack et al. (1996) was mediated via an interaction of the proteasome and the PA28 activator (Reinheckel et al., 1998). However, without excluding a possible interaction between the 20S proteasome and the PA28 activator in vivo, our data show that the purified proteasome may be activated by a mild oxidation treatment only, i.e. without H2O2 added (Table V). On the other hand, further oxidation of already oxidized proteasome (i.e. T24-starved proteasome) resulted in partial inactivation of the three peptidic activities (Table V), as observed for proteasomes treated with increasing H2O2 concentrations (Strack et al., 1996; Conconi et al., 1998; Reinheckel et al., 1998), and in a strong increase in carbonyl residues (Fig. 6). This confirms that 20S proteasome is activated by mild oxidative conditions, but inactivated by strong oxidative treatments. Thus, our data suggest that the in vivo oxidation of the 20S proteasome during starvation is mild enough not only to avoid inactivation of its activities, but to stimulate some of its peptidic and caseinolytic activities.
In mammalian cells, the 20S proteasome has been shown to recognize and selectively degrade oxidatively damaged proteins, such as hemoglobin (Fagan and Waxman, 1991; Giulivi et al., 1994), Gln synthetase (Rivett, 1985; Sahakian et al., 1995), superoxide dismutase (Grune et al., 1995), insulin-B chain (Dick et al., 1991), or Glc-6-phosphate dehydrogenase (Friguet et al., 1994), via ATP-independent degradation processes (for review, see Grune et al., 1997). Moreover, we observed a rise in oxidized protein amounts in Glc-starved maize root tips compared with non-starved ones, and we found that oxidized proteins were preferentially degraded by the proteasome in vitro and in vivo (G. Basset, P. Raymond, and R. Brouquisse, unpublished data). Taken together, the data of the literature and the present study suggest that, in vivo, sugar deprivation conditions induce a mild oxidative stress in the cells that leads to the oxidation of the proteins, including the 20S proteasome. Besides its role in the selective 26S proteasome-dependent proteolysis (Vierstra, 1996), oxidatively activated 20S proteasome could be involved in the degradation of nuclear and cytosolic oxidized proteins, thus contributing, on the one hand, to the detoxification of the cell through the elimination of oxidatively damaged proteins, and on the other hand, to the supply of amino acids for the synthesis of new proteins and as respiratory substrate for energy production.
MATERIALS AND METHODS
Plant Materials and Incubation Conditions
Maize (Zea mays L. cv DEA 1992) seeds (Pioneer France Maïs, Toulouse, France) were soaked for 3 h in water and germinated for 3 d on layers of wet filter paper; 3-mm-long primary root tips or 3- to 4-cm-long primary roots were then excised and either immediately used (T0), or incubated for 24 h in the absence (T24-starved) or the presence (T24 + Glc) of 0.2 m Glc. Incubation conditions were essentially as described (Brouquisse et al., 1991) but the incubation medium (medium A) was buffered with 10 mm instead of 100 mm MES (pH 6.0). Incubation medium was renewed every 8 to 12 h.
Preparation of Crude Extracts
Frozen root tissues were crushed at 4°C in a mortar in grinding medium (0.4 mL g fresh weight−1) containing 20 mm HEPES (pH 7.4), 5 mm β-mercaptoethanol, and 0.1% to 0.5% (w/v) insoluble polyvinyl polypyrolidone. The brei was transferred into a 1.5-mL microcentrifuge tube and the mortar was rinsed with the same volume of grinding medium, which was then pooled with the brei. The homogenate was centrifuged at 36,000g for 15 min. The supernatant was used for protein and proteolytic activity measurements, and for immunodetection experiments, as described below.
Purification of the 20S Proteasome from Maize Whole Roots or Root Tips
All steps were performed at 4°C. 20S proteasome was followed through its “chymotrypsine-like” activity.
Step 1
Forty to 60 g of whole maize root were ground in a blender (Waring, New Hartford, CT) with 250 mL of extraction medium (50 mm Tris-HCl, pH 7.5; 5 mm β-mercaptoethanol; and 0.5% [w/v] polyvinyl polypyrolidone). The brei was squeezed through a double layer of Miracloth (Calbiochem, Meudon, France) and centrifuged at 15,000g for 15 min. The supernatant constituted the crude extract. Alternatively, 1,000 maize root tips (2–2.5 g fresh weight) were reduced to powder in liquid nitrogen with a mortar and pestle. Powder was allowed to warm up to 0°C to 4°C, mixed with 6 mL of extraction medium, and centrifuged at 15,000g for 15 min. The pellet was resuspended twice in a 6-mL extraction medium and centrifuged again. The three supernatant fractions were pooled and constituted the crude extract.
Step 2
Crude extract was applied to a 25- × 2.6-cm column of Sepharose-DEAE Fast flow (Pharmacia, Uppsala) equilibrated with 50 mm Tris-HCl, pH 7.5, and 20 mm NaCl (buffer A). The column was washed at 2 mL min−1 with equilibration buffer. When optical density returned to base line, bound proteins were eluted with NaCl gradient from 20 to 500 mm (200 mL), 500 mm to 1 m (60 mL), and up to 1 m NaCl (90 mL). Fractions of 8 mL were collected and those containing chymotrypsin-like activity were pooled and concentrated 2-fold in a stirred cell (Amicon, Beverly, MA) with a 30K Filtron membrane.
Step 3
The concentrated fraction was applied to a Sephacryl S-300 HR (Pharmacia) gel filtration column (80 × 2.6 cm) equilibrated with buffer A, and eluted at 0.5 mL min−1. The fractions (7 mL) corresponding to the first activity peak, containing the 20S proteasome, were pooled (35–42 mL) but not concentrated.
Step 4
The sample was applied, at 0.5 mL min−1, to an FPLC Mono-Q 5/5 HR (Pharmacia) column equilibrated with buffer A. Bound proteins were eluted by increasing NaCl concentration, at the same flow rate, using the following gradient of buffer B (50 mm Tris-HCl, pH 7.5, and 1 m NaCl): (a) 4 mL from 0% to 21% (v/v), (b) 4 mL at 21% (v/v), (c) 7 mL from 21% to 33% (v/v), (d) 2.5 mL from 33% to 75% (v/v), and (e) 5 mL from 75% to 100% (v/v). Active fractions (1 mL) were pooled (3–4 mL) and not concentrated.
Step 5
The sample was brought to 800 mm (NH4)2SO4 and applied to an FPLC Phenyl-Superose HR 5/5 column (Pharmacia) equilibrated with 50 mm Tris-HCl, pH 7.5; 20 mm NaCl; and 800 mm (NH4)2SO4 (buffer C), at 0.25 mL min−1. The column was washed with buffer C (20S proteasome did not bind to the column) and bound proteins were eluted with buffer A. Active fractions (1 mL) were pooled (10–12 mL) and desalted with buffer A using 30K Filtron Microsep.
Determination of Molecular Mass
The molecular mass of the purified 20S proteasome was estimated by gel filtration on Sephacryl S-300 HR column (80 × 2.6 cm), equilibrated with buffer A and calibrated with the following proteins as standards: thyroglobulin (669 kD), apoferritin (443 kD), and α-amylase (200 kD). Blue Dextran (2,000 kD) was used as an exclusion volume marker.
Enzymic Activity Assays
Chymotrypsin-like, trypsin-like, and PGPH activities of the 20S proteasome were measured respectively with Suc-L-L-V-Y-AMC or Suc-A-A-F-AMC, Cbz-G-G-R-βNA, and Cbz-L-L-E-βNA. The assay mixture consisted of 10 μL synthetic substrate (stock solutions at 0.1–5 mm in dimethylformamide) and 100 μL proteasome extract plus buffer mixture (50 mm Tris, pH 8.1 at 37°C, and NaCl 20 mm). After a 15-min incubation, at 37°C, the reaction was stopped with the addition of 100 μL of 10% (w/v) SDS and 2 mL of Tris 100 mm, pH 9. The AMC or NA radicals released were measured fluorometrically (excitation 380 nm/emission 460 nm and excitation 335 nm/emission 410 nm, respectively). Activities were calculated using AMC and NA standard curves made in the same conditions. Unless mentioned in the text, Suc-L-L-V-Y-AMC, Suc-A-A-F-AMC, Cbz-G-G-R-βNA, and Cbz-L-L-E-βNA substrates were routinely used at 100, 100, 170, and 170 μm final concentrations, respectively, in the assays, and dimethylformamide final concentration did not exceed 120 mm (10% [v/v]). 20S proteasome activities against whole protein were measured with iodinated casein. Casein was radiolabeled with 125I by the chloramine-T method (Ciechanover et al., 1980). The initial specific radioactivity of [125I]-casein was approximately 2 × 104 cpm.μg−1, and the protein concentration was 5 μg μL−1. Twenty microliters of purified proteasome (5 μg of protein), 75 μL of buffer A, and 5 μL of [125I]-casein (100,000 cpm μL−1) were incubated for 20 to 30 min at 37°C. The reaction was stopped with 100 μL of 30% (w/v) trichloroacetic acid and the samples were centrifuged for 10 min at 30,000g; 100 μL of supernatant was used for radioactivity counting with a liquid scintillation analyzer (Tri-Carb 2000CA, Packard, Meriden, CT). Linearity of casein degrading activity was checked. Aminopeptidase activities were measured as described by (Sarath et al., 1989) using the β-naphtylamine conjugates of L-Ala, L-Leu, L-Arg, L-Phe, or L-Pro as substrates.
Determination of the Optimum pH
The pH dependence of the proteolytic activity against iodinated casein and synthetic substrates was determined using a three-component buffer mixture (50 mm acetic acid, 50 mm MES, and 100 mm Tris).
Effect of Inhibitors and Activators of the 20S Proteasome
Proteasome inhibitors/activators were prepared as the following stock solutions: E-64 (10 mm), iodoacetamide (0.1 m), ATP-Mg2+ (0.1 m), Na2-EDTA (0.5 m), 10% (w/v) SDS, and 10% (w/v) poly-Lys were in water; hemin (50 mm) was in 0.1 mm NaOH; PMSF (0.2 m) was in ethanol; N-acetyl-leucyl-leucyl-norleucinal (MG-132; 5 mm), Z-Ile-Glu(OtBu)- Ala-Leucinal (PI1; 2 mm), and chymostatin (10 mm) were in dimethylformamide. Inhibitors were first preincubated for 15 min with proteasome prior substrate addition, and activities were measured as described above. Control assays were carried out with the corresponding solvent.
MCO of Purified Proteasome
Oxidative treatment of the proteasome in vitro was adapted from Conconi et al. (1998). Ten micrograms of purified 20S proteasome (60–100 μL) was incubated for 2 to 3 h at 37°C with an equal volume of 20 mm sodium phosphate buffer, pH 7.0, with or without (control) 50 mm ascorbate/200 μm FeSO4. Except when mentioned, no H2O2 was added. After treatment, the peptidic activities of oxidized and nonoxidized proteasomes were measured as described above.
Electrophoresis
Native PAGE and SDS/PAGE were performed with 6% (w/v) and 12.5% (w/v) polyacrylamide gels, respectively, by the procedure of (Laemmli, 1970). Gels were fixed, stained with Coomassie Blue or silver stain, and destained using standard methods. Two-dimensional electrophoreses (isoelectric focusing and SDS/PAGE) were as described by O'Farrel (1975).
Preparation of Antibodies and Western-Blot Analysis
Polyclonal antibodies against maize 20S proteasome were produced by subcutaneous injection of the purified enzyme into a Fauve de Bourgogne rabbit as in James et al. (1996). Serum IgG fraction was purified using 1 mL of HiTrap Protein A column (Pharmacia) as recommended by the manufacturer. Western-blot experiments were as described by James et al. (1996).
Immunoprecipitation of the Proteasome
For immunoprecipitation experiments, 500 μL of crude extracts was desalted through an Econo-pac 10DG (Bio-Rad Laboratories, Hercules, CA) column equilibrated with buffer A. Fractions of 160 μL of desalted crude extract or purified proteasome were incubated for 1 h at 4°C with increasing volumes of purified immune or pre-immune serum. Immune complexes were incubated for 1 h at 4°C with a 2-fold (IgG binding) excess of protein A-agarose (Affi-gel, Bio-Rad) and then centrifuged for 5 min at 10,000g. The various proteasome activities were measured in each supernatant fraction as described above.
Immunochemical Detection of Proteasome Carbonyls
Carbonyl content of purified proteasome was measured by reaction with DNPH. Two to 10 μg of purified proteasome were reacted with 200 μL of 10 mm DNPH (in 2 m HCl) or 2 m HCl (control) for 1 h at 25°C. The proteins were precipitated with 20% (w/v) trichloroacetic acid, centrifuged, and washed three times with ethanol:ethyl acetate (1:1 [v/v]). The final protein pellets were dissolved in 2× Laemmli buffer at 100°C for 15 min. The DNPH-derivatized enzymes were separated by 12.5% (w/v) SDS-PAGE. After electrotransfer of the proteins to nitrocellulose membranes, DNPH moieties were detected with rabbit anti-DNP primary antibodies (dilution 1:5,000 [v/v], Sigma, St. Louis) and goat anti-rabbit-IgG-alkaline phosphatase conjugate (dilution 1:30,000 [v/v], Sigma). Immunosignal was quantified by scanning the blots with an Image Master VDS and using the Image Master 1D software (Pharmacia Biotech).
RNA Extraction and Northern-Blot Analysis
Total RNA from maize root tips was extracted using the hot phenol method as described (Chevalier et al., 1995). For northern-blot experiments, total RNA was size fractionated by 6.6% (v/v) formaldehyde and 1.2% (w/v) agarose gel electrophoresis, transferred to Hybond N+ (Amersham, Buckinghamshire, UK) membranes by capillarity, and hybridized to random-primed labeled probes. Clones 5c02a05 and 5c01h12 cDNA encoded for partial sequence of α3- and β6-subunits of maize 20S proteasome, respectively (Shen et al., 1994). These are similar to PAC1 and PBF1 Arabidopsis components (Fu et al., 1998), and were kindly provided by the Maize RFLP Laboratory (Columbia, MO). Hybridizations were performed at 65°C as described in (Sambrook et al., 1989).
Protein Determination
Proteins were quantified according to the method of Bradford (1976). Bovine γ-globulin was used as the protein standard.
Footnotes
This work was supported by the Institut National de la Recherche Agronomique and by the Ministère de l'Education Nationale, de la Recherche, et de la Technologie (grant to G.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010612.
LITERATURE CITED
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R. Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol. 1996;133:1251–1263. doi: 10.1083/jcb.133.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn RV, Spitz DR, Liu X, Galoforo SS, Sim JE, Ridnour LA, Chen JC, Davis BH, Corry PM, Lee YJ. Metabolic oxidative stress activates signal transduction and gene expression during glucose deprivation in human tumor cells. Free Radic Biol Med. 1999;26:419–430. doi: 10.1016/s0891-5849(98)00217-2. [DOI] [PubMed] [Google Scholar]
- Bose S, Mason GGF, Rivett AJ. Phosphorylation of proteasomes in mammalian cells. Mol Biol Rep. 1999;26:11–14. doi: 10.1023/a:1006969517958. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brouquisse R, Gaudillère JP, Raymond P. Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol. 1998;117:1281–1291. doi: 10.1104/pp.117.4.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, James F, Pradet A, Raymond P. Asparagine metabolism and nitrogen distribution during protein degradation in sugar-starved maize root tips. Planta. 1992;188:384–395. doi: 10.1007/BF00192806. [DOI] [PubMed] [Google Scholar]
- Brouquisse R, James F, Raymond P, Pradet A. Study of glucose starvation in excised maize root tips. Plant Physiol. 1991;96:619–626. doi: 10.1104/pp.96.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, Masclaux C, Feller U, Raymond P. Protein hydrolysis and nitrogen remobilization in plant life and senescence. In: Lea PJ, Morot-Gaudry J-F, editors. Plant Nitrogen, INRA. Paris: Springer-Verlag; 2000. pp. 275–293. [Google Scholar]
- Chao CC, Ma YS, Stadtman ER. Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc Natl Acad Sci USA. 1997;94:2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Bourgeois E, Pradet A, Raymond P. Molecular cloning and characterization of six cDNAs expressed during glucose starvation in excised maize (Zea mays L.) root tips. Plant Mol Biol. 1995;28:473–485. doi: 10.1007/BF00020395. [DOI] [PubMed] [Google Scholar]
- Chevalier C, Lequerrec F, Raymond P. Sugar levels regulate the expression of ribosomal protein genes encoding protein S28 and ubiquitin-fused protein S27a in maize primary root tips. Plant Sci. 1996;117:95–105. [Google Scholar]
- Ciechanover A, Heller H, Elias S, Haas A, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci USA. 1980;77:1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi M, Petropoulos I, Emod I, Turlin E, Biville F, Friguet B. Protection from oxidative inactivation of the 20 S proteasome by heat-shock protein 90. Biochem J. 1998;333:407–415. doi: 10.1042/bj3330407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Dean RT, Fu S, Stocker R, Davies MJ. Biochemistry and pathology of radical-mediated protein oxidation. Biochem J. 1997;324:1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick LR, Moomaw CR, DeMartino GN, Slaughter CA. Degradation of oxidized insulin B chain by the multiproteinase complex macropain (proteasome) Biochemistry. 1991;30:2725–2734. doi: 10.1021/bi00224a022. [DOI] [PubMed] [Google Scholar]
- Dieuaide M, Brouquisse R, Pradet A, Raymond P. Increased fatty acid β-oxidation after glucose starvation in maize root tips. Plant Physiol. 1992;99:595–600. doi: 10.1104/pp.99.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Amrani A, Camara B, Gaudillere JP, Couee I. Accumulation of plastidial alanine-aminopeptidase in relation to plastid damage in cotyledons of dark-grown sugar beet seedlings. Plant Physiol Biochem. 1998;36:263–268. [Google Scholar]
- Fagan JM, Waxman L. Purification of a protease in red blood cells that degrades oxidatively damaged hemoglobin. Biochem J. 1991;277:779–786. doi: 10.1042/bj2770779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Murray P, Giordano CV, Passeron S, Barneix AJ. Purification and characterization of 20S proteasome from wheat leaves. Plant Sci. 1997;125:127–136. [Google Scholar]
- Friguet B, Stadtman ER, Szweda LI. Modification of glucose-6-phosphate dehydrogenase by 4-hydroxy-2-nonenal: formation of cross-linked protein that inhibits the multicatalytic protease. J Biol Chem. 1994;269:21639–21643. [PubMed] [Google Scholar]
- Fu H, Doelling JH, Arendt CS, Hochstrasser M, Vierstra RD. Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics. 1998;149:677–692. doi: 10.1093/genetics/149.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Derevier A, Fleck J. Cell cycle-dependent proteolysis in plants: identification of the destruction box pathway and metaphase arrrest produced by the proteasome inhibitor MG132. Plant Cell. 1998;10:2063–2075. doi: 10.1105/tpc.10.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Jamet E, Philipps G, Parmentier Y, Gigot C, Fleck J. Molecular characterization of a beta-type proteasome subunit from Arabidopsis thaliana co-expressed at a high level with an alpha-type proteasome subunit early in the cell cycle. Plant J. 1994;6:537–546. doi: 10.1046/j.1365-313x.1994.6040537.x. [DOI] [PubMed] [Google Scholar]
- Giulivi C, Pacifici RE, Davies KJA. Exposure of hydrophobic moieties promotes the selective degradation of hydrogen peroxide-modified hemoglobin by the multicatalytic proteinase complex, proteasome. Arch Biochem Biophys. 1994;311:329–341. doi: 10.1006/abbi.1994.1245. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Davies KJA. Degradation of oxidized proteins in mammalian cells. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- Grune T, Reinheckel T, Joshi M, Davies KJA. Proteolysis in cultured liver epithelial cells during oxidative stress. J Biol Chem. 1995;270:2344–2351. doi: 10.1074/jbc.270.5.2344. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hilt W, Wolf DH. Stress-induced proteolysis in yeast. Mol Microbiol. 1992;6:2437–2442. doi: 10.1111/j.1365-2958.1992.tb01419.x. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Ann Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Ichihara A, Tanaka K. Roles of proteasomes in cell growth. Mol Biol Rep. 1995;21:49–52. doi: 10.1007/BF00990970. [DOI] [PubMed] [Google Scholar]
- Irving DE, Hurst PL. Respiration, soluble carbohydrates and enzymes of carbohydrate metabolism in tips of harvested asparagus spears. Plant Sci. 1993;94:89–97. [Google Scholar]
- Ismail I, Debellis L, Alpi A, Smith SM. Expression of glyoxylate cycle genes in cucumber roots responds to sugar supply and can be activated by shading or defoliation of the shoot. Plant Mol Biol. 1997;35:633–640. doi: 10.1023/a:1005840522049. [DOI] [PubMed] [Google Scholar]
- James F, Brouquisse R, Pradet A, Raymond P. Changes in proteolytic activities in glucose-starved maize root tips: regulation by sugars. Plant Physiol Biochem. 1993;31:845–856. [Google Scholar]
- James F, Brouquisse R, Suire C, Pradet A, Raymond P. Purification and biochemical characterization of a vacuolar serine endopeptidase induced by glucose starvation in maize roots. Biochem J. 1996;320:283–292. doi: 10.1042/bj3200283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, Bligny R, Douce R. Biochemical changes during sucrose deprivation in higher plant cells. J Biol Chem. 1986;261:3193–3199. [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Medina R, Wing SS, Goldberg AL. Increase in levels of polyubiquitin and proteasome mRNA in skeletal muscle during starvation and denervation atrophy. Biochem J. 1995;307:631–637. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyasu Y, Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996;111:1233–1241. doi: 10.1104/pp.111.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrel PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ogiso Y, Tomida A, Kim HD, Tsuruo T. Glucose starvation and hypoxia induce nuclear accumulation of proteasome in cancer cells. Biochem Biophys Res Commun. 1999;258:448–452. doi: 10.1006/bbrc.1999.0635. [DOI] [PubMed] [Google Scholar]
- Ozaki M, Fujinami K, Tanaka K, Amemiya Y, Sato T, Ogura N, Nakagawa H. Purification and initial characterization of the proteasome from the higher plant Spinacia oleracea. J Biol Chem. 1992;267:21678–21684. [PubMed] [Google Scholar]
- Peeters KMU, Van Laere AJ. Ammonium and amino acid metabolism in excised leaves of wheat (Triticum aestivum) senescing in the dark. Physiol Plant. 1992;84:243–249. [Google Scholar]
- Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJA, Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J. 1998;335:637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivett J, Savory PJ, Djaballah H. Multicatalytic endopeptidase complex: proteasome. Methods Enzymol. 1994;244:331–350. doi: 10.1016/0076-6879(94)44026-3. [DOI] [PubMed] [Google Scholar]
- Rivett JA. Preferential degradation of the oxidatively modified form of glutamine synthetase by intracellular mammalian proteases. J Biol Chem. 1985;260:300–305. [PubMed] [Google Scholar]
- Sahakian JA, Szweda LI, Friguet B, Kitani K, Levine RL. Aging of the liver: proteolysis of oxidatively modified glutamine synthetase. Arch Biochem Biophys. 1995;318:411–417. doi: 10.1006/abbi.1995.1248. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EJ, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sarath G, De la Motte RS, Wagner FW. Protease assay methods. In: Beynon RJ, Bond JS, editors. Proteolytic Enzymes: A Practical Approach. Oxford: IRL Press; 1989. pp. 25–55. [Google Scholar]
- Schliephacke M, Kremp A, Schmid HP, Köhler K, Kull U. Prosomes (proteasomes) of higher plants. European J Cell Biol. 1991;55:114–121. [PubMed] [Google Scholar]
- Schmidtke G, Kraft R, Kostka S, Henklein P, Frommel C, Lowe J, Huber R, Kloetzel PM, Schmidt M. Analysis of mammalian 20S proteasome biogenesis: The maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis. EMBO J. 1996;15:6887–6898. [PMC free article] [PubMed] [Google Scholar]
- Shen B, Carneiro N, Torres-Jerez I, Stevenson B, McCreery T, Helentjaris T, Baysdorfer C, Almira E, Ferl RJ, Habben JE et al. Partial sequencing and mapping of clones from two maize cDNA libraries. Plant Mol Biol. 1994;26:1085–1101. doi: 10.1007/BF00040691. [DOI] [PubMed] [Google Scholar]
- Skoda B, Malek L. Dry pea seed proteasome. Plant Physiol. 1992;99:1515–1519. doi: 10.1104/pp.99.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack PR, Waxman L, Fagan JM. Activation of the multicatalytic endopeptidase by oxidants: effects on enzyme structure. Biochemistry. 1996;35:7142–7149. doi: 10.1021/bi9518048. [DOI] [PubMed] [Google Scholar]
- Tassi F, Maestri E, Restivo FM, Marmiloni N. The effects of carbon starvation on cellular metabolism and protein and RNA synthesis in Gerbera callus culture. Plant Sci. 1992;83:127–136. [Google Scholar]
- Ullrich O, Reinheckel T, Sitte N, Hass R, Grune T, Davies KJA. Poly-ADP ribose polymerase activates nuclear proteasome to degrade oxidatively damaged histones. Proc Natl Acad Sci USA. 1999;96:6223–6228. doi: 10.1073/pnas.96.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M, Manabe Y, Uchimiya H. Phosphorylation of the C2 subunit of the proteasome in rice (Oryza sativa L.) FEBS Lett. 1997;403:313–317. doi: 10.1016/s0014-5793(97)00073-2. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. Proteolysis in plants: mechanisms and functions. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Webster P, Henry M. Sucrose regulation of protein synthesis in pea root meristem cells. Environ Exp Bot. 1987;27:253–262. [Google Scholar]
- Yang Y, Früh K, Ahn K, Peterson PA. In vivo assembly of the proteasomal complexes, implications for antigen processing. J Biol Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- Zhang H, Olejnicka B, Öllinger K, Brunk UT. Starvation-induced autophagocytosis enhances the susceptibility of insulinoma cells to oxidative stress. Redox Rep. 1996;2:235–247. doi: 10.1080/13510002.1996.11747056. [DOI] [PubMed] [Google Scholar]