Abstract
The sense of smell is arguably our most primal faculty and also the least understood. Even our own olfactorily impaired species is capable of detecting ≈10,000 distinct scents [Buck, L. & Axel, R. (1991) Cell 65, 175–187]. To achieve that amazing diversity, mammals have ≈1,000 olfactory genes, which accounts for ≈3% of their entire genome [Mombaerts, P. (1999) Science 286, 707–711]. The olfactory receptors (ORs) are believed to be seven-helix transmembrane proteins, with an odorant-binding site on the periplasmic domain and a G protein-binding site on the cytoplasmic domain. Odorants first bind to an OR, which then undergoes some structural change that triggers the G protein activation and the following cascade of events leading to nerve cell activity. The structural details of ORs, however, remain to be determined. In this paper, we will describe a hypothesis in which metal ions play an important role for odorant recognition. We analyze the predicted structure and consensus sequence of the ORs and propose a metal-binding site in the loop between fourth and fifth helix (4–5 loop). We have prepared synthetically a pentapeptide that contains this putative binding site and find that it not only has high affinity for binding Cu(II) and Zn(II) ions, but that it also undergoes a dramatic transition to an α-helical structure upon metal ion binding. Based on these observations, we propose a “shuttlecock” mechanism for the possible structural change in ORs upon odorant binding. This mechanism involves membrane penetration of the 4–5 loop after residue charge neutralization by metal ion binding.
Keywords: olfaction‖G protein-coupled receptors‖transmembrane protein
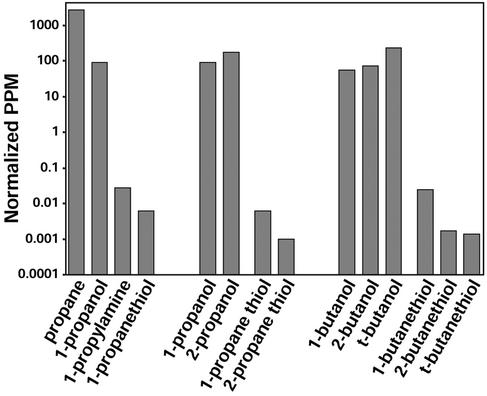
Inorganic chemists know as a rule of thumb that if a volatile compound is a good ligand for metal ion coordination complexes, it probably smells strongly; this observation has lead to recent advances in artificial olfaction (1). The only notable exceptions to this rule are CO and NO, which are produced endogenously as neural messengers (2) and, therefore, elicit no olfactory response. In general, the human olfactory system is extremely sensitive to amines and thiols (good ligands for metal ions) but not to alcohols (which are only weak ligands; ref. 3), as shown in Fig. 1. For example, one can smell methylthiol at less than 1 ppb, methylamine at 18 ppb, but methanol only above 100 ppm, and methane is undetected even at 106 ppm.
Figure 1.
Human olfactory thresholds. Shown are gas phase concentrations at threshold of detection (3); vapors of liquids have been normalized (5) by analyte vapor pressure at 298 K (6).
Odorants needs to bind to an olfactory receptor (OR) to trigger the cascade of events that finally enables us to smell: methylthiol is bound by some OR >1 million times stronger than the OR that responds to methanol, and methylamine is bound >100,000 times stronger than methanol. Differences in hydrogen bonding or van der Waals interactions are insufficient to account for this range, but differences in Lewis basicity to metal ions can. This range of affinities is critical to the pattern recognition processes proposed for neural computation (4).
Thus, the most natural explanation to account for this odorant affinity difference is coordination to a metal ion bound in an OR. Cu(II) or Zn(II) are particularly likely candidates, because they have strong amine- and thiol-binding property and are frequently found in metalloproteins. More than two decades ago, Crabtree (7) prophetically speculated that Cu(I) might be found in ORs, because of the high olfactory sensitivity to amines and thiols.
Consistent with the metalloprotein hypothesis, the ORs also show unusual differences in their shape selectivity for alcohols compared with thiols. Substitution at the α-carbon of alcohols increases the olfactory threshold for alcohols; i.e., greater steric bulk gives weaker binding, as expected for sterically restricted binding sites. In surprising contrast, however, α-substitution of thiols normally decreases the olfactory threshold for thiols. This observation can be explained by OR binding sites that contain a coordinately accessible metal ion, because α-substitution increases the Lewis basicity of the thiol and, hence, its strength of binding to metal ions, as long as the increased steric hindrance is not too great. In keeping with this trend, the human nose is often more sensitive to secondary amines than primary amines; comparisons among amines, however, are complicated by issues of protonation in the olfactory mucosa.
Materials and Methods
Peptide Synthesis.
A five-residue peptide, with sequence HAKCE, was synthesized to study its metal-binding property. The C terminus was amide-capped and the N terminus was capped with an acetyl group to prevent terminus binding to metal ions. The peptide was synthesized by using solid-phase methods and the Fmoc/t-Bu strategy on a Multiple Symphony synthesizer (Protein Technologies, Woburn, MA). Amino acid coupling using N-hydroxybenzotriazole chemistry and Fmoc deprotection was performed by using 20% piperidine in dimethylformamide. The peptide was cleaved with a trifluoroacetic acid/thioanisole/ethanedithiole/anisole/water mixture (82.5/5/2.5/5/5; Reagent K) and purified by reversed-phase C18 HPLC with a linear gradient of acetonitrile and water. The identity and purity of the peptide were confirmed by HPLC and electrospray mass spectroscopy.
CD Spectroscopy.
All CD spectra were measured on a Jasco J-700 spectropolarimeter (Jasco, Easton, MD) at 20°C. All measurements were made on samples whose concentrations were 20 μM (20 mM potassium phosphate, pH 7.4). The binding constants were determined by standard techniques (8).
Molecular Modeling.
Molecular modeling was performed on an Indigo2 Extreme running IRIX V.6.2 with the program MODELER and BIOPOLYMER in INSIGHT-II. (Accelrys, San Diego) Sequence analysis used public domain software: TMHMM V.2.0 (www.cbs.dtu.dk/services/TMHMM).
Results and Discussion
The hidden Markov model (HMM) has been used with high confidence to predict the secondary structure of membrane proteins (9), because residues buried in membrane are normally hydrophobic, and those exposed at loops are hydrophilic, which makes for very distinctive sequence patterns. The ORs are well fit as seven-helix membrane proteins by using this approach (Fig. 2). Structural models have also been calculated from homology modeling (10) by using the structure of bovine rhodopsin, which is also a seven-helix membrane protein. Goddard and coworkers (10) observed in their model that a long narrow pocket is formed in the middle of the membrane helices that seem capable of binding linear alcohols. Their model is consistent with the observed shape selectivity for alcohols, but it does not provide an explanation for the reverse selectivity with substituted thiols.
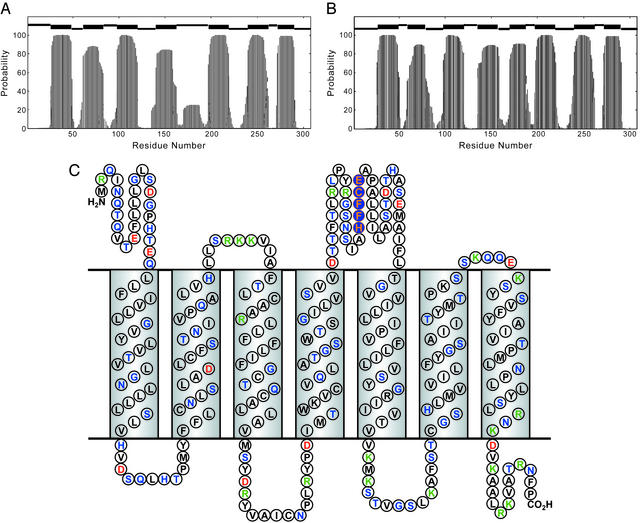
Figure 2.
(A) Secondary structure prediction of hOR o2d2 using the HMM for the native sequence. At the top, the thick bars indicate predicted TM helices and the higher and lower thin bars designate periplasmic and cytoplasmic loops, respectively. (B) Secondary structure prediction of hOR o2d2 using HMM for the charge-neutralized structure (as modeled by Glu-180→Val mutation). (C) Secondary structure prediction with amino acid aligned to the TM helix and loop regions. Coding: hydrophobic residues in black, polar residue in green, positively charged in blue, and negatively charged in red. The consensus metal ion-binding site (conserved in nearly three-quarters of known sequences) is highlighted in orange on blue.
In looking for possible metal ion-binding sites in the genome sequences of ORs, we first made the general observation that zinc and copper metalloproteins usually have clusters of the metal-coordinating amino acids His, Cys, Glu, or Asp: i.e., two or more metal-binding amino acids are generally found to be separated by no more than four amino acids. This proximity is likely a consequence of entropic requirements in the coordination of multiple residues to the metal ion. Keeping that in mind, we immediately find the consensus sequence HXXC[DE] in the 4–5 loop (Fig. 2C) of many OR sequences (11), where Xs generally are hydrophobic residues (especially phenylalanine). More specifically, of the 83 human olfactory receptor (hOR) sequences we extracted from the SWISS-PROT database, 70% (i.e., 58) are found to contain this motif. In a recent study, Zuzulya and coworkers (12) have identified some 347 putative hOR genes; we find the HXXC[DE] consensus sequence in 74% (i.e., 257) of these sequences, as well.
To test this sequence as a possible metal ion-binding site, we synthesized a pentapeptide by standard solid-state techniques. To retain water solubility, it was necessary to set XX to alanine-lysine (AK). HAKCE thus was examined for its metal ion-binding capability. As expected for a short peptide, the CD spectrum of HAKCE shows that it is a random coil (Fig. 3).
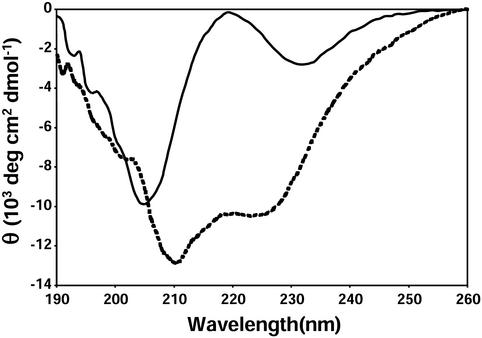
Figure 3.
Far-UV CD spectrum of HAKCE peptide, in the absence of metal ions (solid line) and in the presence of 1.0 equivalent of Cu(II) added (dotted line); 20 mM peptide, pH 7.4 potassium phosphate buffer, 298 K.
Surprisingly, upon addition of 1.0 equivalents of Cu2+, the far-UV CD spectrum changes dramatically to one characteristic of an α-helix. A careful titration with CD monitoring shows 1:1 binding with a dissociation constant between the peptide and Cu2+ of 500 nM. The formation of an α-helix normally requires a relatively lengthy peptide (≈15 residues) to provide sufficient hydrogen bonding to stabilize the helix (13). To the best of our knowledge, no helix-forming oligopeptide as short as five residues has been previously reported. In this special case, however, the chelation of the oligopeptide residues forces its conformation into a helix. Similar helix induction is observed for Zn2+ or Ni2+ (with somewhat larger dissociation constants), but not for Cu+. These observations provide significant evidence for the possible role of Cu(II), Zn(II), or Ni(II) in some ORs, but does not rule out Cu(I), because the real protein environment is much more complex. It is worth noting in passing that one of the distinctive symptoms of dietary zinc deficiency is anosmia (i.e., loss of the sense of smell; ref. 14).
If the active sites of some ORs indeed contain a metal ion, how might an odorant binding create a response mechanism? As a starting point, let us examine the possible structure of these membrane-bound proteins. ORs fall into the broad category of G protein-coupled receptors, which are predicted to be seven-helix membrane proteins and which include bovine rhodopsin, bacteriorhodopsin, halorhodopsin, and sensory rhodopsin. Among those, bacteriorhodopsin is the best understood, and its structure has been available since 1990 (15); high-resolution structures of bacteriorhodopsin (16), bovine rhodopsin (17), halorhodopsin (18), and sensory rhodopsin (19) are also now available. Bacteriorhodopsin is composed of seven-helix bundle protein scaffold covalently linked to all-trans retinal through Lys-216. Absorption of a photon by retinal causes its isomerization to a 13-cis configuration, inducing torsional strain in the chromophore as well as disrupting the Schiff base/Asp-85 ion pair. This isomerization increases the local energy and moves two helices relative to the other five (20), opening a pathway for proton transfer.
Similarly, for a metal ion containing OR, odorants that bind to the metal ion (e.g., thiols, amines, etc.) could disrupt the charge balance by replacing one of the metal-ligated amino acid residues (or a coordinated water or hydroxide ion), resulting in protein structural rearrangement. Alternatively, odorant ligation would also increase the local steric interactions in the active site with a similar effect on protein structure.
Such a mechanism of action for OR–odorant interactions explains the very high sensitivities for metal ion coordinating analytes (e.g., Fig. 1). Metal-ligand bonds range in their bond enthalpies from ≈40 to ≈200 kJ/mol. In contrast, the enthalpy of absorption (e.g., into polymers) relies on weak, van der Waals and dipolar interactions and is only ≈5–20 kJ/mol (i.e., roughly a tenth of a metal bond) for small molecules. Therefore, the equilibrium constant for absorption will typically only be about 5 × 10−5 as large as that for ligation to metal ions.
A more detailed examination of the OR structure leads to a provocative hypothesis concerning the structural change upon odorant binding (Fig. 4). The loop between the putative 4th and 5th transmembrane helices of ORs is uncommonly long and surprisingly hydrophobic. This loop is sufficiently long to be transmembrane helix, but the HMM algorithm does not designate it as such, because of the anionic residue (Asp or Glu) in the middle of the sequence, which would interact unfavorably with the hydrophobic membrane environment. After binding to a metal ion, however, the anionic charge is cancelled. For example, if we replace just that single anionic residue by Val, the HMM algorithm projects an entirely different structure for the specific sequence of hOR-o2d2 (21).
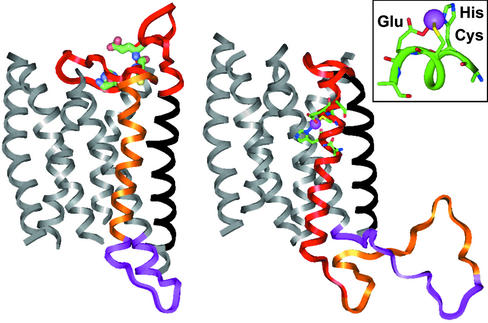
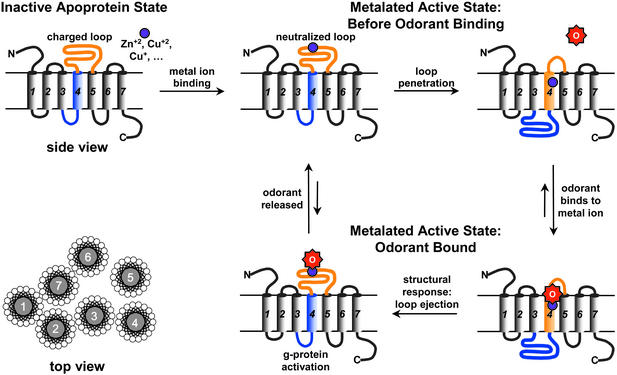
Figure 4.
The homology models of the hOR o2d2 are constructed by using the program MODELER and BIOPOLYMER in INSIGHT-II (Accelrys). (Left) A model of the nonmetalated apoprotein; the predicted hOR transmembrane helix is aligned to the transmembrane helix of bovine rhodopsin (PDB ID code 1F88). (Right) Proposed structure after binding a metal ion (e.g., Cu2+ or Zn2+); the alignment shifts to reflect the speculated 4–5 loop insertion and ejection of the fourth helix. (Inset) Predicted coordination geometry to the metal ion from the glutamate, histidine, and cysteine residues of the consensus sequence HXXC[DE] in the 4–5 loop.
The structure resulting from metal ion binding would be consistent with a new helical region originating from the original 4–5 extramembrane loop; the metal binding site HXXC[DE] is located in the middle of this new helical region. In addition, a tetrahedrally ligating metal ion [e.g., Cu(II) or Zn(II)] can bind to the residues of this new α-helix (to one nitrogen atom from His, one sulfur from Cys, and one oxygen from Glu) without any significant unfavorable steric interaction (Figs. 4 and 5).
Figure 5.
The proposed mechanism of olfactory response via a transmembrane shuttlecock. In the absence of odorant binding, the stable state is the embedded loop conformation (Upper Right). Upon odorant binding, the primary structural response is loop ejection (Lower Left). The G protein activation is a kinetic phenomenon in this mechanism; full equilibration of the OR with odorant leads to no further G protein activation, which may, in part, account for the process of olfactory adaptation (i.e., loss of response after initial exposure to a constant odorant concentration).
The possibility of two different stable conformations for the OR seven-helix bundle leads to an interesting “shuttlecock” hypothesis for metal ion-assisted odorant recognition, as shown in Fig. 5. Initially, the OR is a seven-helix bundle in the conformation currently accepted. Upon metal binding, however, the anionic charge of the 4–5 loop is neutralized, permitting the loop to become helical and penetrate into the membrane, pushing the fourth helix out of the membrane. This configuation, we speculate, is the active form of metal ion-containing ORs.
When an odorant capable of metal ion ligation (e.g., amines, thiols, carboxylic acids, etc.) approaches the active form of the OR, it replaces one of the coordinated amino acids or a bound water or hydroxide, disrupts the local charge and steric balance, and causes the original 4–5 loop to eject from the membrane, permitting the original fourth helix to return into the membrane. This sequence of events causes large structural change in the cytoplasmic domain (Figs. 4 and 5) and activates the G protein. Meanwhile, the metal-binding site will then be exposed to the extracellular water, shifting the equilibrium of ligation and enhancing the departure of the bound odorant. When the odorant leaves the OR, the 4–5 loop again turns into a hydrophobic helix and pushes the fourth helix out of the membrane once again, returning the OR back to its active state.
This hypothesis is testable. Overexpression and purification of olfactory proteins is possible now that their genome has been identified. Identification of metal ion content by trace metal analysis [e.g., using inductively coupled plasma mass spectrometry (ICP-MS)] or, in the case of Cu(II), electron paramagnetic resonance is quite possible. In addition, identification of specific OR sequences that respond strongly to amines and thiols may be possible by using Ca2+-imaging technique and single-cell RT-PCR (22).
There is a possible precedent for the conformational change we are suggesting. Hunt and coworkers (23) discovered that a 36-residue peptide can insert into lipid bilayers as a transmembrane helix after one of its Asp residues is neutralized by lowering the pH. This observation supports our hypothesis that the OR 4–5 loop may turn into a TM helix after its charge is cancelled by ligation to a metal ion. It is also possible that some olfactory proteins are indeed eight-helix bundles after metal ion binding, as predicted by the HMM.
We can use a standard hydrophobicity scale, e.g., Kyte-Doolittle (24), to estimate which helix is likely to eject when the 4–5 loop inserts. For hOR o2d2 (19), the predicted fourth helix has a total hydrophobicity value of 21.9 for residues 140–162; the 4–5 loop has total hydrophobicity value of 21.1 for residues 170–192 (and a total hydrophobicity value of 24.3 after charge-neutralization by Val substitution of Glu); the fifth helix has a total hydrophobicity value of 39.9 for residues 197–219. Because the hydrophobicity of the 4–5 loop and fourth helix are so close, it is very likely that the 4–5 loop can replace the fourth helix when the metal binding cancels the charge on Glu-180 (E180) and decreases polarity on H176. The fifth helix, however, should be more difficult to eject, because it is much more hydrophobic than the 4–5 loop. More generally, the fifth helix is also much more hydrophobic than the fourth helix and the 4–5 loop in the consensus sequence of hOR.
A “shuttlecock” protein motion has not been previously suggested in other membrane bound proteins. Bacteriorhodopsin and bacterial chemotaxis receptors have only small sliding motions, perhaps because their membrane helices are tightly packed (24). Larger motions are observed for acetylcholine receptor and potassium channel (25), where a whole subunit moves to open or close the channel. It will prove interesting to apply atomic force microscopy to ORs and measure the forces necessary to pull helices out of their membrane environments, as has been done recently for bacteriorhodopsin (26).
In conclusion, we have assembled both computer modeling and experimental evidence for the binding of metal ions to a consensus sequence found in ORs. From HMM, we believe that binding of a metal ion precipitates a major rearrangement of the transmembrane helices. Upon odorant binding to the metal ion, a shuttlecock motion that converts loop to helix provides a useful hypothesis for the mechanism of activation of the coupled cytoplasmic G protein. This rearrangement of transmembrane proteins may be generalized to charge-neutralization events other than metal ion coordination: large extramembrane loops may become transmembrane helices upon protonation of anionic residues, deprotonation of cationic residues, or formation of internal salt bridges between oppositely charged residues.
Acknowledgments
We thank Dr. Avijit Sen, Dr. Neal A. Rakow, Prof. K. Schulten, and Prof. P. G. Wolynes for valuable suggestions. This work was supported by National Institutes of Health Grant HL25934.
Abbreviations
- OR
olfactory receptor
- HMM
hidden Markov model
- hOR
human OR
References
- 1.Rakow N A, Suslick K S. Nature. 2000;406:710–714. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]
- 2.Snyder S H, Jaffrey S R, Zakhary R. Brain Res Rev. 1998;26:167–175. doi: 10.1016/s0165-0173(97)00032-5. [DOI] [PubMed] [Google Scholar]
- 3.Devos M, Patte F, Rouault J, Laffort P, Van Gemert L J. Standardized Human Olfactory Thresholds. New York: Oxford Univ. Press; 1990. [Google Scholar]
- 4.Hopfield J J. Proc Natl Acad Sci USA. 1999;96:12506–12511. doi: 10.1073/pnas.96.22.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doleman B J, Severin E J, Lewis N S. Proc Natl Acad Sci USA. 1998;95:5442–5447. doi: 10.1073/pnas.95.10.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaws C L, Lin X, Bu L. Handbook of Vapor Pressure. London: Gulf; 1994. [Google Scholar]
- 7.Crabtree R H. J Inorg Nucl Chem. 1978;40:1453. [Google Scholar]
- 8.Huffman D L, Rosenblatt M M, Suslick K S. J Am Chem Soc. 1998;120:6183–6184. [Google Scholar]
- 9.Eddy S R. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 10.Floriano W B, Vaidehi N, Goddard W A, III, Singer M S, Shepherd G M. Proc Natl Acad Sci USA. 2000;97:10712–10716. doi: 10.1073/pnas.97.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 12.Zuzulya S, Echeverri F, Nguyen T. Genome Biol. 2001;2:18.1–18.12. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrabartty A, Baldwin R L. Adv Protein Chem. 1995;46:141–176. [PubMed] [Google Scholar]
- 14.Ackerman B H, Kasbekar N. Pharmacotherapy. 1997;17:482–496. [PubMed] [Google Scholar]
- 15.Henderson R, Baldwin J M, Ceska T A, Zemlin F, Beckmann E, Downing K H. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 16.Faham S, Bowie J U. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 17.Palczewski K, Kumasaka T, Hori T, Behnke C A, Motoshima H, Fox B A, Le Trong I, Teller D C, Okada T, Stenkamp R E, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 18.Kolbe M, Besir H, Essen L O, Oesterhelt D. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 19.Luecke H, Schobert B, Lanyi J K, Spudich E N, Spudich J L. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramaniam S, Henderson R. Nature. 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 21.Lane R P, Cutforth T, Young J, Athanasiou M, Friedman C, Rowen L, Evans G, Axel R, Hood L, Trask B J. Proc Natl Acad Sci USA. 2001;98:7390–7395. doi: 10.1073/pnas.131215398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malnic B, Hirono J, Sato T, Buck L B. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 23.Hunt J F, Rath P, Rothschild K J, Engelman D M. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 24.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 25.Gerstein M, Chothia C. Science. 1999;285:1682–1683. doi: 10.1126/science.285.5434.1682. [DOI] [PubMed] [Google Scholar]
- 26.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub H E, Muller D J. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]