Abstract
Recent work shows that S-adenosylmethionine (AdoMet) helps maintain normal liver function as chronic hepatic deficiency results in spontaneous development of steatohepatitis and hepatocellular carcinoma. The mechanisms by which these nontraditional functions of AdoMet occur are unknown. Here, we use knockout mice deficient in hepatic AdoMet synthesis (MAT1A−/−) to study the proteome of the liver during the development of steatohepatitis. One hundred and seventeen protein spots, differentially expressed during the development of steatohepatitis, were selected and identified by peptide mass fingerprinting. Among them, 12 proteins were found to be affected from birth, when MAT1A−/− expression is switched on in WT mouse liver, to the rise of histological lesions, which occurs at ≈8 months. Of the 12 proteins, 4 [prohibitin 1 (PHB1), cytochrome c oxidase I and II, and ATPase β-subunit] have known roles in mitochondrial function. We show that the alteration in expression of PHB1 correlates with a loss of mitochondrial function. Experiments in isolated rat hepatocytes indicate that AdoMet regulates PHB1 content, thus suggesting ways by which steatohepatitis may be induced. Importantly, we found the expression of these mitochondrial proteins was abnormal in ob/ob mice and obese patients who are at risk for nonalcoholic steatohepatitis.
S-adenosylmethionine (AdoMet) is an essential metabolite in all cells. AdoMet serves as the methyl donor for many biological methylation reactions, provides the propylamino group for the synthesis of polyamines, and, in the liver, is an intermediary metabolite in the synthesis of glutathione (1–4). Methionine adenosyltransferase (MAT) catalyzes the only known AdoMet biosynthetic reaction from methionine and ATP (5, 6). In mammals, there are three MAT isoforms (MAT I, MAT II, and MAT III) encoded by two distinct genes (MAT1A and MAT2A; refs. 7–11). MAT I and MAT III are a tetramer and a dimer, respectively, of a catalytic α1-subunit encoded by the gene MAT1A (4, 6). A second gene, MAT2A, encodes for a catalytic α2-subunit that forms MAT II (4, 6). The three enzymes catalyze the same reaction, but their cellular distribution, kinetics, and regulation are different. Although MAT II is the only isoform expressed in extrahepatic tissues (refs. 12 and 13; with the exception of the pancreas, which expresses both MAT1A and MAT2A; ref. 14), both MAT III and I, and to a lesser extent MAT II, are expressed in the liver (13). MAT II expression also predominates in fetal liver and is progressively replaced by MAT III and I during liver development (13). In adult liver, increased expression of MAT2A is associated with rapid growth or de-differentiation of the liver, as during liver regeneration induced by partial hepatectomy (15, 16) and in hepatocarcinogenesis (17–19).
MAT1A silencing in extrahepatic tissues and during liver neoplastic transformation correlates with methylation of the gene promoter and its association with nonacetylated histones (20). Seemingly, these epigenetic modifications may lead to a condensed, transcriptionally inactive chromatin, as suggested by the observation that MAT1A expression was up-regulated in HepG2 hepatoma cells in the presence of inhibitors of DNA methylation and histone deacetylation (20). It has been found subsequently that liver samples from cirrhotic patients, who are known to have an impaired methionine metabolism and are at higher risk to develop hepatocellular carcinoma (HCC), show increased methylation of MAT1A promoter, reduced abundance of MAT1A mRNA, and decreased MAT activity (21).
The finding that MAT1A is expressed predominantly in adult liver and that MAT III and I are replaced by MAT II during liver neoplastic transformation suggests that these MAT isoforms have distinct rather than overlapping biological roles. This question has been addressed by the generation of MAT1A knockout (KO; MAT1A−/−) mice (22). Homozygous KO mice were born at normal Mendelian ratios, were fertile, and did not show gross anatomical or behavioral abnormalities (22). In adult KO mice, liver MAT2A expression was up-regulated to compensate for the absence of MAT III and I (22). However, KO mice have hypermethioninemia and reduced hepatic AdoMet content, ≈20% of that in WT animals (22). These results indicate that up-regulation of MAT2A expression in adult MAT1A−/− mouse liver cannot compensate for the absence of MAT III and I in maintaining normal AdoMet levels. This finding is consistent with known differences in kinetics and regulation between MAT isoforms. Whereas MAT II has a low Km for methionine and is inhibited by physiological concentrations of AdoMet (12, 23), MAT III and I have, respectively, a high and intermediate Km for methionine, are activated by methionine and ATP, and are not inhibited by physiological concentrations of AdoMet (6–8, 11, 24, 25).
Cellular content of AdoMet seems to be related to the differentiation status of the hepatocyte (26). In this regard, quiescent and proliferating hepatocytes display different AdoMet levels, being lower in growing (26). This correlation has been observed in rat liver after partial hepatectomy (which triggers the proliferation of the remaining parenchymal cells to divide quickly and restore the original liver mass), where AdoMet levels are dramatically reduced shortly after the intervention coinciding with the onset of DNA synthesis and the induction of the early-response genes (15, 16, 27). When this decrease in AdoMet after partial hepatectomy was prevented by the administration of AdoMet, hepatocyte DNA synthesis was inhibited (19). Recent work shows that AdoMet helps turns off the hepatocyte responses to growth factors, and that without this protection, the uncontrolled growth of cancer takes place (16). Thus, it has been shown that KO mice deficient in AdoMet synthesis (MAT1A−/−) spontaneously develop oxidative stress, nonalcoholic steatohepatitis (NASH), and hepatocellular carcinoma (22, 28), and in hepatoma cell lines, AdoMet inhibits growth and induces apoptosis (18). The medical implication of these observations has emerged by the finding that patients with liver cirrhosis have impaired synthesis of AdoMet (29, 30). The precise mechanisms by which these nontraditional functions of AdoMet take place are unknown. Here, we use MAT1A−/− mice to carry out an extensive proteomic analysis of the liver during the development of NASH. Our results show the existence of a network of mitochondrial proteins regulated by AdoMet that participate in the pathogenesis of the disease.
Materials and Methods
Materials.
The following reagents and materials were used: anti-cytochrome c oxidase (COX) subunits I and II antibodies (Molecular Probes), anti-prohibitin 1 (PHB1) antibody (Calbiochem), anti-ATP synthase β-subunit (Molecular Probes), electrophoresis reagents (Bio-Rad), trypsin (Promega), urea and collagenase (GIBCO/BRL), and thiourea (Merck). All other chemical reagents were from Sigma. Animals were from our inbred colony and were treated humanely, according to our institution's guidelines. The human research review committee of the Hospital Príncipe de Asturias approved this study.
High-Throughput Proteomic Analysis.
Liver samples were homogenized in 20 volumes of lysis buffer containing 7 M urea, 2 M thiourea, 4% (vol/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 1% (vol/vol) DTT, and 0.5% Bio-Lyte 3/10 ampholytes. The homogenates were centrifuged at 100,000 × g for 45 min at 15°C. Protein concentration was determined in the supernatants with the Bradford assay kit (Bio-Rad) by using albumin diluted in lysis buffer as standard. First-dimensional isoelectrofocusing was performed on a Protean IEF cell (Bio-Rad) by using 17-cm ReadyStrips IPG strips (Bio-Rad) with different pH ranges. Sample (300–700 μg of protein) was loaded, and active rehydration was performed for 12 h at 50 V and 20°C. Gels were run for 60,000 V · h by using a progressively increasing protocol implemented by the manufacturer. IPG strips were equilibrated in 50 mM Tris⋅HCl, pH 7.5/6 M urea/30% (vol/vol) glycerol/2% (vol/vol) SDS/2% (vol/vol) DTT and then incubated in the same buffer containing 2.5% iodoacetamide and bromophenol blue, in the absence of DTT. IPG strips were directly loaded onto 12.5% polyacrylamide gels (18 cm × 20 cm × 1 mm) and sealed with low melting point agarose. Second-dimensional SDS/PAGE gels were run for 15 h. Gels were stained with Phastgel Blue R prepared in 65% H2O/25% ethanol/10% acetic acid. Alternatively, silver staining was performed by using the silver staining kit from Amersham Pharmacia. Images were digitized with a Bio-Rad imaging densitometer and analyzed by using PDQUEST software. Qualitative and quantitative differences were detected and were only accepted when at least a twofold change was confirmed in five independent experiments. Gel spots corresponding to proteins differentially expressed were picked up manually and processed on a MassPrep station from Micromass (Manchester, U.K.). Gel specimens were destained with 50 mM ammonium bicarbonate/50% (vol/vol) acetonitrile (Coomassie-stained gels) or 15 mM potassium ferricyanide/50 mM sodium thiosulphate (silver-stained gels). Then, proteins were reduced with 10 mM DTT in 100 mM ammonium bicarbonate and alkylated with 55 mM iodoacetamide in the same buffer. In-gel protein digestion was performed with 6 ng/μl trypsin in 50 mM ammonium bicarbonate for 5 h at 37°C. The resulting peptides were extracted with 1% (vol/vol) formic acid/2% (vol/vol) acetonitrile. Finally, 2-μl samples were mixed with 2 μl of a saturated solution of α-cyano-4-hydroxy-transcinnamic acid in trifluoroacetic acid 0.1%/50% (vol/vol) acetonitrile and spotted into a matrix-assisted laser desorption ionization (MALDI) target plate. Tryptic digests were then analyzed on a MALDI–time-of-flight (TOF) GL-REF mass spectrometer (Micromass). Data processing was performed with MASSLYNX, and database searching (Swiss-Prot, TrEMBL, Ensembl) to identify the proteins of interest from their peptide fingerprint was performed with PROTEINLYNX GLOBAL SERVER (Micromass). Data analysis and clustering was performed with GARBAN. This system is implemented with bioinformatic tools that organize large amounts of genes and proteins into biologically relevant clusters and classify them according to the ontological criteria of the Gene Ontology Consortium (L. A. Martínez-Cruz, A.R., and J.M.M., unpublished work).
Isolation and Culture of Rat Hepatocytes.
Hepatocytes were isolated from male Wistar rats (200–250 g) by collagenase perfusion, as described (31). After isolation, hepatocytes were cultured according to García-Trevijano et al. (19) in the presence or absence of 100 μM methionine/4 mM AdoMet/20 mM cycloleucine (CL) for the indicated periods of time. Cell viability was measured by trypan blue exclusion, and no significant differences were observed at any time between controls and any of the various treatments performed in this study.
Mitochondrial Isolation and Characterization.
An enriched mitochondrial fraction was obtained from 100-mg liver specimens with the Mitochondria Isolation kit from Sigma by two consecutive centrifugation steps (600 × g and 11,000 × g, respectively). The electrochemical proton gradient (Δψ) of the inner mitochondrial membrane was tested by measuring uptake of the fluorescent carbocyanine dye JC-1 (Sigma) into the mitochondria, following the procedure indicated by the manufacturer. Briefly, samples of the mitochondrial suspension containing 5–40 μg of protein were incubated for 7 min in the dark with 0.3 μM JC-1 in the presence of 10 mM sodium succinate. Fluorescence measurements were performed with a Perkin–Elmer LS 50 B spectrofluorimeter.
RNA Isolation and Northern Hybridization Analysis.
Total liver RNA was isolated by the guanidinium thiocyanate method (19). RNA concentration was determined spectrophotometrically before use, and the integrity was checked by electrophoresis with subsequent ethidium bromide staining. Electrophoresis and gel blotting were carried out as described (22). The PHB1 cDNA was cloned by reverse transcriptase-PCR from mouse liver. The Superscript preamplification system (Life Technologies, Grand Island, NY), Taq Long Plus enzyme (Stratagene), and the mouse prohibitin sense 5′-atggctgccaaagtgtttgagtc-3′ antisense 5′-tcactggggaagctggagaagc-3′primers were used. Probes for the ATPase β-subunit and COX subunits I and II have been described (32, 33). Northern hybridization analyses were performed on total RNA by using standard procedures, as described (22). All probes were labeled with [32P]dCTP by using the Rediprime DNA Labeling System (Amersham Pharmacia). To ensure equal loading of RNA samples, membranes were also hybridized with a 32P-labeled 18S rRNA cDNA probe. Autoradiography and densitometry were used to quantitate relative RNA. Results of Northern blot analysis were normalized to 18S rRNA.
DNA Isolation and Southern Blot Analysis.
Total DNA was extracted from the livers of 3-month-old WT and MAT1A−/− mice, as described (33). Total cellular DNA (20 μg) was digested with EcoRI. The digested DNA was resolved on 1% agarose gel, transferred, and fixed onto nylon membranes. The membranes were incubated with a COX II [32P]dCTP-labeled probe. Conditions for hybridization and membrane washing were as described (33).
Western Blot Analysis.
Protein extraction and Western blotting were performed as described (34, 35). Equal amounts of protein (15 μg) were resolved in SDS/12.5% polyacrylamide gels. Proteins were electrophoretically transferred to nitrocellulose membranes. Membranes were probed with anti-MAT (35), anti-PHB1, anti-ATPase β, and anti-COX I and II. A horseradish peroxidase-conjugated secondary antibody was used. Blots were developed by enhanced chemiluminescence (Dupont).
Statistical Analysis.
Data are given as the mean ± SD. For changes in mRNA and protein levels, ratios of the respective mRNA or protein to 18S or red Ponceau staining densitometric values were compared between KO and WT mice by Student's t test. Significance was defined as P < 0.05.
Results
Proteomic Fingerprint of the Pathogenesis of NASH in MAT1A−/− Mouse.
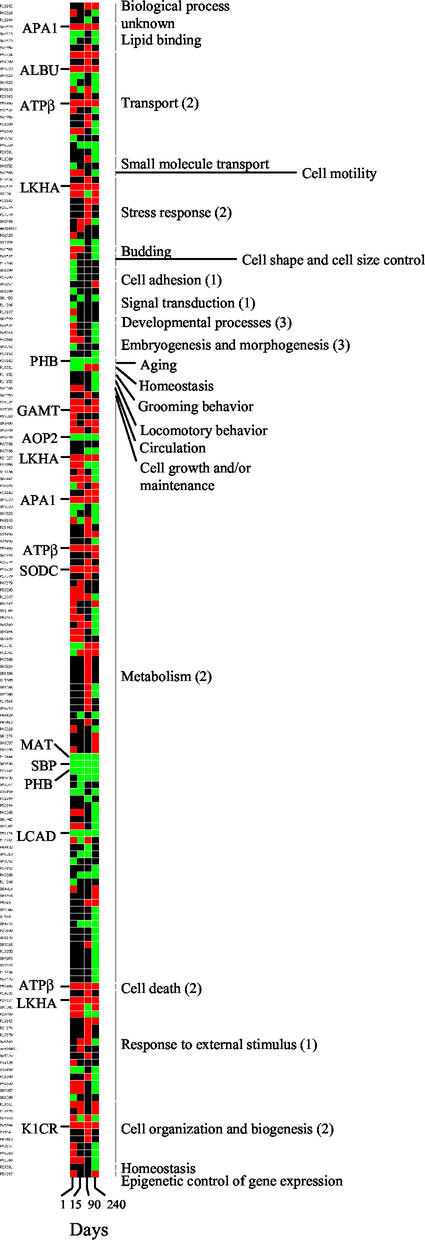
We have investigated the molecular mechanisms involved in the development of NASH in the liver of MAT1A−/− mouse by a high-throughput proteomic approach. Protein expression patterns were obtained by two-dimensional electrophoresis (2-DE) analysis of liver extracts from 1-, 15-, 90-, and 240-day-old WT and MAT1A−/− mice. Five independent experiments were performed by using liver extracts from different animals. Based on the results of the image analysis with PDQUEST (Bio-Rad), an average of 1,500–2,000 spots were visualized, depending on the gel staining procedure. WT and KO gel images (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org) were compared to determine the differences of protein expression, and changes were only accepted when at least a twofold increase or decrease was confirmed in all five experiments. Our analysis revealed that 1 day after birth, 140 differences were already induced in the liver of MAT1A−/− mice, and that changes accumulate with age (242, 259, and 297 differences by 15, 90, and 240 days, respectively; see Fig. 7, which is published as supporting information on the PNAS web site). The distribution between up-regulated and down-regulated proteins is maintained up to 3 months; 53–70% were up-regulated and 30–47% were down-regulated proteins. In contrast, this pattern switches in 8-month-old KO liver, when NASH has already developed, where 27% were up-regulated and 73% were down-regulated proteins (see Fig. 7). From all of the original changes, because of technical reasons, only the most abundant proteins represented in the databases were identified by peptide mass fingerprinting, resulting in 117 proteins successfully identified (see Table 1, which is published as supporting information on the PNAS web site). Consequently, alterations other than those reported in this study might also participate in the development of NASH in MAT1A−/− mice. We have found a linear correlation between the Mr and pI calculated from the sequence of the identified proteins and the experimental relative electrophoretic mobility (Rf) of the corresponding spot, calculated from the 2-DE gels (see Fig. 8, which is published as supporting information on the PNAS web site). This finding further validates the identity of the analyzed spots. Some pI deviations of linearity were observed, which is likely a consequence of posttranslational modifications. The specific protein expression profile of the MAT1A−/− liver provides a proteomic fingerprint of NASH (see Fig. 6). Up- and down-regulated proteins were classified by the biological processes in which they are involved according to the gene ontology criteria and were represented as shown in Fig. 1. Most proteins differentially expressed in the KO liver clustered into three biological processes (Fig. 1): cell communication (group 1), cell growth and/or maintenance (group 2), and developmental processes (group 3). Some of the identified proteins in these clusters are tubulin α 6 chain, tubulin β 5 chain, actin, and PHB1, which control DNA synthesis and regulate cell proliferation. These proteins are involved in embryogenesis, morphogenesis, and aging. Additionally, proteins involved in response to stress, such as leukotriene A4 hydrolase (LKHA), heat shock cognate 71, and heat shock protein 75, are up-regulated. Changes in the expression pattern of proteins involved in oxidative stress were also identified, for example, glutathione peroxidase, antioxidant protein 1 and 2 (AOP2), or Cu2+/Zn2+ superoxide dismutase (SODC). Finally, according to the broad metabolic activity of the liver, 80% of the proteins whose expression changes in KO mouse correspond to metabolic enzymes. Most of the alterations affect carbohydrate, lipid, and amino acid metabolism. For example, fructose 1,6 biphosphatase (up-regulated) and glycerol 3 phosphate dehydrogenase (down-regulated) are key enzymes in gluconeogenesis and glycolysis, respectively. Acyl CoA dehydrogenase long chain specific (ACDL) and Δ3,5 Δ2,4 dienoyl CoA isomerase (both down-regulated) are involved in fatty acid β-oxidation. Apolipoprotein A1 (APA1; up-regulated) and farnesyl pyrophosphate synthetase (down-regulated) participate in cholesterol transport and biosynthesis, respectively, and malate dehydrogenase and isocitrate dehydrogenase (both down-regulated) catalyze two steps of the citrate cycle. In addition, major differences affecting branched, aromatic, and sulfur amino acid metabolism were identified (see Fig. 9, which is published as supporting information on the PNAS web site). A group of 12 proteins changed their expression pattern 1 day after birth, and this alteration is maintained along the progression of NASH in MAT1A−/− liver (Fig. 1). These proteins, which can be considered as early markers of NASH, were identified as APA1, LKHA, selenium binding protein, AOP2, MAT, keratin type 1 cytoskeletal 18, guanidinoacetate methyltransferase, PHB1, SODC, albumin, ACDL, and ATPase β-subunit. Most of these proteins are metabolic enzymes or participate in the antioxidant response of the KO hepatocyte. Interestingly, PHB1 and ATPase β-subunit are mitochondrial proteins, and changes in their expression might compromise mitochondrial function.
Figure 1.
Proteins differentially expressed in the liver of MAT1A−/− mouse during the development of NASH. Liver extracts from 1-, 15-, 90-, and 240-day-old WT and MAT1A−/− mice were analyzed by 2-DE. Proteins differentially expressed were identified by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry and clustered by the biological process in which they are involved according to gene ontology criteria: (i) cell communication, (ii) cell growth and/or maintenance, and (iii) developmental processes. Up-regulated proteins are shown in red and down-regulated proteins are shown in green. Proteins whose up- or down-regulation was constantly detected in the liver of MAT1A−/− mouse from 1 to 240 days are indicated. APA1, ATPβ, LKHA, and PHB1 were clustered in different gene ontology categories.
AdoMet Regulates a Network of Mitochondrial Proteins and Mitochondrial Function.
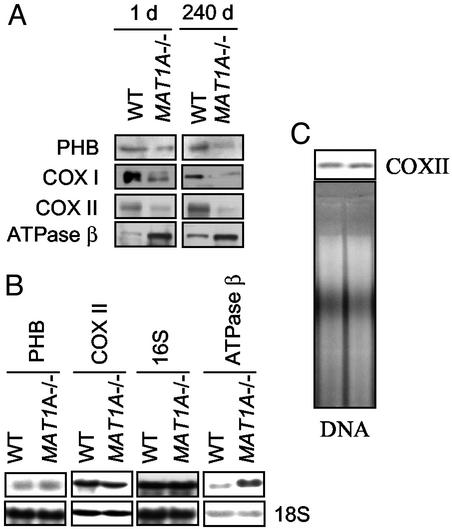
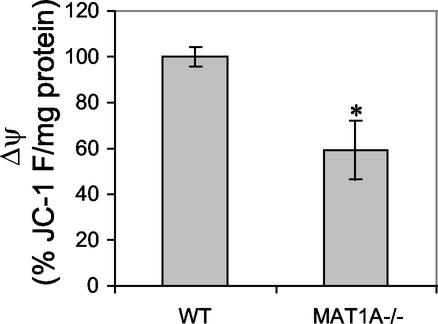
Alteration of mitochondrial function might be a key factor in the progression of NASH in MAT1A−/− mice. Up-regulation (450 ± 37%) of ATPase β-subunit and down-regulation (53 ± 3%) of PHB1 were confirmed by Western blot analysis with specific antibodies (Fig. 2). Additionally, the steady-state levels of subunits I and II of COX were also diminished in KO liver (55 ± 7% and 53 ± 5%, respectively; Fig. 2). These alterations were not observed in other tissues (not shown). The expression of genes encoding for these proteins was also investigated by Northern blot analysis. The mRNA level of ATPase β-subunit is increased twofold in MAT1A−/− liver (Fig. 2). In contrast, mRNA levels of PHB1 and COX II, as well as 16S rRNA, were similar to those found in the WT animals (Fig. 2). These observations indicate that the down-regulation of PHB1 and COX subunits by AdoMet involves posttranscriptional mechanisms. Whereas PHB1 is the product of a nuclear gene, COX subunits are encoded by mitochondrial genes. Additionally, no changes on mitochondrial DNA content were observed in MAT1A−/− liver (Fig. 2). Changes on the expression profile of PHB1, COX, and ATPase proteins suggest a deficiency in mitochondrial respiration. The mitochondrial inner membrane potential was measured in an enriched mitochondrial fraction purified from liver extracts of WT and MAT1A−/− mice. A 40% reduction of the electrochemical proton gradient was assessed in the KO (Fig. 3), indicating a deficiency of the mitochondrial inner membrane integrity.
Figure 2.
(A) Hepatic steady-state PHB1, COX I and II, and ATP β-protein levels in MAT1A−/− and WT liver. Liver extracts (15 μg/lane) from 1- and 240-day-old mice were analyzed by Western blotting. Equal protein loading was assured by red Ponceau staining of membranes (not shown). ATPaseβ was 484 ± 51% and 432 ± 33%, up-regulated by 1 and 240 days, respectively. PHB1 was 62 ± 4% and 54 ± 6%, down-regulated by 1 and 240 days, respectively. COX I was 52 ± 5% and 57 ± 4%, down-regulated by 1 and 240 days, respectively. COX II was 51 ± 6% and 54 ± 4%, down-regulated by 1 and 240 days, respectively. (B) Expression of PHB1, COX II, ATP β, and 16S rRNA in MAT1A−/− and WT liver. Liver RNA samples (30 μg/lane) from 90-day-old mice were analyzed by Northern blot hybridization with specific probes. Blots were hybridized with a probe for 18S rRNA to check for equal loading. (C) mtDNA content in MAT1A−/− relative to WT liver. Total DNA was isolated from 3-month-old mice, cleaved with EcoRI, and used for Southern analysis with a specific probe for COX II. Equivalent amounts of DNA in each lane were ensured by ethidium-bromide staining of the gel. Representative blots from three independent experiments are shown.
Figure 3.
Electrochemical proton gradient of the inner mitochondrial membrane. Mitochondrial functionality was studied by measuring Δψ in an enriched mitochondrial fraction from 3-month-old WT and MAT1A−/− liver. Value of 100% was 175.51 ± 6.69 fluorescence units per mg of protein. The average of three independent experiments is shown. *, P < 0.05 vs. WT by Student's t test.
Regulation of PHB1 by AdoMet.
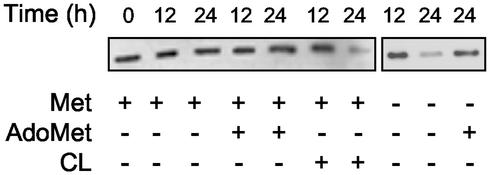
Next, we investigated whether AdoMet regulates PHB1 levels in cultured rat hepatocytes. In the presence of methionine or AdoMet in the culture media, no effect on PHB1 protein levels was detected after a 24-h incubation under standard conditions (Fig. 4). However, impairment of AdoMet synthesis by restriction of methionine or addition of CL, a well known inhibitor of MAT activity (36), to the culture media resulted in the down-regulation of PHB1 (51 ± 4% and 56 ± 8%, respectively; Fig. 4). Restoration of hepatocyte intracellular pool of AdoMet by exogenous addition of this compound after a 12-h culture in the absence of methionine prevented the fall of PHB1.
Figure 4.
Regulation of PHB1 levels by AdoMet in cultured rat hepatocytes. PHB1 levels were measured in rat hepatocytes cultured for 12 or 24 h in the presence or absence of 100 μM methionine/4 mM AdoMet or 20 mM cycloleucine. (Right) AdoMet (4 mM) was added after 12-h culture in a methionine-deficient media. Liver extracts (15 μg per lane) were analyzed by Western blotting by using specific antibodies. PHB1 was down-regulated by 51 ± 4% and 56 ± 8% by the restriction of methionine or the addition of cycloleucine, respectively. Equal protein loading was assured by red Ponceau staining of membranes (not shown). A representative blot from three independent experiments is shown.
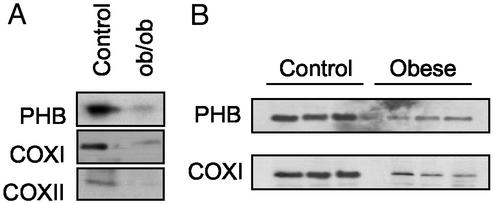
PHB1 and COX I and II Levels in ob/ob Mice and Obese Patients.
To evaluate whether the alterations observed in MAT1A−/− mice provide hitherto unknown mechanisms by which NASH may be induced, the steady-state levels of PHB1 and COX were investigated in liver samples from ob/ob mice and obese patients; individuals with these conditions are prone to the development of NASH. Western blot analysis revealed that PHB1 and COX I and II are down-regulated in ob/ob liver by 76 ± 8%, 81 ± 3%, and 70 ± 5%, respectively. Down-regulation of PHB1 (53 ± 4%) and COX I (82 ± 5%) was also confirmed in the liver of obese patients, although the COX II level was similar to that found in control liver (Fig. 5). Similar to MAT1A−/− mice, the mRNA levels of PHB, COX I, and COX II in ob/ob mice and obese patients are unchanged from normal controls (not shown).
Figure 5.
(A) Hepatic steady-state protein levels of PHB1 and COX I and II in WT and ob/ob mice. A representative blot from three independent experiments is shown. PHB1 and COX I and II are down-regulated in ob/ob liver by 76 ± 8%, 81 ± 3%, and 70 ± 5%, respectively. (B) Hepatic steady-state protein levels of PHB1 and COX I in control and obese patients. Liver extracts (15 μg per lane) were analyzed by Western blotting by using specific antibodies. PHB1 and COX I are down-regulated by 53 ± 4% and 82 ± 5% in the liver of obese patients. Equal protein loading was assured by red Ponceau staining of membranes (not shown).
Discussion
AdoMet generally has been considered as a central intermediary metabolite involved in the synthesis of homocysteine and polyamines, as well as the main cellular methyl group donor (2). However, recent evidence indicates that, in addition to its central metabolic function, AdoMet functions in the liver as an intracellular control switch that regulates essential hepatocyte functions such as proliferation, differentiation, and death (26). To better understand the mechanisms by which these nontraditional functions of AdoMet take place, we studied the pathogenesis of NASH in KO mice deficient in hepatic AdoMet synthesis (MAT1A−/−) by using a high-throughput proteomic approach.
Analysis of the differential protein expression profile of MAT1A−/− and WT mice indicates that a chronic deficiency in AdoMet synthesis has a pleiotropic effect in liver that alters essential hepatic functions. The initial depletion of MAT1A might result in changes in liver cell populations other than hepatocytes, which may also contribute to the development of NASH. We have identified 117 proteins that are differentially expressed in the liver of MAT1A−/− mice during the development of NASH (see Table 1). The global analysis of the observed differences indicates up-regulation of antioxidant proteins (SODC, catalase, glutathione peroxidase) in the MAT1A−/− liver that might reflect an adaptive mechanism to dissipate oxidative stress generated by oxidant genes (22, 28). Major metabolic alterations were also found at protein expression level in the KO liver. Lipid, carbohydrate, and amino acid metabolism is impaired in MAT1A−/− mice from birth, although differences accumulate during the development of NASH. These up- and down-regulated proteins provide a specific proteomic profile that might explain some of the metabolic alterations reminiscent of those found in obesity and other conditions associated with NASH, which lead to development of cirrhosis and HCC (37–39).
The analysis of the 117 proteins differentially expressed on MAT1A−/− liver revealed that most of the changes detected 1 day after birth are not maintained during the development of NASH. These time-dependent differences may reflect the adaptation of the hepatocyte to performing its normal biological function under a chronic deficiency of AdoMet, leading to the accumulation of alterations that condition the development of the disease. However, 12 proteins change their expression pattern after birth, when MAT1A is switched on in WT mice, and this alteration is maintained to the onset of histological lesions. Among these early changes are the up-regulation of SODC and AOP2, which was deduced from our two-dimensional electrophoresis (2-DE) gels in accordance with earlier evidence (40), as well as the down-regulation of ACDL and up-regulation of APA1. These alterations agree with the implication of oxidative stress and abnormal lipid metabolism in the pathogenesis of fatty liver disease. Interestingly, four proteins involved in mitochondrial function were also identified, ATPase β, COX I and II, and PHB1. Transcriptional up-regulation of ATPase β has been implicated in mitochondrial maturation and in cell neoplastic transformation (41, 42), which is consistent with the proliferative and de-differentiated state of MAT1A−/− hepatocytes (22, 28). Translational down-regulation of COX indicates a deficient transference of electrons to oxygen, the last step of the mitochondrial electron transfer chain, and therefore provides a molecular explanation for the oxidative stress found in MAT1A−/− liver (22, 28, 43). The fall in PHB1 steady-state levels might explain the down-regulation of COX. PHB1 is the product of a nuclear gene that is targeted to the inner mitochondrial membrane (44). It has been proposed recently that PHB1 is a chaperone-like protein that participates in the correct folding and assembly of some of the components of the mitochondrial respiratory chain (45, 46). According to this hypothesis, a deficiency in PHB1 may impair the native and functional organization of respiratory proteins that are subsequently degraded by mitochondrial proteases, compromising mitochondrial functionality (45, 46). Therefore, the decrease of PHB1 might induce a reduction of COX, with the concomitant loss of mitochondrial function in MAT1A−/− hepatocytes.
Previous evidence correlates down-regulation of PHB1 gene expression with accelerating aging and age-related degenerative diseases (46, 47). Our results indicate that AdoMet regulates PHB1 levels, likely through a posttranslational mechanism, as deduced from the observation that mRNA content of PHB1 remains normal in liver under a chronic deficiency of AdoMet. The correlation between a deficiency of AdoMet synthesis and down-regulation of PHB1 found in MAT1A−/− mice was confirmed by the in vitro experiments on isolated rat hepatocytes. Reduction of AdoMet synthesis by using a culture media without methionine or in the presence of CL, an inhibitor of MAT activity (36), results in the down-regulation of PHB1. Replenishment of hepatocyte AdoMet content prevented the fall of PHB1. The mechanism by which AdoMet regulates PHB1 levels requires further investigation. However, phosphorylation of PHB1 has been reported in human fibroblasts (48) and in hormone-treated ovarian cells (49), and the ability to posttranslationally phosphorylate PHB1 is lost during the process of cellular aging (48).
PHB1 steady-state levels are also decreased in the liver of ob/ob mice and in obese patients who are predisposed to develop NASH. NASH is a chronic disorder with an increasing prevalence in the population and is fast becoming one of the most significant and common disorders of clinical hepatology (37). The pathogenesis of NASH is poorly understood, but present evidence supports mitochondrial alterations correlating with oxidative stress as one of the most important factors (37, 50). Down-regulation of PHB1 and COX induced by a chronic deficiency of hepatic AdoMet may provide new molecular mechanisms involved in the pathogenesis of NASH. The results of our experiments with MAT1A−/− mice indicate that PHB1 and COX fell long before any histological manifestation of the disease was detected. Furthermore, our data also suggest one of the key mechanisms of AdoMet's hepatoprotective action may be to restore the expression of PHB1. Whether AdoMet might be of benefit in the treatment of NASH should be investigated further.
In summary, our results provide a mechanism by which a deficiency of AdoMet impairs mitochondrial function and generates oxidative stress in liver. The fall of PHB1 leading to a mitochondrial failure, together with abnormal lipid, carbohydrate, and amino acid metabolism might explain, at least in part, the pathogenesis of NASH.
Supplementary Material
Acknowledgments
We thank Carmen Miqueo, Carolina Gómara, and Estefanía Fernández for technical support. This work was supported by National Institutes of Health National Center for Complementary and Alternative Medicine Grant R01 AT-1576 (to S.C.L., J.M.M., M.A.A., and F.J.C.), National Institute on Alcohol Abuse and Alcoholism Grant R0I AA-12677 (to S.C.L., J.M.M., and M.A.A.), National Institutes of Health Grant R01 AA13847 (to S.C.L., J.M.M., and F.J.C.), Plan Nacional de I+D Grant 99/0038 (to J.M.M.), Ministerio de Sanidad y Consumo Grant FIS 01/0712 (to M.A.A. and F.J.C.), Fundación Iñigo Alvarez de Toledo (to M.A.A. and F.J.C.), Gobierno de Navarra Grants 5697/1999, 681/2000, and 394/2001 (to F.J.C., J.M.M., and M.A.A., respectively), and National Institutes of Health Grant R01 DK-51719 (to S.C.L.). E.S. is a fellow of the University of Navarra. M.U.L. is a fellow of the Spanish Ministerio de Ciencia y Tecnología.
Abbreviations
- AdoMet
S-adenosylmethionine
- MAT
methionine adenosyltransferase
- NASH
nonalcoholic steatohepatitis
- PHB1
prohibitin 1
- COX
cytochrome c oxidase
- KO
knockout
- APA1
apolipoprotein A1
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Tabor C W, Tabor H. Annu Rev Biochem. 1976;45:285–306. doi: 10.1146/annurev.bi.45.070176.001441. [DOI] [PubMed] [Google Scholar]
- 2.Cantoni G L. Annu Rev Biochem. 1975;44:435–441. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- 3.Mudd S H, Poole J R. Metabolism. 1975;29:707–720. [Google Scholar]
- 4.Mato J M, Alvarez L, Ortiz P, Pajares M A. Pharmacol Ther. 1997;73:265–280. doi: 10.1016/s0163-7258(96)00197-0. [DOI] [PubMed] [Google Scholar]
- 5.Cantoni G. J Biol Chem. 1953;204:403–416. [PubMed] [Google Scholar]
- 6.Kotb M, Mudd S H, Mato J M, Geller A M, Kredich N M, Chou J Y, Cantoni G L. Trends Genet. 1997;13:51–52. doi: 10.1016/s0168-9525(97)01013-5. [DOI] [PubMed] [Google Scholar]
- 7.Okada G, Teraoka H, Tsukada K. Biochemistry. 1981;20:934–940. doi: 10.1021/bi00507a045. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan D M, Hoffman J L. Biochemistry. 1983;22:1636–1641. doi: 10.1021/bi00276a017. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman J L. Methods Enzymol. 1983;94:223–228. doi: 10.1016/s0076-6879(83)94038-7. [DOI] [PubMed] [Google Scholar]
- 10.Kotb M, Kredich N M. J Biol Chem. 1985;260:3923–3930. [PubMed] [Google Scholar]
- 11.Cabrero C, Puerta J, Alemany S. Eur J Biochem. 1987;170:299–304. doi: 10.1111/j.1432-1033.1987.tb13699.x. [DOI] [PubMed] [Google Scholar]
- 12.Kotb M, Geller A M. Pharmacol Ther. 1993;59:125–143. doi: 10.1016/0163-7258(93)90042-c. [DOI] [PubMed] [Google Scholar]
- 13.Gil B, Casado M, Pajares M A, Bosca L, Mato J M, Martin-Sanz P, Alvarez L. Hepatology. 1996;24:876–881. doi: 10.1002/hep.510240420. [DOI] [PubMed] [Google Scholar]
- 14.Lu S C, Gukovsky I, Lugea A, Reyes C N, Huang Z Z, Chen L, Mato J M, Bottiglieri T, Pandol S J. FASEB J. 2003;17:56–58. doi: 10.1096/fj.01-0752fje. [DOI] [PubMed] [Google Scholar]
- 15.Huang Z Z, Mato J M, Kanel G, Lu S C. Hepatology. 1999;29:1471–1478. doi: 10.1002/hep.510290525. [DOI] [PubMed] [Google Scholar]
- 16.Latasa M U, Boukaba A, García-Trevijano E R, Torres L, Rodriguez J L, Caballeria J, Lu S C, Lopez-Rodas G, Franco L, Mato J M, Avila M A. FASEB J. 2001;15:1248–1250. doi: 10.1096/fj.00-0556fjev1. [DOI] [PubMed] [Google Scholar]
- 17.Cai J, Sun W M, Hwang J J, Stain S C, Lu S C. Hepatology. 1996;24:1090–1097. doi: 10.1002/hep.510240519. [DOI] [PubMed] [Google Scholar]
- 18.Cai J, Mao Z, Hwang J J, Lu S C. Cancer Res. 1998;58:1444–1450. [PubMed] [Google Scholar]
- 19.García-Trevijano E R, Latasa M U, Carretero M V, Berasain C, Mato J M, Avila M A. FASEB J. 2000;14:2511–2518. doi: 10.1096/fj.00-0121com. [DOI] [PubMed] [Google Scholar]
- 20.Torres L, Avila M A, Carretero M V, Latasa M U, Caballeria J, Lopez-Rodas G, Boukaba A, Lu S C, Franco L, Mato J M. FASEB J. 2000;14:95–102. doi: 10.1096/fasebj.14.1.95. [DOI] [PubMed] [Google Scholar]
- 21.Avila M A, Berasain C, Torres L, Martín-Duce A, Corrales F J, Yang H, Prieto J, Lu S C, Caballeria J, Rodes J, Mato J M. J Hepatol. 2000;33:907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 22.Lu S C, Alvarez L, Huang Z Z, Chen L, An W, Corrales F J, Avila M A, Kanel G, Mato J M. Proc Natl Acad Sci USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotb M, Kredich N M. Biochim Biophys Acta. 1990;1039:253–260. doi: 10.1016/0167-4838(90)90193-j. [DOI] [PubMed] [Google Scholar]
- 24.Pajares M A, Duran C, Corrales F, Pliego M M, Mato J M. J Biol Chem. 1992;267:17598–17605. [PubMed] [Google Scholar]
- 25.Sánchez del Pino M M, Corrales F J, Mato J M. J Biol Chem. 2000;275:23476–23482. doi: 10.1074/jbc.M002730200. [DOI] [PubMed] [Google Scholar]
- 26.Mato J M, Corrales F J, Lu S C, Avila M A. FASEB J. 2002;16:15–26. doi: 10.1096/fj.01-0401rev. [DOI] [PubMed] [Google Scholar]
- 27.Horikawa S, Ozasa H, Ito K, Katsuyama I, Tsukada K, Sugiyama T. Biochem Mol Biol Int. 1996;40:807–814. doi: 10.1080/15216549600201413. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Chantar M L, Corrales F J, Martinez-Cruz L A, García-Trevijano E R, Huang Z Z, Chen L, Kanel G, Avila M A, Mato J M, Lu S C. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 29.Cabrero C, Duce A M, Ortiz P, Alemany S, Mato J M. Hepatology. 1988;8:1530–1534. doi: 10.1002/hep.1840080610. [DOI] [PubMed] [Google Scholar]
- 30.Martín-Duce A, Ortiz P, Cabrero C, Mato J M. Hepatology. 1988;8:65–68. doi: 10.1002/hep.1840080113. [DOI] [PubMed] [Google Scholar]
- 31.Avila M A, Carretero M V, Rodriguez E N, Mato J M. Gastroenterology. 1998;114:364–371. doi: 10.1016/s0016-5085(98)70489-5. [DOI] [PubMed] [Google Scholar]
- 32.Izquierdo J M, Cuezva J M. Mol Cell Biol. 1997;17:5255–5268. doi: 10.1128/mcb.17.9.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otero G, Avila M A, de la Pena L, Emfietzoglou D, Cansado J, Popescu G F, Notario V. Carcinogenesis. 1997;18:1569–1575. doi: 10.1093/carcin/18.8.1569. [DOI] [PubMed] [Google Scholar]
- 34.Avila M A, Velasco J A, Cho C, Lupu R, Wen D, Notario V. Oncogene. 1995;10:963–971. [PubMed] [Google Scholar]
- 35.Ruiz F, Corrales F J, Miqueo C, Mato J M. Hepatology. 1998;28:1051–1057. doi: 10.1002/hep.510280420. [DOI] [PubMed] [Google Scholar]
- 36.Lombardini J B, Talalay P. Adv Enzyme Regul. 1970;9:349–384. doi: 10.1016/s0065-2571(71)80054-7. [DOI] [PubMed] [Google Scholar]
- 37.Reid A E. Gastroenterology. 2001;121:710–723. doi: 10.1053/gast.2001.27126. [DOI] [PubMed] [Google Scholar]
- 38.Angulo P. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 39.Clark J M, Brancati F L, Diehl A M. Gastroenterology. 2002;122:1649–1657. doi: 10.1053/gast.2002.33573. [DOI] [PubMed] [Google Scholar]
- 40.Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, Lunardi J. J Biol Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 41.Izquierdo J M, Ricart J, Ostronoff L K, Egea G, Cuezva J M. J Biol Chem. 1995;270:10342–10350. doi: 10.1074/jbc.270.17.10342. [DOI] [PubMed] [Google Scholar]
- 42.de Heredia M L, Izquierdo J M, Cuezva J M. J Biol Chem. 2000;275:7430–7437. doi: 10.1074/jbc.275.10.7430. [DOI] [PubMed] [Google Scholar]
- 43.Sohal R S. Free Radical Biol Med. 1993;14:583–588. doi: 10.1016/0891-5849(93)90139-l. [DOI] [PubMed] [Google Scholar]
- 44.Ikonen E, Fiedler K, Parton R G, Simons K. FEBS Lett. 1995;358:273–277. doi: 10.1016/0014-5793(94)01444-6. [DOI] [PubMed] [Google Scholar]
- 45.Nijtmans L G, de Jong L, Artal Sanz M, Coates P J, Berden J A, Back J W, Muijsers A O, van der Spek H, Grivell L A. EMBO J. 2000;19:2444–2451. doi: 10.1093/emboj/19.11.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nijtmans L G, Artal S M, Grivell L A, Coates P J. Cell Mol Life Sci. 2002;59:143–155. doi: 10.1007/s00018-002-8411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coates P J, Nenutil R, McGregor A, Picksley S M, Crouch D H, Hall P A, Wright E G. Exp Cell Res. 2001;265:262–273. doi: 10.1006/excr.2001.5166. [DOI] [PubMed] [Google Scholar]
- 48.Liu X T, Stewart C A, King R L, Danner D A, Dell'Orco R T, McClung J K. Biochem Biophys Res Commun. 1994;201:409–414. doi: 10.1006/bbrc.1994.1716. [DOI] [PubMed] [Google Scholar]
- 49.Thompson W E, Sanbuissho A, Lee G Y, Anderson E. J Reprod Fertil. 1997;109:337–348. doi: 10.1530/jrf.0.1090337. [DOI] [PubMed] [Google Scholar]
- 50.Yang S, Zhu H, Li Y, Lin H, Gabrielson K, Trush M A, Diehl A M. Arch Biochem Biophys. 2000;378:259–268. doi: 10.1006/abbi.2000.1829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.