Abstract
Modulation of the structure of a leader RNA to control formation of an intrinsic termination signal is a common mechanism for regulation of gene expression in bacteria. Expression of the S box genes in Gram-positive organisms is induced in response to limitation for methionine. We previously postulated that methionine availability is monitored by binding of a regulatory factor to the leader RNA and suggested that methionine or S-adenosylmethionine (SAM) could serve as the metabolic signal. In this study, we show that efficient termination of the S box leader region by bacterial RNA polymerase depends on SAM but not on methionine or other related compounds. We also show that SAM directly binds to and induces a conformational change in the leader RNA. Both binding of SAM and SAM-directed transcription termination were blocked by leader mutations that cause constitutive expression in vivo. Overproduction of SAM synthetase in Bacillus subtilis resulted in delay in induction of S box gene expression in response to methionine starvation, consistent with the hypothesis that SAM is the molecular effector in vivo. These results indicate that SAM concentration is sensed directly by the nascent transcript in the absence of a trans-acting factor.
A variety of mechanisms for control of gene expression by premature termination of transcription have been uncovered in bacteria (1, 2). Genes regulated in this way contain a transcription termination signal in the mRNA region upstream of the coding sequence of the regulated gene. The activity of this terminator can be controlled by modification of the activity of RNA polymerase (RNAP), blocking access of transcription-termination factor Rho, or by modulation of the leader RNA structure, commonly through alternate folding patterns. RNA folding can be controlled in turn through interaction with some regulatory factor, such as a translating ribosome, as in the Escherichia coli trp operon, or an RNA binding protein, as in the Bacillus subtilis trp operon or the E. coli bgl system. Modulation of RNA structure by an effector, in the absence of accessory proteins, has been demonstrated for the T box system, in which uncharged tRNA interacts directly with the leader RNA to promote antitermination (3, 4). Similar regulation by small molecules has recently been demonstrated for riboflavin and thiamin biosynthesis genes, by using flavin mononucleotide and thiamin pyrophosphate, respectively (5–7).
The S box regulatory mechanism represents a system in which synthesis of the full-length transcript is determined by controlling whether the leader folds into the stem loop of an intrinsic terminator or a competing antiterminator structure (ref. 8 and Fig. 1). Formation of the antiterminator is inhibited by an alternative helical structure, which functions as an anti-antiterminator. An anti-antiterminator element is also found in the B. subtilis pyr operon, in which leader RNA structure is controlled by binding of a regulatory protein (9). We initially identified the S box family by recognizing a highly conserved set of sequence and structural features in the leader regions of 18 transcriptional units involved in methionine metabolism in Gram-positive bacteria; the alternate structures are always found and are supported by covariation within helical domains. Multiple genes in this family, including B. subtilis yitJ, ykrT, ykrW, yjcI, yxjG, and yxjH, have been shown to be repressed at the level of transcription termination during growth in the presence of methionine, whereas limitation for methionine induces terminator read-through (refs. 8, 10, and 11 and unpublished results).
Figure 1.
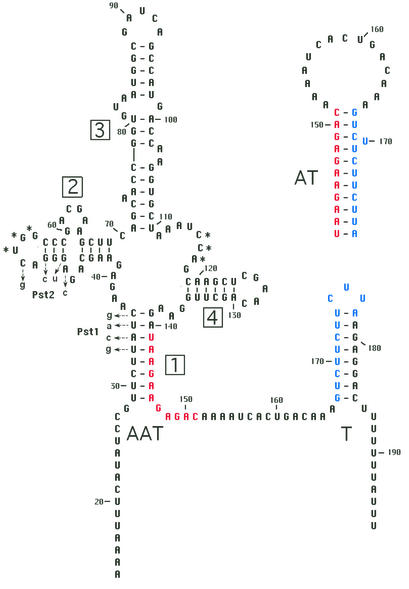
B. subtilis yitJ leader structural model. The structural model is based on phylogenetic analyses (8) and is shown in the terminator conformation. Red and blue residues indicate the alternate pairing for formation of the antiterminator, shown above the terminator. Boxed numbers indicate helices 1–4; T, terminator; AT, antiterminator; AAT, anti-antiterminator. Numbering of residues is relative to the predicted transcription start point. Sequence changes in the Pst-1 and Pst-2 alleles are shown with lowercase letters.
Genetic analysis of the B. subtilis yitJ leader supported a simple model for transcription termination control (8). Disruption of the leader region terminator resulted in high, constitutive expression, consistent with the idea that regulation is at the level of premature termination of transcription. Mutations that prevent formation of the antiterminator resulted in very low expression even when cells were starved for methionine, indicating that the terminator is always functional in the absence of antiterminator formation. Disruption of the anti-antiterminator element (helix 1; Fig. 1) resulted in constitutive read-through, suggesting that the antiterminator competes very efficiently with the terminator helix and must be destabilized by formation of the anti-antiterminator to allow termination. Mutations in conserved elements in the helix 1–4 junction region outside the anti-antiterminator itself also resulted in read-through of the terminator during growth in the presence of methionine, suggesting that this region plays a role in sensing methionine availability. We postulated that the helix 1–4 region of the RNA could serve as the binding site for a regulatory factor (8). In this study, we demonstrate that transcription of S box leaders in a purified in vitro system with either B. subtilis or E. coli RNAP resulted in efficient read-through of the leader region terminators. Addition of S-adenosylmethionine (SAM) at physiologically relevant concentrations stimulated efficient termination and caused a structural change in the anti-antiterminator domain. We also show that the leader RNA binds tightly and specifically to SAM and that sequence alterations that result in constitutive expression in vivo block SAM binding and SAM-dependent transcription termination in vitro. These results indicate that SAM is the effector in this system, and that it is monitored directly by the leader RNA during transcription.
Materials and Methods
Bacterial Strains and Growth Conditions.
B. subtilis strain BR151MA (lys-3 trpC2) was used as the source of chromosomal DNA for amplification by PCR. Strains 168 (trpC2) and SA29 [trpC2 metK1 sacB∷φPvegmetK+ (12), obtained from J. Pero (OmniGene Bioproducts, Cambridge, MA); metK was formerly designated metE] were used to test expression of gene fusions. The yitJ-lacZ and ykrW-lacZ transcriptional fusions were transferred into strains 168 and SA29 by transduction with SPβ prophages, as described (8, 10). Strains containing fusions were grown in the presence of chloramphenicol (5 μg/ml). Cells were propagated on tryptone blood agar base medium (Difco) or in Spizizen minimal medium (13).
In Vitro Transcription Assays.
Templates for in vitro transcription by B. subtilis or E. coli RNAP were generated by PCR using oligonucleotide primers (Integrated DNA Technologies, Coralville, IA) that contained the glyQS promoter sequence and hybridized within the leader region of the target gene, to generate a transcription start site ≈20 nt upstream of the start of helix 1 (Fig. 1). The promoter sequences were designed to allow initiation with a dinucleotide corresponding to the +1/+2 positions of the transcript and a halt early in the transcript by omission of a single nucleotide; the position of the halt ranged from 15 to 25 nt for different S box genes. The PCR fragments were ≈400 bp in length and included 50–150 bp downstream from the transcription terminator to allow resolution of terminated and read-through products. PCR products were purified with a Qiagen PCR cleanup kit (Qiagen, Chatsworth, CA). Templates for yitJ leader region variants were generated by PCR with plasmid templates containing the appropriate alleles (8).
Single-round transcription reactions were carried out as described (4) by using template DNA (10 nM), His-tagged B. subtilis RNAP (6 nM) purified as described by Qi and Hulett (14), or E. coli RNAP (10 nM) purified as described by Hager et al. (15). The initiation reactions contained 1× transcription buffer (16), the appropriate dinucleotide (150 μM; Sigma), UTP (0.75 μM), [α-32P]UTP (800 Ci/mmol; 1 Ci = 37 GBq) at 0.25 μM, and the remaining two NTPs required for elongation to the halt at 2.5 μM. The initiation reaction mixtures were incubated at 37°C for 15 min and were then placed on ice. Heparin (20 μg/ml; Sigma) was added to block reinitiation, and elongation was triggered by the addition of NTPs to 10 μM in the presence of other reagents as indicated. Elongation reactions were incubated at 37°C for an additional 15 min and were terminated by extraction with phenol. Transcription products were resolved by denaturing PAGE and visualized by PhosphorImager (Molecular Dynamics) analysis.
RNase H cleavage experiments were carried out with two ykrW RNAs, both of which were uniformly labeled by transcription in the presence of [α-32P]UTP. The longer RNAs (154 nt) ended at the 3′ side of the antiterminator and were generated by E. coli RNAP in the presence or absence of SAM (150 μM). Shorter RNAs (142 nt), ending in the loop of the antiterminator, were generated by T7 RNAP transcription by using a Mega Shortscript T7 kit (Ambion, Austin, TX) and were heated to 65°C and slow-cooled to 40°C (1.25°C/min) before addition of SAM. Antisense oligonucleotides were added at 100-fold molar excess to E. coli RNAP transcription complexes or T7 RNAP-generated RNA (40 nM), and the reactions (10 μl) were incubated at 30°C for 5 min before digestion at 30°C for 10 min with RNase H (Roche; 0.6 units). Products were resolved on high-resolution denaturing polyacrylamide gels.
SAM Binding Assays.
Template DNAs for T7 RNAP transcription were generated by PCR using a primer containing a T7 promoter initiating with tandem G residues fused to positions +2 or +14 for the ykrW or yitJ leaders, respectively. The endpoint of the PCR products corresponded to the position just 5′ to the start of the terminator helix. RNA (8 μM) in 1× transcription buffer was heated to 65°C for 5 min, then slow-cooled to 40°C before addition of [methyl-14C]-SAM [ICN; 52 mCi/mmol (1.92 GBq/mmol); 8 μM final concentration], and incubated at room temperature for 5 min. Unlabeled SAM or S-adenosylhomocysteine (SAH) was added at 400 μM. Samples were filtered through a Nanosep 3K Omega filter microconcentrator (Pall) and washed with 1× transcription buffer. Material retained by the filter was mixed with Packard BioScience Ultima Gold scintillation fluid and counted in a Packard Tri-Carb 2100TR liquid scintillation counter.
β-Galactosidase Measurements.
Cells containing lacZ fusions were grown in Spizizen minimal medium containing methionine (50 μg/ml) until early exponential growth and then were harvested by centrifugation and resuspended in the same medium in the presence or absence of methionine. Samples were collected for β-galactosidase measurements and were assayed as described by Miller (17) by using toluene permeabilization of the cells.
Results
In Vitro Transcription of S Box Genes.
Initial attempts at single-round transcription assays of the B. subtilis yitJ and ykrW genes in a purified system resulted in weak transcription by B. subtilis RNAP, consistent with the poor adherence of the sequence in the −10 regions of these promoters to the consensus for B. subtilis EσA RNAP. Transcription of the ykrW gene with E. coli RNAP, which is less stringent in promoter recognition, resulted in efficient terminator read-through (Fig. 2A, lane 1). To allow analysis of these genes with B. subtilis RNAP, we took advantage of our previous demonstration that replacement of the yitJ promoter had no effect on the response to methionine limitation in vivo (8). We generated constructs in which the promoters of S box genes were replaced with the strong B. subtilis glyQS promoter and, where necessary, modified the region immediately downstream of the transcription start site to allow a halt early in the transcription reaction by omission of a single nucleotide from the initiation reaction. Transcription of the Pgly-ykrW construct resulted in very efficient terminator read-through with either B. subtilis or E. coli RNAP (Fig. 2A, lanes 3 and 7). Transcription of the yitJ leader under control of either the B. subtilis tyrS promoter (Ptyr-yitJ) or glyQS promoter (Pgly-yitJ) also gave high read-through, although the yitJ terminator was somewhat more active with B. subtilis RNAP (Fig. 2A, lanes 5 and 9). These results are consistent with the in vivo data demonstrating that read-through is the default state of the S box leaders (8).
Figure 2.
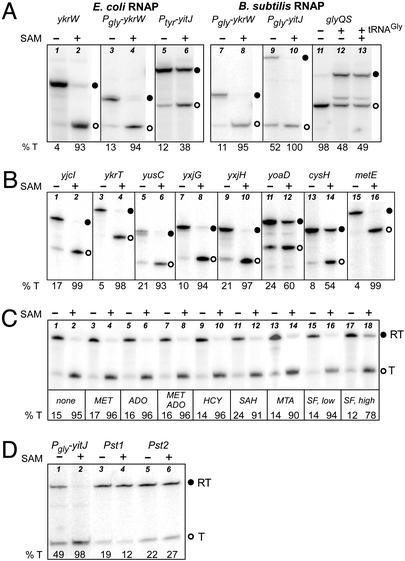
In vitro transcription of S box genes. ○, Terminated transcript; ●, read-through transcript. Percent termination (% T) is shown at the bottom of each lane. (A) SAM-dependent transcription termination. DNA templates were transcribed with E. coli (lanes 1–6) or B. subtilis (lanes 7–13) RNAP. Lanes 1 and 2, ykrW DNA; lanes 3, 4, 7, and 8, Pgly-ykrW DNA; lanes 5 and 6, Ptyr-yitJ DNA; lanes 9 and 10, Pgly-yitJ DNA; lanes 11–13, glyQS DNA. SAM was added at 150 μM where indicated (lanes 2, 4, 6, 8, 10, and 13). tRNAGly was added at 70 nM for lanes 12 and 13. The Pgly-yitJ constructs contain additional sequences downstream of the terminator, resulting in a larger read-through product than is observed with Ptyr-yitJ DNA. (B) SAM-dependent transcription termination of multiple B. subtilis S box leaders. DNA templates were transcribed with B. subtilis RNAP in the presence (lanes 2, 4, 6, 8, 10, 12, 14, and 16) or absence (lanes 1, 3, 5, 7, 9, 11, 13, and 15) of SAM (150 μM). Gene names are shown above each pair of lanes. (C) Specificity of SAM-dependent transcription termination of the B. subtilis ykrW leader. The ability of SAM-related compounds to stimulate termination by E. coli RNAP, and to block SAM-dependent termination, was tested. MET, methionine; ADO, adenosine; HCY, homocysteine; MTA, methylthioadenosine; SF, sinefungin. All compounds were tested at 1.5 mM except homocysteine (500 μM) and sinefungin, which was tested at both 500 μM (low, lanes 15 and 16) and 1.5 mM (high, lanes 17 and 18). SAM was included at 7.5 μM where indicated. (D) Effect of yitJ leader mutations on SAM-dependent transcription termination. DNA templates were Pgly-yitJ constructs; mutations are shown in Fig. 1. Transcription was with E. coli RNAP, in the presence (lanes 2, 4, and 6) or absence (lanes 1, 3, and 5) of SAM (150 μM). Templates were yitJ (lanes 1 and 2), yitJ-Pst-1 (lanes 3 and 4), and yitJ-Pst-2 (lanes 5 and 6).
SAM-Dependent Transcription Termination.
We tested the effect of addition of methionine, SAM, and related compounds on the activity of the ykrW terminator in vitro. Addition of SAM resulted in ≈95% termination for the Pgly-ykrW template with either B. subtilis or E. coli RNAP (Fig. 2A, lanes 4 and 8); similar results were obtained for transcription of a ykrW template containing its own promoter with E. coli RNAP (Fig. 2A, lane 2), confirming that the effect is independent of the promoter used to initiate transcription. The yitJ leader also exhibited SAM-dependent termination (Fig. 2A, lanes 6 and 10). In contrast, the glyQS leader-region terminator, which is inhibited by tRNAGly via the T box termination control system (4), was unaffected by SAM addition (Fig. 2A, lanes 11–13). This indicates that SAM does not generally promote transcription termination, but acts specifically on S box leader templates in the absence of cellular components other than RNAP.
We further determined the generality of the SAM response by examining eight additional B. subtilis S box templates. As shown in Fig. 2B, all of these leaders exhibited increased termination at the position of the leader-region intrinsic terminator in response to SAM addition, although there was some variability in the efficiency of termination in the absence of SAM, and in the magnitude of the SAM-dependent response. Regulation in vivo has been demonstrated for yitJ, ykrW, ykrT, yjcI, yxjG, and yxjH (refs. 8, 10, and 11 and unpublished results). The B. subtilis cysH operon was reported to be regulated primarily at the level of transcription initiation in response to O-acetyl-serine, with only a small response to methionine limitation (18); it appears that the cysH leader is capable of a response to SAM, at least in vitro. The yoaD leader, which exhibited the weakest response to SAM, is unique among the S box leaders in that it contains only a tetraloop at the top of helix 3 and has an extra stem loop inserted between the base of helix 1 and the terminator (8).
Titration of the SAM concentration required for ykrW termination activity in vitro indicated that 1.6 μM gave 60% termination, and 100 μM was a saturating level, resulting in >95% termination (data not shown). SAM pools in B. subtilis have been reported at 400 μM for cells grown in the presence of methionine (repressing conditions for S box gene expression) and at 80 μM for prototrophic cells grown in the absence of methionine (conditions under which S box genes are expressed at a low level; refs. 8 and 19). The SAM concentration required for transcription termination in vitro is therefore consistent with the physiologically relevant range for B. subtilis.
If SAM is the true molecular effector in vivo, it is essential for the system to respond specifically to SAM, but not to related compounds normally present in the cell. Methionine, homocysteine, adenosine, SAH, and methylthioadenosine (a breakdown product of SAM generated during polyamine biosynthesis) all failed to promote transcription termination of the ykrW leader (Fig. 2C), indicating that the SAM effect is specific. These compounds also failed to inhibit SAM-directed termination when SAM was used at 7.5 μM (nonsaturating) and the test compounds were added at a high molar excess to SAM. Sinefungin, an analog of SAM produced by Streptomyces sp. in which methionine is replaced by ornithine (20), acts as a competitive inhibitor of many SAM-dependent methyltransferase reactions (21). Sinefungin failed to promote ykrW transcription termination, but increased read-through in the presence of SAM ≈4-fold when added at a 200-fold molar excess (Fig. 2C, lane 18). This suggests that sinefungin may interact with the ykrW leader, but with a much lower affinity than SAM. Together, these results indicate that SAM recognition is highly specific, consistent with the requirement for discrimination from related compounds in vivo.
Effect of Leader Region Mutations on Termination Activity in Vitro.
Initial studies of the effect of yitJ leader region mutations on expression in vivo provided the basis for the regulatory model (8). Both the Pst-1 variant, in which alterations in the 5′ side of helix 1 disrupt formation of the anti-antiterminator but leave the antiterminator intact, and the Pst-2 variant, which contains an intact antiterminator and anti-antiterminator but has alterations in conserved residues in helix 2, resulted in high expression during growth in methionine, suggesting that these regions are required for terminator activity. Other mutations in the helix 1–4 junction region had similar effects (ref. 22 and unpublished results). As shown in Fig. 2D, the Pst-1 and Pst-2 variants exhibited high read-through in vitro in the presence or absence of SAM, in agreement with the in vivo results. It therefore appears that at least some of the same leader determinants are required for regulation in vivo and the response to SAM in vitro.
Binding of 14C-SAM to S Box Leader RNAs.
Specific binding of 14C-SAM to yitJ and ykrW leader RNAs containing the helix 1–4 region was tested by using a size-exclusion Nanosep 3K filter. Both RNAs bound 14C-SAM efficiently (Fig. 3A); addition of unlabeled SAM at a 50-fold molar excess blocked retention of 14C-SAM, whereas addition of unlabeled SAH had no effect. The RNA–SAM interaction appears to be very stable, because label was retained after repeated washing of the filter; very little 14C-SAM was retained by the filter in the absence of RNA. Binding of 14C-SAM to yitJ RNA containing the Pst-2 allele was reduced 20-fold, consistent with the observation that this variant resulted in constitutive read-through both in vivo and in vitro. The residual binding of 14C-SAM to the mutant RNA is specific, because it was prevented by addition of unlabeled SAM but not SAH. These results demonstrate that SAM binds directly to the helix 1–4 region of the leader RNA, and binding exhibits leader RNA requirements similar to those for transcription termination.
Figure 3.
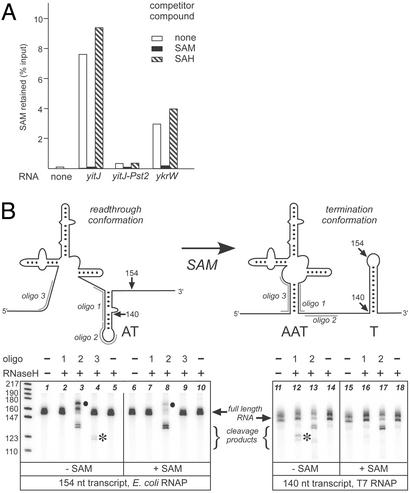
Interaction of SAM with S box leader RNAs. (A) Binding of 14C-SAM. 14C-SAM (8 μM) was incubated with T7 RNAP-transcribed leader RNA (8 μM) in 1× transcription buffer in the presence or absence of unlabeled SAM or SAH (400 μM). SAM binding is expressed as the fraction of 14C-SAM retained after filtration through a Nanosep 3K filter and washing with 1× transcription buffer relative to the amount added to the binding reaction. (B) Antisense oligonucleotide-dependent RNase H cleavage. The cartoon shows the structural models in the absence or presence of SAM. T, terminator; AT, antiterminator; AAT, anti-antiterminator. Positions of pairing of oligonucleotides 1–3 are shown. The 154-nt RNA was generated by transcription with E. coli RNAP (lanes 1–10) in the presence or absence of SAM (150 μM). The 140-nt RNA was generated by T7 RNAP transcription, heated to 65°C and slow-cooled to 40°C before addition of SAM (lanes 11–18). Oligonucleotides were added at a 100-fold molar excess before digestion with RNase H, which specifically targets RNA–DNA hybrids. Positions of the uncut RNA and RNase H cleavage products are indicated by brackets. Cleavage products that change in response to SAM are marked with asterisks. The higher-molecular-weight band observed with E. coli RNAP in the presence of oligo 2 (●, lanes 3 and 8) is likely to be the result of template switching during transcription, because it was absent when oligo 2 was added to premade RNA (lanes 13 and 17). The leftmost lane contains 32P-labeled pBR322 DNA digested with MspI used as a molecular weight standard.
SAM Causes a Transition in Leader RNA Structure.
The model for S box gene regulation predicts that the anti-antiterminator (helix 1) is stabilized in the presence of SAM (Fig. 3B). We probed the structure of helix 1 by using oligonucleotides complementary to the 5′ and 3′ sides of this helix and detected annealing by sensitivity to cleavage with RNase H, which only cleaves RNA–DNA hybrids. Transcripts generated by in vitro transcription with E. coli RNAP (Fig. 3B, lanes 1–10) included sequences necessary for formation of the antiterminator but lacked the 3′ portion of the terminator. Oligo 2, which hybridizes to the region downstream of the 3′ side of helix 1, triggered RNase H-catalyzed cleavage of transcripts generated in the presence or absence of SAM. Cleavage was stimulated by transcription in the presence of SAM, consistent with the prediction that the region targeted by this oligonucleotide includes sequences predicted to be sequestered in the antiterminator structure in the absence of SAM. Oligo 1, which is complementary to the 3′ side of helix 1, did not promote cleavage regardless of the presence of SAM, consistent with the prediction that this region is paired in both the anti-antiterminator and antiterminator conformations and is therefore not available for hybridization. Oligo 3, which hybridizes to the 5′ side of helix 1, directed cleavage of the transcript only in the absence of SAM, as expected if SAM promotes formation of the anti-antiterminator. We also used T7 RNAP transcription to generate a shorter RNA lacking the 3′ sequences of the antiterminator (Fig. 3B, lanes 11–18). When denatured and allowed to refold in the presence or absence of SAM, this RNA exhibited cleavage directed by oligo 1 only in the absence of SAM, consistent with the prediction that helix 1 is destabilized in the absence of SAM, and the 3′ region of helix 1 is available for hybridization with oligo 1 only when antiterminator formation is also prevented. These results are consistent with the proposed SAM-dependent structural transition in the leader RNA.
Overexpression of metK Delays Induction in Vivo.
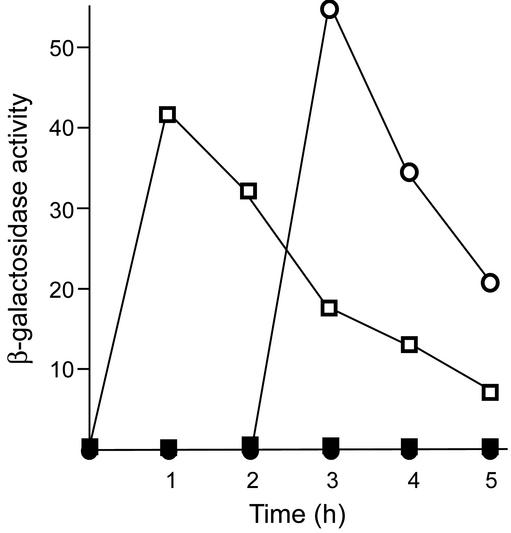
Because the in vitro transcription analyses suggested that SAM promotes transcription termination, we attempted to modulate SAM pools in vivo by overexpression of SAM synthetase. In E. coli, elevation of SAM synthetase activity results in increased SAM levels during growth in the presence of methionine (23). Yocum et al. (12) generated B. subtilis strain SA29, in which the metK coding sequence is expressed under the control of the strong Pveg promoter, and showed that SAM synthetase activity was elevated 6-fold relative to the wild-type strain during growth in minimal medium in the presence of methionine. In the wild-type strain, expression of a yitJ-lacZ transcriptional fusion was rapidly induced after cells were deprived of methionine, then gradually stabilized to the level normally observed during steady-state growth under these conditions (Fig. 4 and data not shown). Induction was delayed in strain SA29, containing increased SAM synthetase activity, consistent with the idea that the cells contained elevated SAM pools during growth in methionine and required a longer period of growth without methionine before SAM pools were depleted and induction could be observed. Both strains exhibited strong repression of yitJ expression during growth in methionine, indicating that the wild-type level of SAM synthetase is sufficient for full repression. Similar results were obtained with a ykrW-lacZ fusion (data not shown).
Figure 4.
Expression of a yitJ-lacZ transcriptional fusion. Fusions were introduced into strains 168 (wild type; squares) and SA29 (metK overexpression strain; circles). Cells were grown in defined medium containing methionine, collected by centrifugation, and resuspended in the same medium (filled symbols) or in medium without methionine (open symbols). Samples were taken at 1-h intervals and assayed for β-galactosidase activity, expressed in Miller units.
Discussion
Since the demonstration by Yanofsky of attenuation in the E. coli trp operon, many variations on alternate RNA structures as a means of transcriptional control have been described (reviewed in refs. 1 and 2). The regulation of T box genes in many organisms by direct binding of tRNA (3, 4) established that effectors can be measured directly by a leader RNA without a protein factor or ribosome. Recent studies demonstrated that RNA structure can be modulated by binding of other small molecules, without a requirement for a regulatory protein (5–7). Our discovery of the S box regulatory system suggested that many methionine-related genes in B. subtilis are coordinately controlled at the level of premature termination of transcription (8). The current study demonstrates that SAM controls the activity of S box leader-region terminators in vitro by binding directly to the leader RNA and altering its structure.
Yocum et al. (12) reported that overproduction of SAM synthetase results in methionine auxotrophy in B. subtilis and suggested that SAM could be the molecular effector controlling methionine biosynthesis gene expression. Mutants in which SAM synthetase activity was reduced exhibited both a decrease in SAM pools and an increase in intracellular methionine pools during growth in the absence of methionine (19), consistent with this idea. SAM is the major effector controlling methionine gene regulation in E. coli, acting at the level of transcription initiation (24). The results presented here provide strong evidence that SAM is the key effector in B. subtilis but acts at the level of transcription termination rather than transcription initiation. It therefore appears that many organisms see SAM as the crucial end-product of sulfur metabolism, and these organisms regulate methionine biosynthesis pathways by monitoring SAM concentration.
Altered RNA folding patterns have been proposed to control gene expression by sequestration of the translation start site in the mRNA, or by controlling formation of the helical region of an intrinsic terminator. In this study, we demonstrated that SAM does not generally promote transcription termination, because it had no effect on the glyQS terminator, but instead acts specifically on S box leader region terminators. It was especially important to demonstrate altered function for a bacterial RNAP termination site by using bacterial RNAP rather than bacteriophage T7 RNAP, which does not generally recognize termination signals for bacterial RNAPs (25). We found that both B. subtilis and E. coli RNAP exhibit a similar response in vitro, despite the fact that S box leaders are absent from E. coli. We also demonstrated that SAM could interact with preformed RNAs generated by transcription with bacterial RNAP or T7 RNAP. The utilization of SAM by the S box leader represents an example of direct modulation of RNA structure by an effector molecule. It is interesting to note that several genes controlled by the S box mechanism in Bacillus, Clostridium, and related genera are instead regulated by the T box mechanism in other organisms such as Enterococcus sp., by using tRNAMet charging rather than SAM as the physiological signal (26, 27). It therefore appears that these mechanisms represent alternative solutions to the same regulatory problem. S box leaders generally exhibit higher primary sequence conservation than T box leaders; this observation is consistent with the utilization of a single effector, SAM, for the S box system, whereas T box leaders must respond individually to the cognate uncharged tRNA and therefore must discriminate against noncognate tRNA species.
The secondary structure model of the leader RNA shown in Fig. 1 was derived from phylogenetic analysis of the initial 18 S box leaders (8). Identification of 60 additional leaders from multiple genera has further supported this model (unpublished results). Covariation of residues in the helix 2 loop and the unpaired region between helices 3 and 4 was proposed to indicate a possible tertiary interaction between these domains, suggesting that the structure of the conserved region at the junction of helices 1–4 may be even more complex (8). The arrangement of conserved elements in helix 2 follows the pattern for a kink-turn motif, and this prediction was supported by mutational studies (22), providing an additional indication that the RNA folds into a complex three-dimensional structure. We have also used nuclease digestion assays to detect a SAM-induced structural rearrangement in this region, consistent with the proposed stabilization of helical domains in the presence of SAM (data not shown). Because mutations disrupting these conserved elements result in loss of repression during growth in methionine, loss of SAM binding to the leader RNA, and loss of SAM-directed transcription termination in vitro, these conserved elements are important for interaction of the leader RNA with SAM.
Selection for SAM-binding RNAs in vitro yielded RNAs that recognize the adenosine moiety, and not the entire molecule (28). Aptamers selected for binding to SAH and counterselected with SAM recognized both the adenosine and the sulfur/thioether moiety; these RNAs exhibited ≈2.5-fold preference for SAH over SAM or adenosine (29). The in vitro selections are limited in the number of nucleotides that can be randomized to generate a suitably diverse pool of RNAs. Biological RNAs, like the S box leaders, face no such limitations. The more complex S box leader RNAs are highly specific to SAM, because only SAM could promote transcription termination, and related compounds were also unable to act as competitive inhibitors of SAM-directed termination. The weak inhibition by sinefungin, a potent inhibitor of methyltransferases, suggests that the S box leader RNAs are more selective than certain SAM-binding proteins. The ability of the S box leaders to effectively discriminate against related metabolites is essential for an appropriate regulatory response in vivo. Uncovering the determinants for the specific recognition of SAM by the RNA will be a key step in understanding the molecular basis for this regulatory mechanism.
Our previous genetic studies indicated that the default state of the S box leaders is antitermination and that termination requires some regulatory element to prevent formation of the antiterminator structure. The results reported here demonstrate that SAM interacts directly with the leader RNA, in the absence of any regulatory protein, and blocks antiterminator formation by stabilization of the anti-antiterminator helix. The termination/read-through decision must be made before the elongating RNAP leaves the leader region termination site. It is therefore likely that the rate of transcription through the leader is a key parameter of this regulatory system, and pausing at discrete sites during transcription in vivo may be required to allow an opportunity for SAM to bind and the appropriate structure to be established in the nascent transcript. The in vitro transcription experiments described herein used low NTP concentrations to artificially slow the rate of transcription, whereas experiments by using T7 RNAP-generated RNA were designed to permit refolding of the RNA in the presence of SAM. Other cellular factors may play a role in controlling the rate of transcription in vivo. It is also possible that although the leader RNA is capable of a productive interaction with SAM under the conditions tested, there may be additional factors that modulate this interaction or control leader RNA folding in vivo.
Acknowledgments
We thank F. M. Hulett for providing the strain for production of B. subtilis RNAP; J. Pero for providing strain SA29; S. Lehman for technical assistance; M. Ibba for assistance with SAM-binding assays; and M. Ibba and V. Svetlov for comments on the manuscript. This work was supported by National Institutes of Health Grant GM63615 (to T.M.H.) and National Institutes of Health Predoctoral Fellowship F31 GM20923 (to B.A.M.M.).
Abbreviations
- RNAP
RNA polymerase
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
References
- 1.Henkin T M. Curr Opin Microbiol. 2000;3:149–153. doi: 10.1016/s1369-5274(00)00067-9. [DOI] [PubMed] [Google Scholar]
- 2.Henkin T M, Yanofsky C. BioEssays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 3.Grundy F J, Henkin M. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 4.Grundy F J, Winkler W C, Henkin T M. Proc Natl Acad Sci USA. 2002;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mironov A S, Gusarov I, Rafikov R, Lopez L E, Shatalin K, Kreneva R A, Perumov D A, Nudler E. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 6.Winkler W, Nahvi A, Breaker R R. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 7.Winkler W C, Cohen-Chalamish S, Breaker R R. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundy F J, Henkin T M. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 9.Switzer R L, Turner R J, Lu Y. Prog Nucleic Acid Res Mol Biol. 1999;62:329–367. doi: 10.1016/s0079-6603(08)60512-7. [DOI] [PubMed] [Google Scholar]
- 10.Murphy B A, Grundy F J, Henkin T M. J Bacteriol. 2002;184:2314–2318. doi: 10.1128/JB.184.8.2314-2318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auger S, Yuen W H, Danchin A, Martin-Verstraete I. Microbiology. 2002;148:507–518. doi: 10.1099/00221287-148-2-507. [DOI] [PubMed] [Google Scholar]
- 12.Yocum R R, Perkins J N, Howitt C L, Pero J. J Bacteriol. 1996;178:4604–4610. doi: 10.1128/jb.178.15.4604-4610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anagnostopoulos C, Spizizen J. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Y, Hulett F M. Mol Microbiol. 1998;28:1187–1197. doi: 10.1046/j.1365-2958.1998.00882.x. [DOI] [PubMed] [Google Scholar]
- 15.Hager D A, Jin D J, Burgess R R. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 16.Grundy F J, Moir T R, Haldeman M T, Henkin T M. Nucleic Acids Res. 2002;30:1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 18.Mansilla M C, Albanesi D, de Mendoza D. J Bacteriol. 2000;182:5885–5892. doi: 10.1128/jb.182.20.5885-5892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wabiko H, Ochi K, Nguyen D M, Allen E R, Freese E. J Bacteriol. 1988;170:2705–2710. doi: 10.1128/jb.170.6.2705-2710.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boeck L D, Clem G M, Wilson M M, Westhead J E. Antimicrob Agents Chemother. 1973;3:49–56. doi: 10.1128/aac.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schluckebier G, Kozak M, Bleimling N, Weinhold E, Saenger W. J Mol Biol. 1997;265:56–67. doi: 10.1006/jmbi.1996.0711. [DOI] [PubMed] [Google Scholar]
- 22.Winkler W C, Grundy F J, Murphy B A, Henkin T M. RNA. 2001;7:1165–1172. doi: 10.1017/s1355838201002370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posnick L M, Samson L D. J Bacteriol. 1999;181:6756–6762. doi: 10.1128/jb.181.21.6756-6762.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene R C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtis R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaecter A, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 542–560. [Google Scholar]
- 25.MacDonald L E, Durbin R K, Dunn J J, McAllister W T. J Mol Biol. 1994;238:145–158. doi: 10.1006/jmbi.1994.1277. [DOI] [PubMed] [Google Scholar]
- 26.Grundy F J, Henkin T M. In: Bacillus subtilis and Its Closest Relatives: From Genes to Cells. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 245–254. [Google Scholar]
- 27.Grundy F J, Henkin T M. Front Biosci. 2003;8:d20–d31. doi: 10.2741/908. [DOI] [PubMed] [Google Scholar]
- 28.Burke D H, Gold L. Nucleic Acids Res. 1997;25:2020–2024. doi: 10.1093/nar/25.10.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gebhardt K, Shokraei A, Babaie E, Lindqvist B H. Biochemistry. 2000;39:7255–7265. doi: 10.1021/bi000295t. [DOI] [PubMed] [Google Scholar]