Abstract
The lipooligosaccharides (LOS) of Haemophilus ducreyi are highly sialylated, a modification that has been implicated in resistance to host defense and in virulence. In previous work, we demonstrated that H. ducreyi scavenges sialic acid from the extracellular milieu and incorporates those residues into LOS. Here we report that H. ducreyi can use unnatural sialic acids bearing elongated N-acyl groups from three to seven carbon atoms in length, resulting in outer membrane presentation of unnatural sialyl-LOS. The unnatural variant comprises ≈90% of cell surface sialosides when exogenous substrates are added to the media at micromolar concentrations, despite the availability of natural sialic acid in the growth media. Although they represent the majority of cell surface sialosides, analogs with longer N-acyl groups diminish the overall level of LOS sialylation, culminating in complete inhibition of LOS sialylation by N-octanoyl sialic acid. Thus, sialylation of H. ducreyi LOS can be modulated with respect to the structure of the terminal sialic acid residue and the extent to which the LOS acceptor is modified by supplying the bacteria with various sialic acid analogs.
The Gram-negative human mucosal pathogen Haemophilus ducreyi causes chancroid, a sexually transmitted disease prevalent in developing countries (1). In addition, this pathogen poses a significant risk factor for the transmission of HIV (2, 3). The lipooligosaccharide (LOS) of H. ducreyi is considered a major virulence factor and has been implicated in the adherence of H. ducreyi to human foreskin fibroblasts and keratinocytes (4, 5). The recent structural identification of major LOS glycoforms from H. ducreyi has provided insight into their contribution to pathogenesis (6–8). The principal cell surface glycoform expressed by many wild-type strains contains a terminal N-acetyllactosamine (Galβ1–4GlcNAc) unit that is often sialylated to produce a sialyl-N-acetyllactosamine (Siaα2–3Galβ1–4GlcNAc) structure (7, 9). Similar structures are prevalent on human glycoconjugates, and it has been proposed that by mimicking host structures, H. ducreyi can thwart recognition by host immune functions (10).
The ability to perturb sialylated structures on H. ducreyi would facilitate elucidation of the roles of this sugar in adherence, infection, and virulence. In mammalian cells, alterations in the structure of sialic acid can be achieved by metabolic conversion of unnatural substrates (11–14). When fed to cells, unnatural variants of N-acetylmannosamine (ManNAc) are transformed by the sequential action of five enzymes (ManNAc-6-kinase, NeuAc-9-phosphate synthase, NeuAc-9-phosphate phosphatase, CMP-NeuAc synthetase, and a sialyltransferase) to the corresponding unnatural sialosides resident on the cell surface. A variety of mannosamine derivatives successfully navigate this biosynthetic process, including analogs with extended N-acyl groups (11) and analogs modified at the N-acyl position with reactive electrophiles (12–14). Cells bearing modified sialic acids have shown altered profiles of virus binding (15) and altered responses to adhesion molecules and growth factors (16, 17), effects that implicate sialic acid as a determinant of each process. These examples prompted us to consider whether bacterial sialylation pathways are amenable to biosynthetic modulation. If so, the approach would be a powerful tool for elucidating the functions of sialic acid in pathogens such as H. ducreyi, and a potential mechanism for modulating pathogenicity.
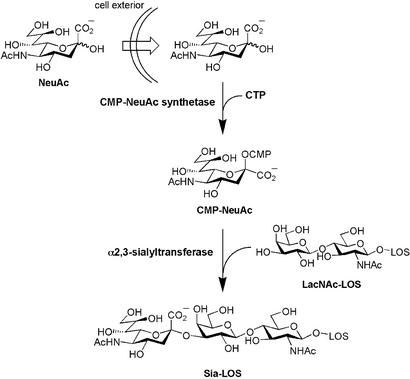
We have previously investigated the biosynthetic origin of sialyl-LOS in H. ducreyi (18). Unlike mammalian cells, which biosynthesize the majority of their sialic acid from the committed precursor N-acetylmannosamine (ManNAc), H. ducreyi must import sialic acid from the extracellular environment; sialyl-LOS are generated exclusively from exogenous sialic acid that has been scavenged, converted to CMP-NeuAc, and transferred onto LOS by a sialyltransferase (Fig. 1). An analogous pathway has been described for Haemophilus influenzae (19) and in Escherichia coli, sialic acid can be imported from the extracellular pool (20) and/or synthesized via a multistep pathway from exogenous ManNAc or GlcNAc (21).
Figure 1.
The proposed sialic acid biosynthetic pathway for H. ducreyi. NeuAc is scavenged from the environment, activated to CMP-NeuAc by CMP-NeuAc synthetase, and transferred onto an acceptor-LOS by a sialyltransferase.
Here we report that the cellular biosynthetic pathway for sialyl-LOS in H. ducreyi is promiscuous for unnatural sialic acid analogs modified at the N-acyl position with elongated alkyl chains. H. ducreyi incubated with unnatural sialic acids displayed the corresponding unnatural sialyl-LOS on the outer membrane, as determined and quantified by using highly sensitive mass spectrometric techniques. Although metabolized by the cells with high efficiency compared with native sialic acid, certain analogs also depressed the overall sialylation of H. ducreyi glycoconjugates. The results indicate that both the structure of the sialic acid residues and the extent of sialylation can be modulated by exposure to unnatural substrates.
Materials and Methods
General.
All reagents used in chemical syntheses were obtained from commercial suppliers and used without further purification unless otherwise noted. Mannosamine hydrochloride was from Pfanstiehl Laboratories (Waukegan, IL). Sialic acid, anhydrous hydrazine and hemin chloride were purchased from Sigma. [1-13C]-Sialic acid was from Omicron Biochemicals (South Bend, IN). Sialic acid aldolase was from Toyobo (catalog no. NAL-301, Shinko America, New York). GC Medium Base, brain heart infusion, and hemoglobin were from Difco. IsoVitaleX and petri dishes were purchased from Becton Dickinson; dish diameter: 60 mm for 10 ml volume of solid media or dish diameter: 100 mm for 20 ml volume of solid media. Distilled H2O was used in all manipulations.
Synthesis of N-Acylmannosamine Compounds.
The majority of N-acylmannosamines were prepared by reacting the appropriate symmetrical anhydride with mannosamine generated by neutralization of mannosamine hydrochloride with NaOMe in MeOH. N-Octanoylmannosamine was prepared by reaction of neutralized mannosamine hydrochloride with octanoyl isobutylchloroformate, generated from octanoic acid by using a procedure similar to that previously reported (12). For all compounds, purification was accomplished by silica gel chromatography eluting with a gradient of 20:1 to 4:1 CHCl3/MeOH.
Enzymatic Synthesis of Unnatural Sialic Acids.
Unnatural sialic acids were synthesized enzymatically by using sialic acid aldolase to condense pyruvate with the unnatural mannosamines. In a typical procedure, 0.88 mmol of N-acylmannosamine was dissolved in 8.8 ml of 0.05 M KH2PO4 (pH 7.2) along with 8.8 mmol of pyruvic acid, 440 μl of a 1% NaN3 solution, and 20 units of sialic acid aldolase (Toyobo lot no. 85211). The reaction was incubated at 37°C with shaking for 12 h, after which 1H NMR analysis indicated the reaction was complete. The resulting sialic acid was purified by anion exchange chromatography (Bio-Rad AG1-X2, formate form), eluting with a gradient of 1–2.5 M formic acid at 1 ml/min. Fractions were analyzed for the presence of sialic acid by using the periodate-resorcinol method (22). Fractions containing the desired compound were collected and concentrated in vacuo, redissolved in H2O and lyophilized. 1H and 13C NMR spectral data for all mannosamine and sialic acid analogs are provided in the supporting information, which is published on the PNAS web site, www.pnas.org.
Preparation of Chocolate Agar Plates.
Media for solid chocolate agar plates contained GC Medium Base, 1% (wt/vol) hemoglobin, and 1% (vol/vol) IsoVitaleX (23). The solution for the solid media was autoclaved, cooled to 45°C, and subsequently poured into petri dishes to obtain regular chocolate agar plates. For incorporation studies, sialic acids were dissolved in water, sterile filtered, and added to the autoclaved media, which had been cooled to 45°C. The warm media was then poured into petri dishes. Usually, a total of 10 ml of media containing the sialic acid analogs was prepared, yielding sialic acid concentrations ranging from 0.05 to 1 mM.
Incorporation of Sialic Acid Analogs into H. ducreyi LOS.
H. ducreyi strain 35000 was grown directly on chocolate agar plates that contained sialic acids (incubation in a candle jar at 34°C for 2 d). As control experiments, bacteria were also cultivated on media lacking added sialic acids (“blank”). Bacteria were harvested for isolation and analysis of LOS.
Phenol Extraction of LOS.
The harvested bacterial cells obtained from plates were washed with 0.5–1 ml of saline PBS (pH 7.4) containing 0.15 mM CaCl2 and 0.5 mM MgCl2. The washed cells were resuspended in 0.5 ml of H2O and transferred into a heat-resistant Eppendorf tube. An equal volume of a 90% phenol/H2O solution (wt/vol) was added and the tube was heated at 65°C for 35 min (vigorous stirring every 10 min) (24). Subsequently, the mixture was centrifuged at 4°C (12,000 × g) for 30 min. The phases were separated and the phenol phase was re-extracted with 0.5 ml of H2O. The combined water extracts were concentrated to a volume of 100 μl. The LOS was precipitated by addition of 100 μl of ethanol and cooling at −20°C overnight. The LOS pellet was obtained by centrifugation at 4°C (8,500 × g) for 45 min.
Preparation of O-Deacylated LOS.
O-Deacylated LOS (O-LOS) are generally more amenable to mass spectrometric analysis than intact LOS (6). The dried LOS were O-deacylated by incubation with 50 μl of anhydrous hydrazine at 37°C for 35 min (25). The mixture was cooled, and chilled acetone (200 μl) was slowly added. The solution was kept at −20°C for 2 h and then centrifuged at 4°C (12,000 × g, 45 min). The O-LOS precipitate was separated from the supernatant, dried, and finally redissolved in 20 μl of H2O. Aliquots of the sample were desalted by drop dialysis using nitrocellulose membranes (20 μ, Millipore).
Mass Spectrometric Characterization of LOS.
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) MS was performed on a Voyager DE mass spectrometer as described (18). The samples were further analyzed by electrospray ionization MS (ESI-MS) on a quadrupole orthogonal time-of-flight mass spectrometer (QSTAR, MDS Sciex) equipped with a Protana nanospray ion source (18). Mass spectra (ESI-MS) and tandem mass spectra (ESI-MS/MS) of O-deacylated LOS were recorded in positive ion mode, and calibrated as described previously (18). Additional internal calibration by using well characterized O-LOS glycoforms with exact molecular masses M = 2956.0004 ([M + 3H]3+ at m/z 986.3413) and M = 2793.9476 ([M + 3H]3+ at m/z 932.3237) yielded mass accuracies of ±10 ppm. Exact masses of unnatural sialyl-LOS are provided as supporting information (see Table 4, which is published as supporting information on the PNAS web site).
Results
Metabolic Incorporation of Unnatural Sialic Acids into H. ducreyi LOS.
We generated a panel of sialic acid analogs bearing elongated N-acyl groups of three to eight carbon atoms by enzymatic synthesis from the corresponding mannosamine analogs. N-Propanoyl (SiaProp), N-butanoyl (SiaBut), N-pentanoyl (SiaPent), N-hexanoyl (SiaHex), N-heptanoyl (SiaHept), or N-octanoyl (SiaOct) sialic acid (Table 1) was added to the growth media, and its incorporation into LOS was determined by MS. 13C-Sialic acid (13C-NeuAc) was used as a control. The incorporation of unnatural sialic acids into LOS was first determined by MS analysis (MALDI-TOF) of isolated O-LOS from H. ducreyi grown on solid media impregnated with the various synthetic analogs. Under linear time-of-flight conditions, the molecular ions for individual O-LOS species typically appeared as unresolved isotopes whose centroids corresponded to the average mass. Fig. 2 shows an example of a LOS structure and the expected masses of various fragment ions. Sialylated glycoforms that express natural sialic acid generally show singly deprotonated ions [M – H]− at m/z 3,247 and m/z 3,124.
Table 1.
Panel of sialic acid analogs bearing N-acyl groups varying in chain length
Figure 2.
Structure of LOS of H. ducreyi. Expected average masses for the deprotonated molecular ions of O-deacylated LOS recorded in negative ionization mode [M – H]− are shown. Masses in bold represent the LOS containing one phosphoethanolamine (PEA) group (n = 0); masses in gray represent the LOS containing two PEA groups (n = 1). All heptoses are l-glycero-d-manno-heptoses except for the branch heptose (d-glycero-d-manno-heptose).
MALDI mass spectra of O-LOS isolated from H. ducreyi grown in media supplemented with 1 mM 13C-NeuAc, SiaProp, SiaBut, and SiaPent are shown in Fig. 3 A–D, respectively. Previously, we reported the incorporation of both natural and/or 13C-isotopically labeled sialic acid under identical conditions as described here. The MALDI-MS spectra of those experiments, of which Fig. 3A is an example, showed the efficient incorporation of exogenous sialic acid into LOS glycoforms containing terminal N-acetyllactosamine (18). The unsialylated glycoforms terminating in N-acetyllactosamine (Gal-GlcNAc-Gal-Hep-Glc-Hep3(PEA)0,1-KdoP(PEA)-Lipid A, where PEA = phosphoethanolamine) yielded deprotonated molecular ions [M – H]− at m/z 2,956 and 2,833 (Fig. 2). The peaks at m/z 2,794 and m/z 2,671 arose from the additional loss of a terminal galactose residue. The peak at m/z 3,159 probably corresponds to a glycoform generated by the addition of a GlcNAc residue to the lactosamine acceptor (Fig. 2). In addition, some minor ion species were observed that resulted from salt adducts, such as Na+ and Fe+3, as reported (6). Fig. 3A shows LOS glycoforms that have incorporated isotopically labeled sialic acid (13C-NeuAc) with [M – H]− at m/z 3,125 (1 PEA) and m/z 3,248 (2 PEA), revealing a 1-Da shift compared with natural, nonlabeled NeuAc. When H. ducreyi were incubated with SiaProp, no glycoforms modified with natural sialic acid were detected. Instead, exclusive incorporation of SiaProp was observed as [M – H]− at m/z 3,138 (1 PEA) and m/z 3,261 (2 PEA) (Fig. 3B). Similarly, LOS isolated from bacteria that were grown with SiaBut or SiaPent expressed sialylated LOS with molecular ions [M – H]− at m/z 3,275 (SiaBut-LOS, Fig. 3C) and m/z 3,289 (SiaPent-LOS, Fig. 3D).
Figure 3.
Negative-ion MALDI-TOF mass spectrum showing [M – H]− ions for O-deacylated LOS isolated from H. ducreyi. Bacteria were grown on solid media containing 1 mM 13C-NeuAc (A), SiaProp (B), SiaBut (C), and SiaPent (D). Mass peaks representing LOS sialylated with the corresponding sialyl analog are labeled in bold. Asterisk refers to m/z at 3,159.9.
To confirm the structures of unnatural sialyl-LOS, we performed ESI-MS/MS. LOS samples were O-deacylated and subsequently analyzed in positive-ionization mode. Both doubly and triply charged molecular ions for each glycoform were observed with mass accuracies of <20 ppm relative to their calculated values. In ESI-MS/MS these O-LOS molecular ions generated carbohydrate sequence ions resulting from cleavage of glycosidic bonds. Ion nomenclature is that proposed by Domon and Costello (26). As recently reported, sialyl-N-acetyllactosamine fragment ions, so called B3 ions, can be used to identify the structural context and identity of the sialic acid residue (18). Fig. 4 shows ESI-MS/MS spectra of O-LOS from bacteria treated with the various unnatural sialic acids, up to SiaHept. In each case, fragment ions corresponding to unnatural sialyl-N-acetyllactosamine (B3), sialyl-Gal (B2) and sialic acid (B1) were observed. Bacteria treated with SiaOct showed no detectable mass peak corresponding to the SiaOct-LOS. These results indicate that unnatural sialic acids with N-acyl chains shorter than eight carbon atoms are metabolized by H. ducreyi and incorporated into LOS.
Figure 4.
Positive-ion ESI-MS/MS spectra of O-deacylated LOS. Triply or doubly charged molecular ions were selected as precursor ions and subsequently fragmented. Sialylated glycoforms show the characteristic fragment ions B1, B2, B3, and dehydrated B1 (B1 – H2O) corresponding to the incorporated sialic acid analog SiaProp (A), SiaBut (B), SiaPent (C), SiaHex (D), and SiaHept (E). Incorporation of SiaProp into LOS yielded a B3 ion at m/z 671.2 (A), SiaBut-LOS revealed a B3 ion at m/z 685.3 (B), SiaPent-LOS revealed a B3 ion at m/z 699.3 (C), SiaHex-LOS revealed a B3 ion at m/z 713.3 (D), and SiaHept-LOS revealed a B3 ion at m/z 727.3 (E). Similar to observed B3 fragment ions, B1 ions representing the sialic acid fragment and the corresponding dehydrated B1 ions (B1 – H2O) can also be used to confirm the incorporation of a modified sialic acid residue. SiaProp-, SiaBut-, SiaPent-, SiaHex-, and SiaHept-containing LOS revealed ion pairs at m/z 306.1/288.1, m/z 320.1/302.1, m/z 334.2/316.1, m/z 348.2/330.2, and m/z 362.2/344.2, respectively.
Quantitation of Sialyl-LOS by MS.
In previous reports we have demonstrated that MS can be used to determine both molecular weights and relative abundances of LOS glycoforms from complex mixtures. LOS preparations from commercial Salmonella typhimurium (standard for evaluation purpose) and from pathogenic Haemophilus and Neisseria species were analyzed by ESI-MS (27) and by matrix-assisted laser desorption ionization MS (6). A preliminary study addressing quantification of sialic acid from LOS mixtures has been reported†† and we have added to these data here by analyzing 10 defined mixtures of sialylated LOS mixing a non-sialylated bacterial strain with a strain overexpressing sialylated glycoforms. The LOS mixtures were analyzed repeatedly by three independent methods, SDS/PAGE, MALDI-MS, and ESI-MS (see supporting information). As described in more detail in the supporting information, each individual method showed a good linear correspondence between amount of sialylated-LOS present in the glycoform mixture based on densitometry reading (SDS/PAGE) and ion abundance as determined by MS. Thus, we are confident that ion abundance provides an accurate representation of the actual abundance of the LOS glycoform.
In this sialic acid incorporation study, the percentage of total sialic acids represented by the unnatural variant (SiaX) and the percent sialylation of acceptor (i.e., the percent of acceptor LOS bearing a sialic acid residue relative to those capable of sialylation) were determined for each unnatural analog. Relative abundances were determined by peak height (ESI-MS) or by integrating the area (MALDI-MS) of the molecular ion peaks for each relevant glycoform (see Table 2). When 13C-NeuAc was added to the media at a concentration of 1 mM, 38% of the LacNAc-containing acceptor LOS chains were found to be sialylated exclusively with the isotopically labeled sugar (Table 2). In the absence of added 13C-NeuAc, only 13% of acceptor LOS were sialylated, the result of metabolism of endogenous sialic acid present in the media at 0.5 μM (18). We have previously shown that numerous H. ducreyi strains synthesize sialylated LOS glycoforms when sialic acid is present in the growth media, but only 30–40% of available LacNAc-terminated LOS acceptor molecules are sialylated under those circumstances (7, 9). In fact, for strain 35000 we have determined that the sialylation of acceptor LOS reaches a plateau level of 38% with increasing concentrations of exogenous natural sialic acid (% sialylation = Sia-LOS/(Sia-LOS + LacNAc-LOS) where 100% sialylation corresponds to the case in which no LacNAc-terminated LOS is detectable; ref. 18). The observation of incomplete LOS sialylation, even in the presence of high exogenous sialic acid concentrations, may reflect a limiting enzymatic activity in the pathway that converts sialic acid to CMP-NeuAc, or limiting sialyltransferase activity in the bacterial cells.
Table 2.
Incorporation of sialic acid analogs into H. ducreyi LOS as determined by MS
| Sialyl substrate* | Concentration, mM | % SiaX-LOS of total sialylated LOS† | % Sialylation of acceptor with SiaX substrate‡ |
|---|---|---|---|
| No added substrate | 0.0005* | – | 13* |
| 13C-NeuAc | 1 | 100§ | 38 |
| SiaProp | 1 | >95 | 36 |
| SiaBut | 1 | >89 | 19 |
| SiaPent | 1 | >90 | 8 |
| SiaHex | 1 | >87 | 8 |
| SiaHept | 1 | >75 | 8 |
| SiaOct | 1 | – | – |
Approximately 0.5 μM endogenous NeuAc exist in the growth media, which leads to 13% sialylation of the lactosamine-bearing LOS (acceptor).
Percent of total sialylated LOS containing SiaX analog: [SiaX-LOS/(SiaX-LOS + Sia-LOS)] (competition with 0.5 μM endogenous NeuAc).
Percent of substrate SiaX-LOS relative to the combined populations of SiaX-LOS plus unmodified LacNAc-bearing acceptor LOS: [SiaX-LOS/(SiaX-LOS + LacNAc-LOS)].
One hundred percent incorporation of 13C-NeuAc was observed.
SiaProp was incorporated into LOS with the same efficiency and overall sialylation level as natural sialic acid (13C-NeuAc) (Table 2). SiaBut was also incorporated into LOS at high efficiency (>89% of total sialic acids were represented by the unnatural variant), but the overall level of LOS sialylation diminished to 19% of total LOS (Sia-LOS + LacNAc-LOS). This trend was mirrored by SiaPent, SiaHex, and SiaHept. In these three cases, the unnatural sialic acid dominated over natural sialic acid (>75% representation), but the overall level of sialylation decreased to 8% of total LOS. As mentioned above, no incorporation of SiaOct into LOS was observed. Interestingly, this unnatural analog decreased modification of LOS with natural sialic acid to undetectable levels.
Competition of Natural Sialic Acid with SiaProp.
To determine whether the sialic acid biosynthetic pathway of H. ducreyi had a preference for natural sialic acid compared with SiaProp, we performed competition experiments. Bacteria were grown in the presence of varying concentrations of both SiaProp and 13C-NeuAc, and the efficiency of utilization was quantified by ESI-MS analysis. Table 3 shows the ratio of SiaProp to total sialic acids added to the media, and the observed incorporation of SiaProp (% SiaProp-LOS of total sialylated LOS). At each concentration, SiaProp was underrepresented within LOS, suggesting a preference by the biosynthetic machinery for the natural sugar by ≈3-fold.
Table 3.
Efficiency of SiaProp incorporation into LOS in the presence of 13C-NeuAc
| SiaProp, mM | 13C-NeuAc, mM | % SiaProp-LOS of total sialylated LOS* | % Total sialylation of acceptor† |
|---|---|---|---|
| 0.1 | 0.0 | >95 | 32 |
| 0.1 | 0.05 | 31 | 38 |
| 0.1 | 0.1 | 20 | 35 |
| 0.05 | 0.1 | 9 | 36 |
| 0.1 | 0.5 | 6 | 37 |
| 0.0 | 0.1 | 0‡ | 40 |
Percent of total sialylated LOS containing SiaProp: [SiaProp-LOS/(SiaProp-LOS + Sia-LOS)].
Percent of total sialylated LOS relative to the combined populations of total sialylated LOS plus unmodified LacNAc-bearing acceptor LOS: [(SiaProp-LOS + Sia-LOS)/(SiaProp-LOS + Sia-LOS + LacNAc-LOS)].
One hundred percent incorporation of 13C-NeuAc was observed.
Discussion
In this study, we showed that H. ducreyi is capable of incorporating unnatural sialic acid analogs into LOS. Sialic acid analogs extended at the N-acyl position with chains up to seven carbon atoms in length were incorporated into LOS at significant levels. This observation indicates that the biosynthetic enzymes are tolerant of unnatural substrates. Furthermore, if a permease is responsible for the uptake of these sialic acid substrates, our results suggest that it also tolerates modifications at the N-acyl position. A combination of mass spectrometric techniques was applied for structural characterization and identification of the engineered glycoforms. Unlike other previously described techniques used for the analysis of unnatural cell surface sialic acids (i.e., flow cytometry), MS provides both quantitation and structural confirmation.
Although the enzymes are successful in metabolizing the analogs, extension of the chain length causes a precipitous drop in the percent sialylation of LOS acceptors. In fact, when the N-acyl group bears eight carbon atoms, no sialylation of LOS with either SiaOct or NeuAc was detectable by MS. It is possible that SiaOct is not recognized by the permease and therefore excluded from the cell. In this case, however, one would expect the LOS to be normally sialylated with NeuAc. The complete absence of natural NeuAc residues within the LOS suggests that SiaOct inhibits one or more enzymes involved in the sialoside biosynthetic pathway. In preliminary work we have excluded the putative permease as a point of inhibition, because radiolabeled NeuAc is taken up by H. ducreyi at normal levels in the presence of high concentrations of SiaOct (see supporting information). Identification of the mechanism by which the unnatural sialic acid analogs effect the percent sialylation of LOS acceptors will require more detailed analysis of their activities with isolated enzymes in the pathway.
The ability of H. ducreyi to import and metabolize sialic acid analogs provides a new avenue for studying the roles of sialyl-LOS in infection and immunological recognition. By altering the structure of the sialic acid (via SiaProp through SiaHept), or by deleting it altogether (via SiaOct), one may be able to perturb sialic acid-dependent processes in vitro or in vivo. Furthermore, H. ducreyi bearing altered sialic acids can be probed for resistance/susceptibility to immunological recognition and the bactericidal effects of human serum, processes that are modulated by similar sialyl oligosaccharides in humans. Because unnatural sialic acids can elicit a vigorous immune response in mammals (28, 29), their incorporation into H. ducreyi may provide an avenue for vaccine development. Given the similarity of the H. ducreyi sialoside biosynthetic pathway with the analogous pathway in other bacteria, unnatural sialic acid metabolism may be a broadly applicable mechanism for modulating bacterial outer membrane structure. Finally, “cell surface remodeling” experiments that exploit reactive electrophiles within unnatural sialosides might be performed on H. ducreyi to further alter the structures of their LOS (13, 14).
Supplementary Material
Acknowledgments
We thank Dr. Nicole M. Samuels for experimental assistance, Applied Biosystems (Framingham, MA) for generous support of the MALDI-TOF instrumentation in our laboratory (B.W.G.), and the Arnold and Mabel Beckman Foundation for funding. This work was supported by National Institutes of Health Grants AI31254 (to B.W.G.) and GM 58867 (to C.R.B.), and by the Director, Office of Science, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering and the Office of Energy Biosciences of the U.S. Department of Energy under Contract DE-AC03-76SF00098.
Abbreviations
- LOS
lipooligosaccharides
- MALDI-TOF
matrix-assisted laser desorption ionization–time-of-flight
- ESI-MS
electrospray ionization MS
Footnotes
Tullius, M. V. & Gibson, B. W. (1998) Proceedings of the 46th ASMS Conference on Mass Spectrometry and Allied Topics, May 31–June 4, Orlando, FL (abstr.), p. 399.
References
- 1.Jessamine P G, Ronald A R. Med Clin North Am. 1990;74:1417–1431. doi: 10.1016/s0025-7125(16)30488-6. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Morbid Mortal Wkly Rep. 1998;47:1–24. [Google Scholar]
- 3.Spinola S M, Orazi A, Arno J N, Fortney K, Kotylo P, Chen C Y, Campagnasi A A, Hood A F. J Infect Dis. 1996;173:394–402. doi: 10.1093/infdis/173.2.394. [DOI] [PubMed] [Google Scholar]
- 4.Alfa M J, DeGagne P. Microb Pathog. 1997;22:39–46. doi: 10.1006/mpat.1996.0089. [DOI] [PubMed] [Google Scholar]
- 5.Gibson B W, Campagnari A A, Melaugh W, Phillips N J, Apicella M A, Grass S, Wang J, Palmer K, Munson R S J. J Bacteriol. 1997;179:5062–5071. doi: 10.1128/jb.179.16.5062-5071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson B W, Engstrom J, John C M, Hines W, Falick A M. J Am Soc Mass Spectrom. 1997;8:645–658. [Google Scholar]
- 7.Melaugh W, Phillips N J, Campagnari A A, Tullius M V, Gibson B W. Biochemistry. 1994;33:13070–13078. doi: 10.1021/bi00248a016. [DOI] [PubMed] [Google Scholar]
- 8.Schweda E K, Sundstrom A C, Eriksson L M, Jonasson J A, Lindberg A A. J Biol Chem. 1994;269:12040–12048. [PubMed] [Google Scholar]
- 9.Melaugh W, Campagnari A A, Gibson B W. J Bacteriol. 1996;178:564–570. doi: 10.1128/jb.178.2.564-570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandrell R E, McLaughlin R, Kwaik Y A, Lesse A, Yamasaki R, Gibson B, Spinola S M, Apicella M A. Infect Immun. 1992;60:1322–1328. doi: 10.1128/iai.60.4.1322-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keppler O T, Horstkorte R, Pawlita M, Schmidt C, Reutter W. Glycobiology. 2001;11:11R–18R. doi: 10.1093/glycob/11.2.11r. [DOI] [PubMed] [Google Scholar]
- 12.Mahal L K, Yarema K J, Bertozzi C R. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 13.Yarema K J, Mahal L K, Bruehl R E, Rodriguez E C, Bertozzi C R. J Biol Chem. 1998;273:31168–31179. doi: 10.1074/jbc.273.47.31168. [DOI] [PubMed] [Google Scholar]
- 14.Saxon E, Bertozzi C. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 15.Keppler O T, Stehling P, Herrmann M, Kayser H, Grunow D, Reutter W, Pawlita M. J Biol Chem. 1995;270:1308–1314. doi: 10.1074/jbc.270.3.1308. [DOI] [PubMed] [Google Scholar]
- 16.Raimund Wieser J, Heisner A, Stehling P, Oesch F, Reutter W. FEBS Lett. 1996;395:170–173. doi: 10.1016/0014-5793(96)01029-0. [DOI] [PubMed] [Google Scholar]
- 17.Collins B E, Fralich T J, Itonori S, Ichikawa Y, Schnaar R L. Glycobiology. 2000;10:11–20. doi: 10.1093/glycob/10.1.11. [DOI] [PubMed] [Google Scholar]
- 18.Schilling B, Goon S, Samuels N M, Gaucher S P, Leary J A, Bertozzi C R, Gibson B W. Biochemistry. 2001;40:12666–12677. doi: 10.1021/bi0107849. [DOI] [PubMed] [Google Scholar]
- 19.Vimr E, Lichtensteiger C, Steenbergen S. Mol Microbiol. 2000;36:1113–1123. doi: 10.1046/j.1365-2958.2000.01925.x. [DOI] [PubMed] [Google Scholar]
- 20.Martinez J, Steenbergen S, Vimr E. J Bacteriol. 1995;177:6005–6010. doi: 10.1128/jb.177.20.6005-6010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringenberg M, Lichtensteiger C, Vimr E. Glycobiology. 2001;11:533–539. doi: 10.1093/glycob/11.7.533. [DOI] [PubMed] [Google Scholar]
- 22.Jourdian G W, Dean L, Roseman S. J Biol Chem. 1971;246:430–435. [PubMed] [Google Scholar]
- 23.Palmer K L, Goldman W E, Munson R S., Jr Med Microbiol. 1996;21:13–19. doi: 10.1046/j.1365-2958.1996.00615.x. [DOI] [PubMed] [Google Scholar]
- 24.Inzana T J. J Infect Dis. 1983;148:492–499. doi: 10.1093/infdis/148.3.492. [DOI] [PubMed] [Google Scholar]
- 25.Helander I M, Nummila K, Kilpelainen I, Vaara M. Prog Clin Biol Res. 1995;392:15–23. [PubMed] [Google Scholar]
- 26.Domon B, Costello C E. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
- 27.Gibson B, Melaugh W, Phillips N, Apicella M, Campagnari A, Griffiss J. J Bacteriol. 1993;175:2702–2712. doi: 10.1128/jb.175.9.2702-2712.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Guo Z, Yang Q, Sad S, Jennings H J. J Biol Chem. 2000;275:32832–32836. doi: 10.1074/jbc.C000573200. [DOI] [PubMed] [Google Scholar]
- 29.Lemieux G A, Bertozzi C R. Chem Biol. 2001;8:265–275. doi: 10.1016/s1074-5521(01)00008-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.