Abstract
The mRNA and protein expression in Saccharomyces cerevisiae cultured in rich or minimal media was analyzed by oligonucleotide arrays and quantitative multidimensional protein identification technology. The overall correlation between mRNA and protein expression was weakly positive with a Spearman rank correlation coefficient of 0.45 for 678 loci. To place the data sets in a proper biological context, a clustering approach based on protein pathways and protein complexes was implemented. Protein expression levels were transcriptionally controlled for not only single loci but for entire protein pathways (e.g., Met, Arg, and Leu biosynthetic pathways). In contrast, the protein expression of loci in several protein complexes (e.g., SPT, COPI, and ribosome) was posttranscriptionally controlled. The coupling of the methods described provided insight into the biology of S. cerevisiae and a clustering strategy by which future studies should be based.
Although quantitative and global analysis of mRNA expression has provided comprehensive access to the transcriptome of a cell (1), posttranscriptional regulatory mechanisms (2) can result in discordant mRNA and protein abundances (3, 4) or mRNA and protein expression changes resulting from a stimulus (5, 6). To date, the largest study analyzing the correlation between protein and mRNA expression was of 245 loci from which a weakly positive correlation between mRNA and protein expression ratios was determined (6). In each analysis described so far, the correlation of mRNA and protein abundance or expression of a limited number of highly abundant proteins has been discussed. Current quantitative proteomic analyses have not gathered comprehensive enough data sets to detail patterns in the correlation of mRNA and protein expression and develop strategies by which to organize the correlated expression data sets.
Metabolic labeling by growth of cells in media with either 14N or 15N as the sole nitrogen source has been demonstrated as a potential method for quantitative proteomics in Saccharomyces cerevisiae (7, 8). In the current paper we compared the mRNA and protein expression changes of the S. cerevisiae strain S288C when cultured on either rich media (yeast extract/peptone/dextrose, YEPD) or minimal media with either 14N or 15N ammonium sulfate as the sole nitrogen source by using oligonucleotide array analysis (9) and quantitative (8) multidimensional protein identification technology (MudPIT) (10–12). The control of amino acid biosynthesis in S. cerevisiae has been reviewed (13), transcriptional analyses of 3-aminotriazole-induced amino acid starvation by cDNA array analysis (14), and oligonucleotide array analysis of culturing in rich or minimal media (9) have been carried out. Despite the previous analyses of amino acid biosynthesis, the coupled mRNA and protein expression analyses by oligonucleotide arrays and quantitative proteomics described herein provided novel insight into the biology of S. cerevisiae. To determine the effect of culturing S. cerevisiae on rich or minimal media, we clustered the mRNA and protein expression data set based on biochemically characterized protein pathways and complexes described in the literature and accessed via the Yeast Proteome Database (15) and MIPS (16) because a comparative assessment of the two global protein complexes analysis in S. cerevisiae postulated that >50% of the data sets are spurious (17).
Materials and Methods
Materials.
Ammonium-15N sulfate (99 atom %) and ammonium-14N sulfate (99.99 atom %) were products of Aldrich (Milwaukee, WI). Difco bacto peptone, dextrose, yeast extract, and yeast nitrogen base without amino acids or ammonium sulfate were products of Becton Dickinson Microbiology Systems (Sparks, MD). Glacial acetic acid and HPLC-grade acetonitrile (ACN) and HPLC-grade methanol was purchased from Fischer Scientific. Heptafluorobutyric acid (HFBA) was obtained from Pierce. All other chemical reagents were obtained from Sigma.
Growth of S. cerevisiae
After preparing overnight cultures in identical pH-controlled medias, S. cerevisiae strain S288C was grown to mid log phase (OD600 = 0.6) in YEPD (10 g of Bacto yeast extract, 20 g of Bacto peptone, and 20 g of dextrose per liter), 14N or 15N minimal media (1.7 g of yeast nitrogen base without amino acids and ammonium sulfate, 20 g of dextrose, and 5 g of either ammonium sulfate per liter) at 30°C followed by centrifugation at 1,000 × g. The pellets were washed three times with 1× PBS (1.4 mM NaCl/0.27 mM KCl/1 mM Na2HPO4/0.18 mM KH2PO4, pH 7.4). After washing, samples were prepared as described below for oligonucleotide array analysis or MudPIT. S. cerevisiae was cultured in each type of media three independent times, and each set of three cultures was independently analyzed by oligonucleotide array analysis (Affymetrix, Santa Clara, CA) (9) and quantitative MudPIT (8) as described below.
Oligonucleotide Array Analysis of mRNA Expression.
Total yeast RNA was isolated by hot phenol extraction, and all array hybridizations were carried out at 45°C for 16 h in duplicate as described (9). Microarray analysis was performed according to the manufacturer's instructions and as described (9). Briefly, 5 μg total RNA was converted to cDNA and used as a template to generate biotinylated cRNA. After fragmentation, cRNA was hybridized to an Affymetrix S98 Yeast arrays as described in the standard protocol outlined in the GeneChip Expression Analysis Technical Manual (Affymetrix). After sample hybridization, arrays were washed and scanned at a resolution of 3 μM by using a commercially available confocal laser scanner (Affymetrix). Scanned image files were visually inspected for artifacts and analyzed with GENECHIP 3.1 (Affymetrix). The data were normalized by setting the mean hybridization signal for each sample equal to 200. Initial data processing was accomplished with Affymetrix genechip software.
Preparation of Samples for Quantitative Proteomic Analysis.
The soluble portion of the proteome of S. cerevisiae was prepared by sodium carbonate extraction as described (8). The protein content of the supernatant from each of the three cell lysates was determined by the microBCA protein assay (Pierce). Two samples were then generated for subsequent analysis. First, the control sample contained equal protein amounts from the supernatant of the lysis of cells grown in 14N or 15N minimal media. Second, the experimental sample contained equal protein amounts from the supernatant of the lysis of cells grown in 15N minimal media or YPD media. After mixing, each of the samples was brought to 8 M urea and the pH was adjusted to pH 8.5. From this point forward, each sample was prepared for MudPIT analysis as described (8).
Multidimensional Protein Identification Technology Analysis of Quantitative Complex Peptide Mixtures.
A Finnigan DECA ion trap mass spectrometer (Finnigan MAT, San Jose, CA) was interfaced with a quaternary HP 1100 series HPLC pump (Agilent Technologies, Palo Alto, CA) and a fritless capillary fused silica microcolumn (100 μm i.d. × 365 μm o.d.) packed with reverse phase (5-μm Zorbax Eclipse XDB-C18, Agilent Technologies) and strong cation exchange (5 μm Partisphere, Whatman, Clifton, NJ) packing materials as described (11, 18). A fully automated 13-cycle chromatographic run was carried out on each sample by using the four buffer solutions used for the chromatography consisting of 5% ACN/0.012% HFBA/0.5% acetic acid, 80% ACN/0.012% HFBA/0.5% acetic acid, 250 mM ammonium acetate/5% ACN/0.012% HFBA/0.5% acetic acid, and 500 mM ammonium acetate/5% ACN/0.012% HFBA/0.5% acetic acid (8).
With the resulting data set collected, the SEQUEST algorithm (19) was used to interpret the tandem mass spectra generated as described (8). Briefly, the SEQUEST (19) algorithm was run two separate times on each of the three data sets against the yeast_orfs.fasta database from the National Center for Biotechnology Information. Each sample had to be run twice to separately detect and identify peptides from the 14N minimal media sample and from the 15N minimal media sample by using two separate SEQUEST parameters files where the masses of each amino acid was set to the corresponding growth conditions and nitrogen content therein. In both the resulting 14N and 15N data sets, a list of positive peptide identifications was determined by filtering the SEQUEST results based on the charge state of the peptide, the ΔCn value of the SEQUEST result, the tryptic nature of the peptide, and the Xcorr value of the SEQUEST result as described (10–12). The relative abundances of peptides detected and identified in any given MudPIT analysis were computationally determined as described previously (8).
Results and Discussion
Global mRNA and Protein Expression Analysis.
S. cerevisiae was cultured in each type of media (YEPD, 14N minimal media, and 15N minimal media) three independent times, and each set of three cultures was independently analyzed by oligonucleotide arrays (9) and quantitative MudPIT (8) as described below. At both the level of mRNA and protein, the 14N minimal vs. 15N minimal control comparisons demonstrated that the usage of 15N ammonium sulfate did not result in any mRNA (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org) or protein expression changes (data not shown). For example, an ANOVA analysis of the oligonucleotide array data set demonstrated that there was no statistically significant 2-fold or greater difference in mRNA expression (P < 0.05) between the 14N minimal and 15N minimal data sets of loci with intensities >50 counts (Table 2). Therefore, the use of different mass labels has negligible effects on mRNA or protein expression patterns in yeast. The carbonate-extracted proteome of S. cerevisiae cultured in rich media was compared with that of cells grown on 15N minimal media, and a total of 688 proteins and 1,889 peptides were detected, identified, and quantified in at least two independent MudPIT analyses from three independent cultures (Table 3, which is published as supporting information on the PNAS web site). The full data set of loci for which both mRNA and protein expression ratios were determined of S. cerevisiae cultured on rich and minimal media is available in Table 3.
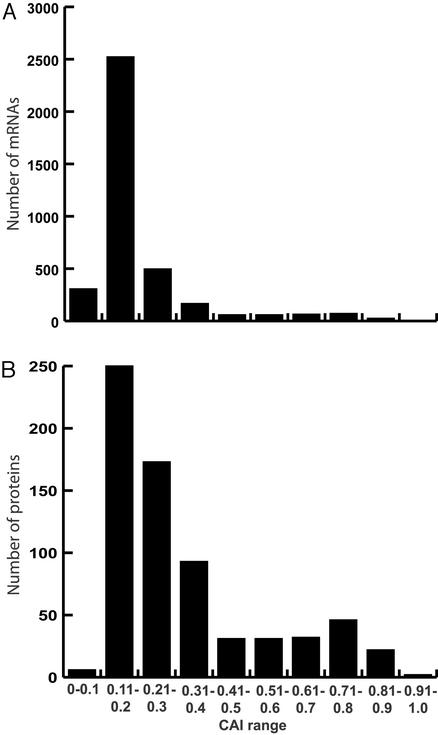
A key requirement of a quantitative proteomic analysis is the ability to reproducibly detect, identify, and quantify low-abundance proteins. The codon adaptation index (CAI) has been shown to be a positive predictor of mRNA abundance in S. cerevisiae (20) and is used as a potential predictor of protein abundance where CAI values <0.2 represent low abundance proteins. Proteins with CAI values <0.2 are generally difficult to detect and identify via two-dimensional polyacrylamide gel electrophoresis (21). The CAI distribution of both the mRNA and protein expression ratio data sets from the current analysis is shown in Fig. 1. The oligonucleotide arrays (Fig. 1A) provided a much larger data set than the quantitative proteomics analysis (Fig. 1B). However, quantitative MudPIT reproducibly detected, identified, and quantified the protein expression ratios for 77 loci (72 of which had CAI values <0.2) whose mRNA intensities in both minimal media and rich media were <50 and were not detectable by oligonucleotide arrays (Table 3).
Figure 1.
CAI distribution of identified mRNA and protein products from S. cerevisiae loci. (A) The 3,776 loci for which the mRNA had an intensity of at least 50 counts, as detected by Affymetrix oligonucleotide array analysis, are shown as a function of the CAI of the given loci. The soluble portion of the protein from each sample was analyzed by means of quantitative multidimensional protein identification technology as ratio mixtures of 14N or 15N proteins from cells grown in YEPD or minimal media. (B) The CAI distribution of the 688 loci for which the protein product was reproducibly detected, identified, and quantified from the YEPD vs. 15N minimal media sample, which was composed of a 1:1 mix of proteins from the soluble extract of cells grown in each of these media.
Correlation of mRNA and Protein Expression.
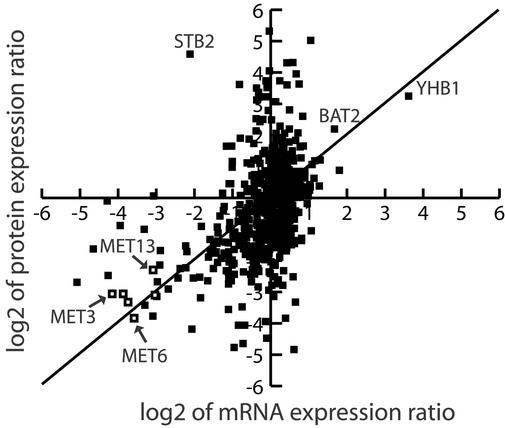
The Spearman rank correlation coefficient (Sr) has been used previously in an attempt to correlate mRNA abundance to protein abundance (3, 4) and mRNA expression ratios to protein expression ratios (6). The Sr for the 678 loci with positive oligonucleotide array intensities was 0.45, a weakly positive correlation between mRNA and protein expression ratios. A scatter plot comparing the mRNA and protein expression ratios for all loci characterized is shown in Fig. 2. A majority of the data points deviating from the perfect positive correlation line shown fall on the y axis indicating that more loci had altered protein expression and unchanged mRNA expression than loci having altered mRNA expression and unchanged protein expression. Furthermore, several loci from the methionine biosynthetic pathway had a near perfect correlation between mRNA and protein expression (Fig. 2), indicating that correlation of mRNA and protein expression should be analyzed at not only the loci by loci level but also at the protein pathway level.
Figure 2.
Scatter plot representation of the correlation between mRNA and protein expression ratios. The log2 value of the mRNA and protein ratios of expression in YEPD media vs. 15N minimal media for each locus was calculated and plotted for the 678 loci for which positive oligonucleotide array intensities and protein expression ratios were determined. A solid line is shown to represent the perfect positive correlation line. In addition, the open boxes shown indicate a grouping of each data point from loci in the methionine biosynthetic pathway from Table 3. The names of additional selected loci are also shown.
Expression Clustering by Protein Pathways and Complexes.
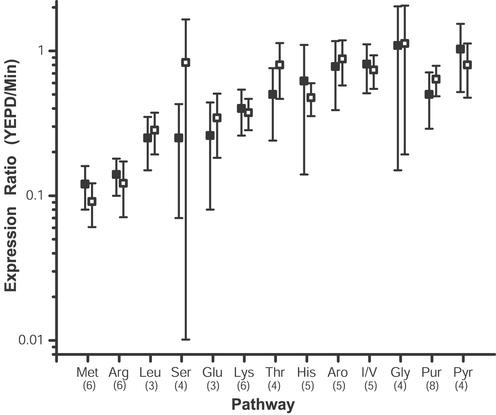
The major challenge facing large-scale mRNA and protein expression analysis is how to interpret the correlated data set. Our strategy for the contextual analysis of integrated mRNA and protein expression data sets was based on the presence of loci analyzed by both methods in protein pathways and protein complexes. The mRNA and protein expression ratios for the components of amino acid and purine biosynthetic pathways (Table 3) were in agreement with the previous analysis of mRNA expression alterations induced by histidine biosynthesis inhibition (14) and induced by culturing of S. cerevisiae on minimal media (9). As shown in Fig. 2 with methionine biosynthetic pathway components, in the current analysis not only did the mRNA and protein expression changes of single loci correlate, but the mRNA and protein expression changes of entire pathways correlated (Fig. 3). For example, the mRNA and protein expression ratios of six loci of the methionine and arginine biosynthetic pathways had ≈10-fold inductions at both the level of mRNA and protein, and the expression ratios correlated not only for individual loci (Table 3) but also for the entire pathway (Fig. 3). Indeed, the probability that mRNA and protein up-regulated in minimal media were unrelated to those with a MIPS (16) functional classification of amino acid metabolism was 3.74 E − 37 for up-regulated mRNA and 3.32 E −21 for up-regulated protein (22). The pattern of the mRNA and protein expression correlating for an entire pathway could be misleading. In the aromatic and histidine biosynthetic pathways there appeared to be no pathway expression changes but the protein expression of individual loci were overexpressed in minimal media (Fig. 3 and Table 3). In general, the mRNA and protein expression ratios correlated for individual loci and entire pathways involved in amino acid and nucleotide biosynthesis demonstrating that the protein expression levels for the entire pathways and subsets of pathways were controlled at the level of transcription.
Figure 3.
Correlation of mRNA and protein expression of amino acid and nucleotide biosynthetic pathway components. The mRNA and protein expression ratios of loci from each of the biosynthetic pathways shown were detected, identified, and quantified as described in Materials and Methods (Table 3). For each pathway that was represented in Table 3 by at least three loci, the average mRNA and protein expression ratio for the whole pathway was determined and plotted. The three-letter code for each pathway shown is of standard nomenclature except for that representing the biosynthetic pathways of the aromatic amino acids (Aro), the shared isoleucine/valine pathway (I/L), and the purine (Pur) and pyrimidine (Pyr) nucleotide biosynthetic pathways. The number in parentheses below each three-letter code represents the number of loci for which both the mRNA and protein expression was determined for the given pathway. The open boxes represent the average mRNA expression ratio of a given pathway, and the filled boxes represent the average protein expression ratio of a given pathway. The error bars represent one standard deviation of the data. Annotation of each identified loci was carried out by accessing the Yeast Proteome Database (15) and MIPS (16).
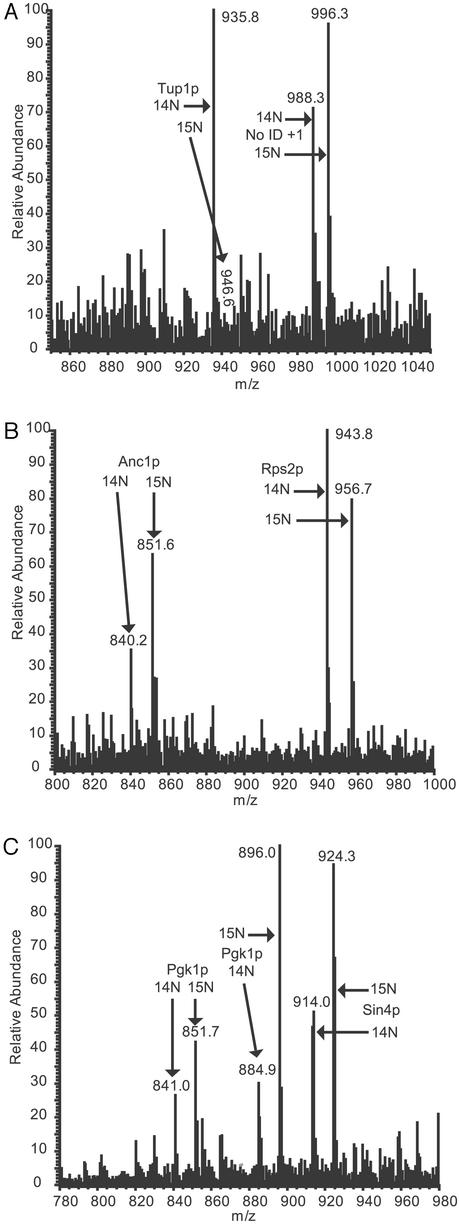
The mRNA and protein expression ratio changes for several protein complexes and regulatory factors are shown in Table 1. Proteins involved in RNA polymerase II transcription (holoenzyme, SPT complex, and histone modification) were overexpressed in minimal media (Table 1). The protein expression changes of the transcriptional repressor Tup1p (23), and the RNA polymerase II holoenzyme components Anc1p and Sin4p (24, 25) are shown in Fig. 4. Tup1p is a general repressor of RNA polymerase II transcription (23), which was 8-fold overexpressed in rich media when compared with minimal media (Fig. 4A), and this protein expression change was not detectable at the level of mRNA (Table 1). Both Anc1p and Sin4p were overexpressed in 15N minimal media as indicated in Fig. 4 B and C, and these changes were not detectable at the level of mRNA (Table 1). The protein levels of additional loci that are involved in RNA polymerase II transcription were overexpressed in minimal media including Eaf3p, Sin3p, Spt4p, Spt6p, Ssu72p, and Stb2p (Table 1). In the analysis by Wodicka et al. (9), 140 mRNAs had at least a 5-fold increase in expression in minimal media, and 36 mRNAs had at least a 5-fold increase in expression in rich media. In the current study a similar global pattern was seen at the level of mRNA where the mRNA expression of 80 loci were overexpressed by at least 5-fold in minimal media whereas the mRNA expression of 32 loci were overexpressed by at least 5-fold in rich media (data not shown). A dramatic decrease in repression and increase in RNA polymerase II machinery provided a potential mechanistic explanation for the observations of Wodicka et al. (9) and those seen in our data where the mRNA of many more loci were overexpressed in cells cultured in minimal media than cells cultured in rich media.
Table 1.
Expression ratios of protein complex components
| Protein complex name (ref.) | Loci characterized* | mRNA expression† | Protein expression† |
|---|---|---|---|
| RNA Polymerase II | |||
| Holoenzyme (24, 25, 31) | ANC1, SIN4, SSU72‡ | 0.94 ± 0.26 | 0.30 ± 0.10 |
| SPT complex (26) | SPT4 | 0.77 ± 0.75 | 0.24 ± 0.06 |
| SPT6 | 1.17 ± 1.19 | 0.58 ± 0.32 | |
| SPT5 | 1.94 ± 0.75 | 1.28 ± 0.35 | |
| Histone modifications (32–34) | EAF3, SIN3, CPR1§, YIll112W‡ | 1.10 ± 0.21 | 0.35 ± 0.17 |
| Repression (23) | TUP1 | 1.10 ± 0.27 | 8.39 ± 0.53 |
| SIN3 binding (35) | STB2‡ | 0.23 ± 1.8 | 24.0 ± 3.6 |
| Cytochrome bc1 (36) | COR1, OCR2, 7 | 0.67 ± 0.09 | 0.42 ± 0.14 |
| COPI complex (37) | COP1, SEC27, 28 | 1.16 ± 0.10 | 0.42 ± 0.10 |
| GTPases (38) | ARF1§ | 1.14 ± 0.17 | 2.11 ± 0.26 |
| ARF2 | 0.66 ± 0.20 | 1.19 ± 0.82 | |
| GEFs (39, 40) | GEA2‡ | 0.79 ± 0.39 | 0.33 ± 0.23 |
| GEA1 | 0.94 ± 0.47 | 0.72 ± 0.29 | |
| SYT1 | 1.25 ± 0.55 | 0.89 ± 0.22 | |
| COPII complex (27) | SEC13 | 1.19 ± 0.50 | 0.58 ± 0.19 |
| SEC23 | 1.33 ± 0.45 | 0.46 ± 0.23 | |
| SEC24 | 1.18 ± 0.30 | 0.97 ± 0.52 | |
| GTPase | SAR1 | 0.81 ± 0.40 | 0.96 ± 0.10 |
| Vacuolar H(+) ATPase (28) | VMA1, 4§, 6, 13, VPH13§ | 0.93 ± 0.10 | 0.48 ± 0.09 |
| VMA24 | 1.01 ± 0.10 | 1.72 ± 0.44 | |
| VMA5 | 0.89 ± 0.22 | 0.99 ± 0.15 | |
| Proteasome (41) | PRE5, 6, PUP3, RPN1, 8 | 0.97 ± 0.12 | 0.94 ± 0.10 |
| PRE6 | 1.10 ± 0.38 | 1.85 ± 1.39 | |
| Chaperonin—T (42, 43) | CCT2, 5, 8 | 1.45 ± 0.18 | 1.65 ± 0.21 |
| Ribosome (44) | 40S (N = 28) | 1.33 ± 0.15 | 1.41 ± 0.27 |
| 60S-1 (N = 28) | 1.31 ± 0.15 | 1.37 ± 0.37 | |
| 60S-2 (N = 8)¶ | 1.39 ± 0.18 | 3.06 ± 0.94 |
Both the mRNA and the protein product of each loci presented in this column were detected, identified, and quantified by using the methods described in Materials and Methods. Annotation of each identified loci was carried out by accessing the Yeast Proteome Database (15) and MIPS (16).
† The average and standard deviation is determined by using the average mRNA or protein expression ratio (YEPD vs. 15N minimal media) of each locus from multiple data sets as described in Table 3.
‡ The average mRNA intensity value of a given loci as measured by oligonucleotide array analysis was below 50 counts as shown in Table 3.
§ The average mRNA intensity value of a given loci as measured by oligonucleotide array analysis was above 2000 counts as shown in Table 3.
¶ The mRNA for all eight of the subset of the 60S ribosome had intensities above 2000. The eight loci in this subset were RPL5, 6B, 8A, 10, 13A/B, 21A/B, 31A/B, and 33A/B.
Figure 4.
Alteration of protein expression of the RNA polymerase II transcriptional regulatory factors Tup1p, Anc1p, and Sin4p. (A) Data from one of the identifications of Tup1p, which was overexpressed in YEPD media by 8.4-fold as compared with minimal media in the combined data set, is shown. A 200-amu window around the MS elution profile for the 14N (M + 2H)2+ peptide VCFSPDGKFLATGAEDR from Tup1p is shown at m/z 935.8. The 15N (M + 2H)2+ version of this peptide from cells grown in 15N minimal media is barely visible at m/z 946.6. In addition, the 14N and 15N peak pair of an unidentified + 1 peptide is shown at m/z 988.3 and 996.3, respectively. (B) Data from one of the identifications of Anc1p, which was overexpressed in minimal media by 2.7-fold as compared with YEPD media in the combined data set, is shown. A 200-amu window around the MS elution profile for the 14N and 15N (M + 2H)2+ peptide VIYHLHPTFANPNR from Anc1p is shown at m/z 840.2 and 851.6, respectively. In addition, a portion of the elution profile of the 14N and 15N peak pair of a peptide from Rps2p is seen at m/z 943.8 and 956.7, respectively. (C) Data from one of the identifications of Sin4p, which was overexpressed in minimal media by 5-fold as compared with YEPD media in the combined data set, is shown. A 200 amu window around the MS elution profile for the 14N and 15N (M + 2H)2+ peptide FKNIIASPLSAGFNYGK from Sin4p is shown at m/z 914.0 and 924.3, respectively. In addition, portions of the elution profiles of the 14N and 15N peak pairs of two unique peptides from Pgk1p are seen at m/z 841.0/851.7 and 884.9/896.0, respectively.
The protein expression levels of several loci in protein complexes were not uniformly overexpressed in either media. Of the three loci in the SPT complex (26), the expression levels of Spt4p and Spt6p were overexpressed in minimal media, whereas the Spt5p was unchanged (Table 1). In the COPII complex (27), Sec24p expression was unchanged, whereas Sec13p and Sec23p were each overexpressed by 2-fold in minimal media (Table 1). The protein expression of five loci of the vacuolar H(+) ATPase complex (28) was increased in minimal media whereas two loci had unchanged protein expression (Table 1). Lastly, a subset (8 loci) of the 60S ribosomal particle was overexpressed in YEPD, whereas the protein expression of the 40S ribosomal particle and the remainder of the 60S ribosomal particle were unchanged in either media (Table 1). Overall repression of the majority of ribosomal genes under conditions of amino acid starvation has been previously seen (14), but this is the first time such an effect has been seen at the protein level and of a distinct subset of only one of the ribosomal particles. These results suggest that there was subcomplex posttranscriptional control of protein expression levels that may have led to altered protein complex stoichiometry.
Conclusion
The methods described in this work demonstrated the ability of quantitative MudPIT to detect, identify, and quantitate the protein expression of loci whose mRNA was unchanged. However, the challenge was to determine the biological context of the changed and unchanged loci. Proteins function in complexes and pathways, and clustering of an integrated mRNA and proteomic data set based on this simple fact provided the context to interpret the biology of the system. At the protein pathway level we determined that the increase of mRNA and protein expression in S. cerevisiae cultured in minimal media for several amino acid (Met, Arg, Glu, and Lys) and the purine biosynthetic pathways not only correlated on a loci by loci basis but also at the whole pathway level. In two specific cases, that of the aromatic amino acid and histidine biosynthetic pathways, whereas the mRNA and protein expression ratios correlated at the pathway level and showed no overall change in pathway expression, the mRNA and protein expression ratios of individual components in each pathway were overexpressed in minimal media. These results suggested that there were pathway and subpathway levels of transcriptional control of protein expression levels.
Clustering of the data set by protein complex yielded novel insight into the biology of S. cerevisiae cultured in rich and minimal media by determining the expression change of protein complexes that have not been previously described (9, 14) and were not detectable by oligonucleotide array analysis of mRNA expression. Several protein complexes or subsets of protein complexes were overexpressed in cells cultured in either media, and in general the mRNA and protein expression for these individual loci and whole complexes did not correlate. The clustering of mRNA and protein expression data by protein complex suggested that posttranscriptional regulatory mechanisms (2) functions at the level of whole complexes and the subcomplex level. In the SPT complex (1 of 3 loci), COPII complex (1 of 3 loci), vacuolar (H)+ ATPase complex (2 of 7 loci), and the ribosome (8 of 36 loci of the 60S subunit), at least one loci known to be in each complex had unchanged protein expression, whereas the protein product of other loci were either overexpressed in minimal or rich media. When the protein expression of a locus in the complex was overexpressed in either media the mRNA expression was unchanged, indicating that posttranscriptional control of protein expression functions at the protein complex and subcomplex level. There are two possible explanations for the results seen in these four complexes. One, the protein stoichiometry in each complex is different in cells cultured in either media. Two, the protein stoichiometry in each complex is the same in cells cultured in either media and the additional protein expression of certain loci from each complex is being used in a different protein complex. Although it is possible that only one of these explanations holds true for all four complexes, it is more likely that both explanations were in effect although we have no data to support either case.
As proteomic technologies approach the capabilities of cDNA array and oligonucleotide array analyses, data clustering methodologies will need to be developed and implemented. Although temporal proteomic analyses will likely require mathematical clustering algorithms (29), clustering by protein pathways and complexes will be essential. The results described herein clearly demonstrate the importance of protein complex clustering because a locus whose protein expression does not change is as important as those that do change. Future correlated large-scale mRNA and protein expression analyses will likely determine similar complex patterns of transcriptional and posttranscriptional control as long as data clustering is based on the fact that proteins function in pathways and complexes. As metabolic labeling has been demonstrated to be a viable quantitative proteomic method in mammalian tissue culture (30), one could envision a target discovery project in a variety of systems where the same analytical and clustering methods are applied to determine which protein pathways and complexes are altered in expression to obtain a more complete and complex biological picture.
Supplementary Material
Acknowledgments
We thank Majid Ghassemian for valuable discussions regarding the manuscript.
Abbreviations
- YEPD
yeast extract/peptone/dextrose
- MudPIT
multidimensional protein identification technology
- ACN
acetonitrile
- HFBA
heptafluorobutyric acid
- CAI
codon adaptation index
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lockhart D J, Winzeler E A. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy J E. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gygi S P, Rochon Y, Franza B R, Aebersold R. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Futcher B, Latter G I, Monardo P, McLaughlin C S, Garrels J I. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ideker T, Thorsson V, Ranish J A, Christmas R, Buhler J, Eng J K, Bumgarner R, Goodlett D R, Aebersold R, Hood L. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 6.Griffin T J, Gygi S P, Ideker T, Rist B, Eng J, Hood L, Aebersold R. Mol Cell Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 7.Oda Y, Huang K, Cross F R, Cowburn D, Chait B T. Proc Natl Acad Sci USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Washburn M P, Ulaszek R, Deciu C, Schieltz D M, Yates J R., III Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 9.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 10.Link A J, Eng J, Schieltz D M, Carmack E, Mize G J, Morris D R, Garvik B M, Yates J R., III Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 11.Washburn M P, Wolters D, Yates J R., 3rd Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 12.Wolters D A, Washburn M P, Yates J R., 3rd Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 13.Hinnebusch A G. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 319–414. [Google Scholar]
- 14.Natarajan K, Meyer M R, Jackson B M, Slade D, Roberts C, Hinnebusch A G, Marton M J. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costanzo M C, Crawford M E, Hirschman J E, Kranz J E, Olsen P, Robertson L S, Skrzypek M S, Braun B R, Hopkins K L, Kondu P, et al. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mewes H W, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Mering C, Krause R, Snel B, Cornell M, Oliver S G, Fields S, Bork P. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 18.Gatlin C L, Kleemann G R, Hays L G, Link A J, Yates J R., III Anal Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- 19.Eng J K, McCormack A L, Yates J R I. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 20.Coghlan A, Wolfe K H. Yeast. 2000;16:1131–1145. doi: 10.1002/1097-0061(20000915)16:12<1131::AID-YEA609>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 21.Gygi S P, Corthals G L, Zhang Y, Rochon Y, Aebersold R. Proc Natl Acad Sci USA. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tavazoie S, Hughes J D, Campbell M J, Cho R J, Church G M. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 23.Smith R L, Johnson A D. Trends Biochem Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Bjorklund S, Jiang Y W, Kim Y J, Lane W S, Stillman D J, Kornberg R D. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gustafsson C M, Myers L C, Beve J, Spahr H, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. J Biol Chem. 1998;273:30851–30854. doi: 10.1074/jbc.273.47.30851. [DOI] [PubMed] [Google Scholar]
- 26.Hartzog G A, Wada T, Handa H, Winston F. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bednarek S Y, Orci L, Schekman R. Trends Cell Biol. 1996;6:468–473. doi: 10.1016/0962-8924(96)84943-9. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Leng X H, Newman P R, Vasilyeva E, Kane P M, Forgac M. J Biol Chem. 1997;272:11750–11756. doi: 10.1074/jbc.272.18.11750. [DOI] [PubMed] [Google Scholar]
- 29.Quackenbush J. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 30.Conrads T P, Alving K, Veenstra T D, Belov M E, Anderson G A, Anderson D J, Lipton M S, Pasa-Tolic L, Udseth H R, Chrisler W B, et al. Anal Chem. 2001;73:2132–2139. doi: 10.1021/ac001487x. [DOI] [PubMed] [Google Scholar]
- 31.Wu W H, Pinto I, Chen B S, Hampsey M. Genetics. 1999;153:643–652. doi: 10.1093/genetics/153.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisen A, Utley R T, Nourani A, Allard S, Schmidt P, Lane W S, Lucchesi J C, Cote J. J Biol Chem. 2001;276:3484–3491. doi: 10.1074/jbc.M008159200. [DOI] [PubMed] [Google Scholar]
- 33.Kadosh D, Struhl K. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pijnappel W W, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart A F. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasten M M, Dorland S, Stillman D J. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure Fold Des. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 37.Gaynor E C, Graham T R, Emr S D. Biochim Biophys Acta. 1998;1404:33–51. doi: 10.1016/s0167-4889(98)00045-7. [DOI] [PubMed] [Google Scholar]
- 38.Poon P P, Cassel D, Spang A, Rotman M, Pick E, Singer R A, Johnston G C. EMBO J. 1999;18:555–564. doi: 10.1093/emboj/18.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson C L, Casanova J E. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 40.Jones S, Jedd G, Kahn R A, Franzusoff A, Bartolini F, Segev N. Genetics. 1999;152:1543–1556. doi: 10.1093/genetics/152.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finley D, Tanaka K, Mann C, Feldmann H, Hochstrasser M, Vierstra R, Johnston S, Hampton R, Haber J, McCusker J, et al. Trends Biochem Sci. 1998;23:244–245. doi: 10.1016/s0968-0004(98)01222-5. [DOI] [PubMed] [Google Scholar]
- 42.Li W Z, Lin P, Frydman J, Boal T R, Cardillo T S, Richard L M, Toth D, Lichtman M A, Hartl F U, Sherman F, et al. J Biol Chem. 1994;269:18616–18622. [PubMed] [Google Scholar]
- 43.Stoldt V, Rademacher F, Kehren V, Ernst J F, Pearce D A, Sherman F. Yeast. 1996;12:523–529. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C523::AID-YEA962%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 44.Planta R J, Mager W H. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.